Abstract
Semantic-based sublanguage grammars have been shown to be an efficient method for medical language processing. However, given the complexity of the medical domain, parsers using such grammars inevitably encounter ambiguous sentences, which could be interpreted by different groups of production rules and consequently result in two or more parse trees. One possible solution, which has not been extensively explored previously, is to augment productions in medical sublanguage grammars with probabilities to resolve the ambiguity. In this study, we associated probabilities with production rules in a semantic-based grammar for medication findings and evaluated its performance on reducing parsing ambiguity. Using the existing data set from 2009 i2b2 NLP (Natural Language Processing) challenge for medication extraction, we developed a semantic-based CFG (Context Free Grammar) for parsing medication sentences and manually created a Treebank of 4,564 medication sentences from discharge summaries. Using the Treebank, we derived a semantic-based PCFG (probabilistic Context Free Grammar) for parsing medication sentences. Our evaluation using a 10-fold cross validation showed that the PCFG parser dramatically improved parsing performance when compared to the CFG parser.
Keywords: natural language processing, parsing, probabilistic context free grammar, sublanguage grammar
1. INTRODUCTION
In the past decade, Electronic Health Records (EHRs) systems have been rapidly adopted in the healthcare industry, resulting in more and more clinical data available in electronic formats [1]. Much of the detailed patient information in EHRs is stored in narrative text documents entered by healthcare providers, and it is not directly accessible to other computerized applications, such as clinical decision support systems. Natural Language Processing (NLP) technologies, which can convert clinical text into structured data, have received great attention in the medical domain. Diverse NLP applications have been used for various clinical tasks [2] including decision support [3, 4], encoding [5, 6], data mining [7], and clinical research [8, 9].
Many clinical NLP systems are focused information extraction (IE) systems, which are developed for specific applications and have demonstrated good performance on designated tasks [10–12]. However, general and comprehensive medical language processing systems are more difficult to build, as they require significant resources to develop and implement; but they often provide more insights to understanding the medical language itself [2]. Different methods have been used to build general clinical NLP systems. A number of systems, including MetaMap [13], KnowledgeMap [14], HiTEXT [15], and cTAKES [16], identify clinical concepts (usually in noun phrases) from text first. Then they integrate other modules to detect contextual features, such as NegEx [17] for detecting negations and ConText [18] for broader types of modifiers. SymText (Symbolic Text processor) is a system developed at University of Utah, which combines syntactic and probabilistic semantic analysis based on Bayesian networks [19]. Some clinical NLP systems have focused on semantic relation extraction. For example, SemRep [20] is a rule-based symbolic NLP system developed to extract semantic predication from Medline citations, which has been used for different applications including clinical guideline development [21]. Two systems of particular interests to us are the MLP (Medical Language Processor) [22] and MedLEE (Medical Language Extraction and Encoding System) [23], which are both based on the sublanguage theory by Zellig Harris [24, 25]. The sublanguage theory states that the structure and regularity of languages from specialized domains can be delineated in the form of a sublanguage grammar, which not only specifies well-formed syntactic structures as in English grammar, but also incorporates domain-specific semantic information and relationships. MLP was the product of the Linguistic String Project (LSP) led by Dr. Sager at New York University, which was the first large-scale study in clinical text processing [22, 26–28]. The system contains a complicated sublanguage grammar that considers both syntactic and semantic patterns of clinical text. Inspired by LSP, Friedman et al. [23, 29] developed MedLEE, a mainly semanticly-driven system for clinical text processing. MedLEE has been shown to be as accurate as physicians at extracting clinical concepts from chest radiology reports [14;15]. It was originally designed for radiology reports of the chest but has been successfully extended to other domains, such as mammography reports [16] and discharge summaries [17]. The success of MedLEE indicates the effectiveness of the semantic-based sublanguage grammar approach for clinical text processing and inspires our work in this study.
One of the important components for systems based on sublanguage grammars such as MedLEE is the parsing step, which determines a grammatical structure of sentences (called a parse tree) with respect to a given grammar (e.g., a Context-Free Grammar - CFG). The biggest challenge in parsing is the problem of ambiguity: there is often more than one possible parse tree for a sentence with a given CFG. Current systems such as MedLEE rely on highly specific rules obtained from careful manual analysis to reduce ambiguity and generate correct parse trees. However, an interesting alternative solution is to associate probabilities with CFG rules – the Probabilistic Context Free Grammar (PCFG), in which each production rule is augmented with a probability that is usually obtained from annotated training data. When there are multiple possible parse trees for a given sentence, the final selection can be determined by the overall probability of a parse tree, which is the product of probabilities of all production rules used to expand each of non-terminal nodes in the parse tree. Many studies on PCFG, such as [30–34], have shown its capability to solve ambiguity in parsing of general English text using syntactic grammars. However, to the best of our knowledge, there is no published study applying PCFG to semantic sublanguage grammars used for medical text processing. In this study, we hypothesized that PCFG could improve parsing of clinical sentences for medical language processing systems that use semantic-based sublanguage grammar.
To study the effect of PCFG on sublanguage grammars that cover all types of clinical entities is a huge project that requires large amounts of efforts on grammar development and corpus annotation. As a first step toward that goal, in this study, we investigated the use of PCFG for parsing medication-related sentences in clinical text. Medications and their related signature information (e.g. dose, frequency, or route of administration) are one of the most important types of clinical information for research that uses EHR data [35]. Sometimes medication sentences in clinical notes can be complex, as drug signatures can be repetitive or even nested (e.g., “Coumadin 2.5mg po dly except 5mg qTu,Th” and “Coumadin 6mg po 4x week, 4mg po 3x week”). Moreover, sentences containing multiple drugs are even more ambiguous because signature modifiers can be linked to one or more drugs in the sentence. For example, consider the sentence, “Therefore, their recommendation was to start the patient on Lovenox for the duration of this pregnancy, which adjusted for her weight was a dose of 90 mg daily, followed by a transition to Coumadin postpartum, to be continued for likely long-term, possibly lifelong duration.” In this example, it is challenging for an NLP system to determine if the duration phrase “lifelong duration” is to modify “Coumadin” only, or to modify both “Lovenox” and “Coumadin”. In previous studies, we have developed a medication information extraction system called MedEx [36]. MedEx uses a semantic grammar and a CFG parser to determine the structure of medications and their modifiers within a sentence. The parsing component of MedEx provides a good start for investigating PCFG in semantic parsing of clinical text. In addition, the 2009 i2b2 NLP challenge provides a semantically annotated corpus of discharge summaries for medication names and their modifiers, from which an annotated data set of parse trees can be developed with relatively less effort.
In the next section we describe the data sets and methods for the PCFG implementation within a medication extraction system. The subsequent section shows results of parsing with and without PCFG implementation. Finally, we discuss some interesting findings in this study, as well as future work.
2. MATERIAL AND METHODS
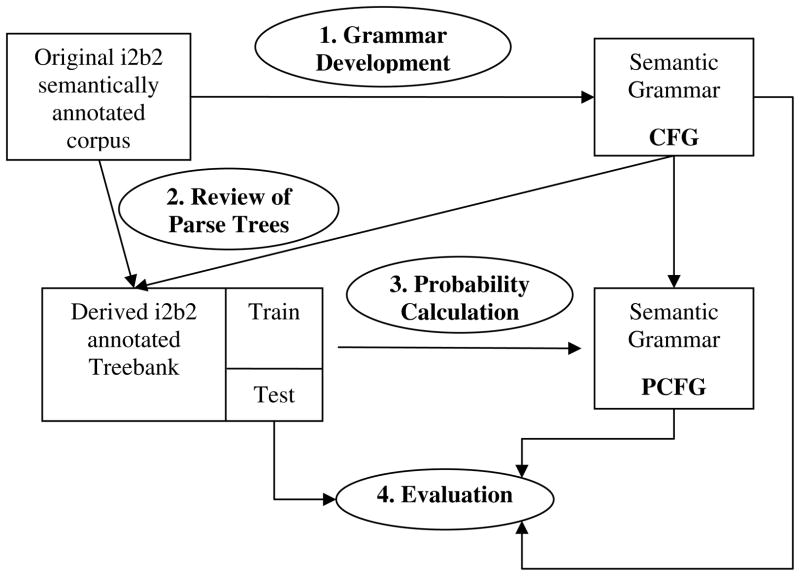
Figure 1 shows an overview of the design of the study. We started with a semantically annotated data set of medication findings from the 2009 i2b2 NLP challenge [37], from which a sublanguage grammar - a CFG that delineates semantic relations and structures of medication findings was developed. By applying the CFG to i2b2 data set, we generated all possible parse trees for each medication sentence and manually reviewed them to determine the correct parse tree for each sentence, thus building an annotated corpus of parse trees (a “Treebank”) for medication sentences. The annotated parse tree corpus was divided into a training set and a test set. The training set was used to calculate the probability for each production in the CFG, thus to build the PCFG. Finally, we applied both the CFG and PCFG to the test set and evaluated the performance of parsing using the PARSEVAL Evalb program (http://nlp.cs.nyu.edu/evalb/).
Figure 1.
An overview of the design of the study, which consists of four steps: 1) to develop a CFG based on semantic patterns extracted from i2b2 semantically annotated data set; 2) to manually create an annotated Treebank from the original i2b2 data set; 3) to develop a PCFG by calculating the probability of each production in CFG using the training set of parse tree corpus; 4) to evaluate the performance of CFG and PCFG using the test set of parse tree corpus.
2.1 i2b2 Corpus for Medication Findings
The 2009 i2b2 NLP challenge was an information extraction task to extract medications and their associated modifiers from hospital discharge summaries [37]. Based on the annotation guideline from the i2b2 challenge, a medication finding consisted of medication name and its modifiers including dosage, frequency, duration, mode, and reason. Table 1 shows some examples of these six medication-related semantic types. Some medication findings are simple, for example, “NPH insulin 20 units q.d.”, which is composed of the name - “NPH insulin”, dosage - “20 units”, and frequency - “q.d ” (it means once a day). However, sometimes medication modifiers can be repetitive, or even nested, which causes additional ambiguity and makes it difficult to accurately link modifiers to medications. For example, medications can have multiple sets of modifiers, e.g., “Midrin 2 po initial then 1 po q6hrs”, where po means “by mouth” and q6hrs means “every 6 hours”. The i2b2 challenge required systems to output multiple entries for such cases (2 entries in this case): “Midrin 2 po q6hrs” and “Midrin 1 po q6hrs”. In order to do that, an NLP system should interpret the frequency term “q6hrs” as a top-level modifier, which applies to both dosage/mode modifiers “2 po” and “1 po”, instead of linking it to the local modifiers “1 po” only. In addition, one sentence can contain multiple drugs. More ambiguity rises when linking modifiers to multiple drugs within one sentence. As we have demonstrated in our previous study [36], one way to delineate the structure of modifiers to each drug in a sentence is to use a sublanguage grammar (a CFG) that is based on semantic patterns of drug related semantic types. However, this approach also faces the problem of ambiguity – multiple possible parse trees could be generated for one sentence based on the CFG. Therefore we investigated how PCFG could help with the ambiguity problem in this study.
Table 1.
Semantic classes in 2009 i2b2 dataset: names, examples, and descriptions.
| Class | Examples | Description |
|---|---|---|
| Medication | “Lasix”, “Caltrate plus D”, “fluocinonide 0.5% cream”, “TYLENOL (ACETAMINOPHEN)” | Prescription substances, biological substances, over-the-counter drugs, excluding diet, allergy, lab/test, alcohol. |
| Dosage | “1 TAB”, “One tablet”, “0.4 mg” “0.5 m.g.”, “100 MG”, “100 mg × 2 tablets” | The amount of a single medication used in each administration. |
| Mode | “Orally”, “Intravenous”, “Topical”, “Sublingual” | Describes the method for administering the medication. |
| Frequency | “Prn”, “As needed”, “Three times a day as needed”, “As needed three times a day”, “x3 before meal”, “x3 a day after meal as needed” | Terms, phrases, or abbreviations that describe how often each dose of the medication should be taken. |
| Duration | “x10 days”, “10-day course”, “For ten days”, “For a month”, “During spring break”, “Until the symptom disappears”, “As long as needed” | Expressions that indicate for how long the medication is to be administered. |
| Reason | “Dizziness”, “Dizzy”, “Fever”, “Diabetes”, “frequent PVCs”, “rare angina” | The medical reason for which the medication is stated to be given. |
The original i2b2 data set contained 268 annotated discharge summaries from Partners Healthcare System. It had 12,773 medication entries, from 9,689 sentences (based on our sentence boundary program). Each medication entry was also labeled as “Narrative” (from narrative sentences), or “List” (from semi-structured list-type format) by i2b2. As our interest and the primary difficulty here was to parse narrative sentences, we removed sentences with List-like format. This resulted in 4,564 medication sentences, which served as the corpus for this study. As mentioned above, different levels of ambiguity exist for different sentences. We further divided those medication sentences into three categories:
1-Sentences containing Single drug and Single set of modifiers (SS), in which there are only one medication name and one set of modifiers, e.g. “5. NPH insulin 20 units q.d.”.
2-Sentences containing Single drug, but Multiple sets of modifiers (SM), in which there are only one medication name, but multiple sets of modifiers, e.g. “7. Insulin 70/30 65 units q.a.m., 35 units q.p.m”.
3-Sentences with Multiple Drugs (MD), in which there are multiple drugs and each drug may have one or multiple set of modifiers, e.g., “sublingual nitroglycerins p.r.n. chest pain, and Glucotrol 5 mg p.o. q.d.”
Based on the annotation by i2b2, we assigned one of the three labels to each sentence, resulting in 3,378, 106, and 1,080 sentences in SS, SM, and MD categories respectively.
2.2 A Semantic CFG for Parsing Medication Sentences
In our previous work, we have developed a semantic grammar that delineates semantic relations and structure of medication findings and used it in the MedEx system [36]. However, the semantic types defined in the i2b2 data set are not exactly the same as those defined in the MedEx system [37]. For example, we did not consider “reason” in our previous study. Therefore, we developed a new semantic grammar (a CFG) based on i2b2 semantic types only, by analyzing semantic patterns derived from i2b2 annotation and leveraging the grammar used in the MedEx system. When we developed the grammar, we tried to make it general enough so that it could cover all the sentences in the corpus. Figure 2 shows some important production rules in the CFG. According to the grammar in Figure 2, a sentence (S) can contain a list (DG_LIST) of drug findings (DG). A drug finding (DG) can be either a drug with single set of modifiers (DG_S_MOD_SET) or a drug with multiple sets of modifiers (DG_M_MOD_SET). For a drug with single set of modifiers (DG_S_MOD_SET), it can contain a drug name (MED) only, or a drug name accompanied by left modifiers (DG_L_MOD), right modifiers (DG_R_MOD), or both. The rules for drugs with multiple sets of modifiers (DG_M_MOD_SET), such as “Insulin 70/30 65 units q.a.m., 35 units q.p.m”, are similar, but it has to contain a non-termal MOD_SET_LIST, which is composed by at least two repetitive MOD_SETs (a set of modifiers). In the above example, “65 units q.a.m” is one MOD_SET, and “65 units q.a.m” is another MOD_SET; together, they form a MOD_SET_LIST.
Figure 2.
The proposed CFG for medication sentences in i2b2; “->” is a rule/”lead to” operator to indicate that the left-hand-side symbol (non-terminal) may be substituted by right-hand-side symbols (nonterminals or semantic tags); “|” presents the alternative rules of a single left-hand-side. Meanings of nonterminals/tags are as below: S – sentence; DG_LIST – list of drugs; DG – a drug finding; DG_S_MOD_SET – a drug finding with single set of modifiers; DG_M_MOD_SET – a drug finding with multiple sets of modifiers; DG_L_MOD – drug left modifiers; DG_R_MOD – drug right modifiers; MOD_SET – a set of drug modifiers; MOD_SET_LIST – a list of sets of drug modifiers; MED – medication name; DOSE – drug administration dosage; MODE – drug administration mode; FREQ – drug administration frequency; REASON – reason of drug administration; DRT – duration of drug administration.
2.3 Development of a Treebank for Medication Sentences
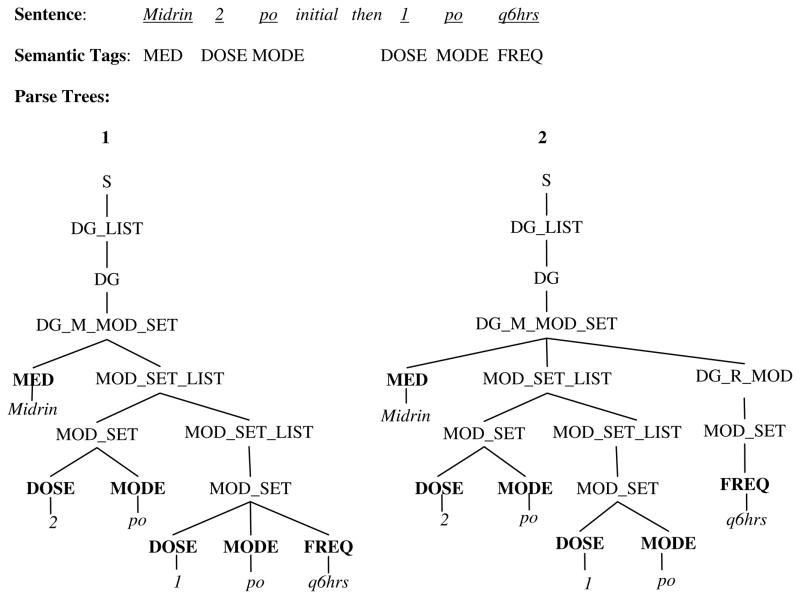
Based on the CFG described above, a medication sentence often can have more than one possible parse tree (ambiguous). Figure 3 shows two possible parse trees for the sentence “Midrin 2 po initial then 1 po q6hrs” using the CFG in Figure 2. The main difference between two parse trees is the position of the “FREQ” modifier: parse tree 1 outputs it as a part of MOD_SET so that it will go with “1 po” only; parse tree 2 outputs it as a top level modifier so that it will modify both “2 po” and “1 po” in the final interpretation. Therefore, in this case, parse tree 2 is the correct one. In this study, we used a Chart parser from NLTK (http://www.nltk.org/) to generate a list of all possible trees for each sentence in the corpus. An annotator who is familiar with medication findings and trained in computational linguistics (SD), manually reviewed outputted parse trees and selected the best parse tree for each sentence. For instances in which he was unsure, the parse trees were also reviewed by other authors (HX, JD) with NLP and clinical experience. By this process, we built a Treebank, which served as the gold standard for the following training and evaluation steps. If the parser failed to generate the best parse of the sentence, the annotator built the best parse tree manually, based on i2b2’s annotation and a simple guideline with examples.
Figure 3.
Two possible parse trees of the sentence “Midrin 2 po initial then 1 po q6hrs” based on the CFG in Figure 1. In this case, parse tree 2 is the correct one.
Generation of the Semantic PCFG for Parsing Medication Sentences
The annotated parse tree corpus (the Treebank) was divided into a training and a test set, using a 10-fold cross validation (CV). Nine folds of data were used as the training set to derive the probability for each production in the CFG, thus generating the PCFG. The left-out fold was used as the test set to evaluate the performance of PCFG. From the Treebank, we computed the probability for each expansion of a non-terminal (α → β) in the CFG by counting the number of times that expansion occurs and normalizing by total count of all expansions of that non-terminal (α), as following:
Figure 4 shows the partial PCFG, where each production rule is associated with a probability calculated from the Treebank.
Figure 4.
Examples of productions with probabilities in the PCFG
Using the PCFG, we can compute a probability of a parse tree T of an input sentence S as:
where P(r) is the probability of any production rule involved in the parse tree. The best parse tree is thus the one that has the highest probability:
2.4 Experiments and Evaluation
After generating the PCFG using the training set in the Treebank, we evaluated and reported the performance of PCFG, as well as CFG, using the test set in the Treebank. Every sentence in the test set was parsed by both a CFG parser and a PCFG parser (namely nltk.parse.chart() and nltk.parse.viterbi(), respectively, in the NLTK package [http://www.nltk.org/]). Both parsers implement the bottom-up Chart Parsing algorithm. The CFG parser outputs parse trees on First-Come-First-Served basis based on the CFG, and we used the first parse tree generated by the Chart Parser as the output. The PCFG parser uses the Viterbi algorithm to determine the path, and it generates parse trees with probabilities. The parse tree with the highest probability was selected as the output, as described above. Parse trees generated from CFG and PCFG parsers for each sentence were compared with the gold standard trees in the Treebank, and the evaluation was done using a package called the PARSEVAL Evalb program (http://nlp.cs.nyu.edu/evalb/), which is a commonly used software for evaluating parsers. The following five PARSEVAL measures were used in this study:
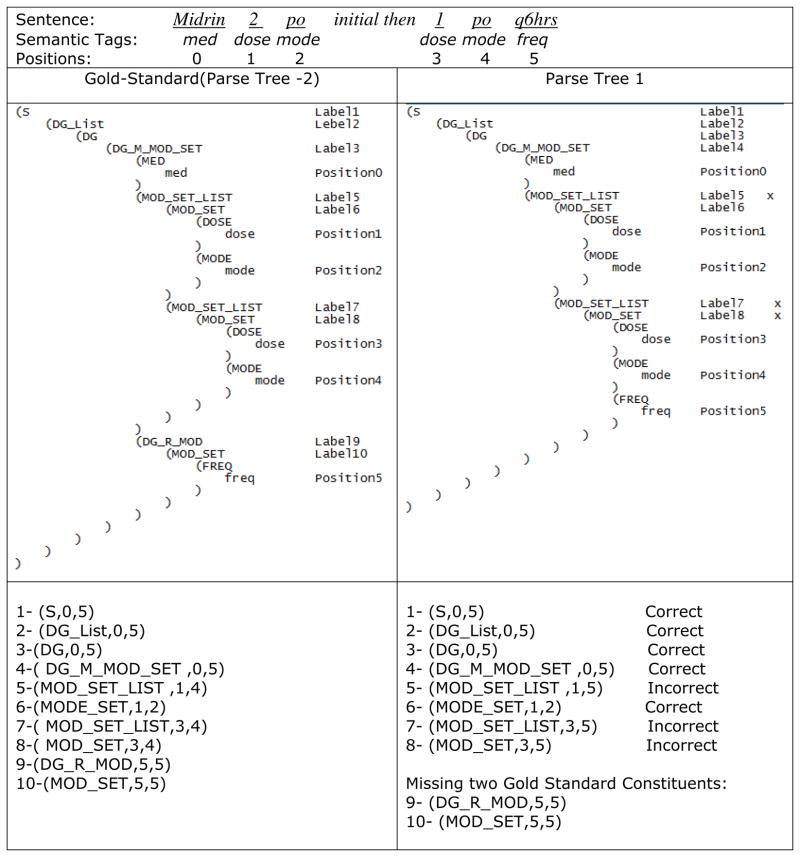
Figure 5 shows the calculation of BR, BP, and BF using the example in Figure 3, where we assume parse tree 2 is the gold standard in the Treebank for that sentence, and we want to measure the BR, BP, and BF for parse tree 1. Only non-terminals were counted as constituents for calculation. A bracketing constituent is defined by its label and its spans (start and stop positions). For example, “(S, 0, 5)”, the first constituent in the gold standard parse tree in Figure 5, is a bracketing constituent having the label S and spans from postion 0 to 5. All three elements in the triple of a constituent must be in the true parse for the constituent to be marked correct. In Figure 5, the fifth constituent in parse tree 1 “(MOD_SET_LIST,1,5)” was wrong, because it did not appear in the gold standard parse tree. Based on such definitions, the results of parse tree 1 were:
Figure 5.
An example of calculation of BR and BP using bracketing consituents. The gold standard (Parse Tree 2) has 10 bracketing constituents, while Parse Tree 1 has only 8 of them, missing the ninth and tenth constituents. Three of the retrieved 8 constituents are wrong, namely the fifth, seventh, and eighth. Therefore BR= 5/10 and BP = 5/8.
A crossing bracket is defined as a bracketed sequence output by the parser that overlaps with one from the treebank but neither is properly contained in the other. In the example in Figure 5, the number of crossing brackets was 2, due to the wrong position of FREQ.
For each sentence, we compared the parse tree generated by a parser with the gold standard parse tree in the treebnk to calculate BR, BP, and BF. Then the averged BR, BP, and BR were reported across all the sentences in the test data set. CM and NC were measured based on all sentences. Whereas CM is used to measure how many sentences parsed completely correctly, NC measures how many constituents parsed correctly in their tree positions.
3. RESULTS
As 10-fold CV was used, results in Tables 2–5 were averages from 10 runs. The overall results for all types of medication sentences showed that PCFG achievements were much better than CFG results in all evaluation metrics (Table 2). PCFG improvements were approximately 20%, 15%, 7%, 14% and 10% in CM, NC, BR, BP and BF measurements respectively. The performance of CFG and PCFG on three different types of medication sentences: SS, SM, and MD are shown in Tables 3, 4, and 5 respectively. In all experiments, the PCFG parser performed better than CFG parser. However, the improvements were different for three different types of medication sentences. SS sentences (Table 2) had highest baseline performance of CFG parser (e.g., 83.26% in CM and 91.26% in BF), and applying PCFG had a reasonable improvement (e.g., 8.84% in CM and 5.37% in BF). SM sentences (Table 4) were difficult for CFG parser (e.g., 35.88% of CM), but PCFG almost doubled the performance with a CM of 76.96%. Table 5 shows the results for MD sentences, for which CFG parser had a reasonable performance (e.g., 82.05% in BF); but the improvement of PCFG was limited (e.g., 3.37% improvement in BF).
Table 2.
Results for all types of sentences
| CM | NC | BR | BP | BF | |
|---|---|---|---|---|---|
| CFG | 60.76 | 77.73 | 83.91 | 79.92 | 81.72 |
| PCFG | 80.45 | 92.73 | 90.77 | 93.15 | 91.94 |
Table 5.
Results for Multiple Drugs (MD) sentences
| CM | NC | BR | BP | BF | |
|---|---|---|---|---|---|
| CFG | 63.15 | 79.28 | 81.97 | 82.15 | 82.05 |
| PCFG | 72.31 | 88.15 | 83.90 | 87.00 | 85.42 |
Table 3.
Results for single drug, single modifier set (SS) sentences
| CM | NC | BR | BP | BF | |
|---|---|---|---|---|---|
| CFG | 83.26 | 98.92 | 90.94 | 91.58 | 91.26 |
| PCFG | 92.10 | 99.97 | 95.58 | 97.70 | 96.63 |
Table 4.
Results for single drug, multiple modifier sets (SM) sentences
| CM | NC | BR | BP | BF | |
|---|---|---|---|---|---|
| CFG | 35.88 | 54.99 | 78.83 | 66.03 | 71.84 |
| PCFG | 76.96 | 90.06 | 92.83 | 94.74 | 93.77 |
4. DISCUSSION
As an initial step of applying PCFG to semantic parsing of clinical text, this study showed that a PCFG parser dramatically improved the performance on parsing medication sentences. The PCFG-based parser achieved increases of 20% in CM, 15% in NC, and 20% in BF, when it was compared with the CFG parser. Such results indicate that PCFG can reduce ambiguity when parsing clinical sentences using a semantic grammar.
As described above, we divided medication sentences into three different categories: SS (Single drug with Single set of modifiers), SM (Single drug with Multiple sets of modifiers), and MD (Multiple drugs), based on different levels of ambiguity. Our expectation was that we would see more improvements from PCFG for SM and MD, as those sentences are more ambiguous than SS sentences. We focused our analysis on Complete Matching (CM) and No Crossing (NC) here, as they assess how the parser achieves the whole parse tree constituents in the order that they should maintain. As expected, PCFG parser almost doubled CM and NC metrics for SM sentences when they were compared with CFG results (Table 4). However, the improvements by PCFG for MD sentences were not high, similar to the improvments on SS sentences. We then looked into the errors in MD sentences by PCFG parser and found that using semantic patterns alone might not be sufficient for delineating the structures of sentences mentioning multiple drugs. For example, it is difficult to determine if the FREQ term (“q.day”) should link to “Aspirin” or “atenolol” in the sentence “Patient’s discharge medications include Aspirin 325 mg p.o. q.day and atenolol 50 mg p.o. daily.”, if only semantic information is used. In such cases, syntactic information such as the coordinating conjunctions (the word “and”) could be helpful to solve the ambiguity here, as they can be used to determine the boundary of medication findings. Combining such syntactic information with semantic patterns into a sublanguage grammar may improve the performance of parsing medication sentences.
It is likely impossible to develop a grammar without ambiguity. In knowledge engineering-based approaches, researchers manually review sentences in a corpus to develop sublanguage grammars and specific rules to reduce ambiguity. However, it is a time consuming task and sometimes such specific grammars will result in unparsed sentences. In our approach, we quickly developed a semantic grammar for medications using pattern extraction methods based on the semantically annotated i2b2 corpus. The grammar reached 100% coverage on medication sentences, as our experiments showed that no sentence had zero Bracketing recall. However, it could contain higher ambiguity than a manually developed grammar. For example, a SS sentence could have multiple parse trees based on our CFG. In the example of “Sevelmar 1600 t.i.d.”, it could be interpreted as a drug with one set of modifiers containing DOSE and FREQ, or as a drug with multiple sets of modifiers (one set with “DOSE” and another set with “FREQ”). However, the PCFG approach can resolve such simple ambiguity easily, as shown in Table 3 that contains results of SS sentences. Efficiently developing good grammars is out of the scope of this study, but clearly very important and highly relevant to parsing methods.
Despite its promising results, this study has limitations. Although we used semantically annotated i2b2 data set, which makes the parse tree annotation relative easy, the Treebank was created primarily by one reviewer, which could lead to some biases. This study investigated PCFG on parsing medication sentences only. Clinical text that contains various types of medical entities such as diseases, tests, and procedures, could be more challenging. Additionally, the evaluation was focused on the parse tree only. How such improvements in parsing can help with real world applications such as information extraction requires further investigation. Finally, and perhaps most importantly, this study relied on an already-annotated corpus from i2b2. Creation of such semantically-tagged corpora is costly but needed to train probabilistic systems such as these. Extension of the PCFG to other clinical semantic grammars would require similar efforts.
5. CONCLUSION
In this study, we applied the PCFG approach to semantic parsing of clinical text, by associating probabilities with productions in a semantic-based grammar for medication sentences. Our initial evaluation using a Treebank of 4,564 medication sentences collected from the 2009 i2b2 NLP challenge showed that the PCFG approach could effectively reduce the ambiguity when parsing medication sentences. Such methods are promising for clinical text processing.
Highlights.
First attempt to apply PCFG to semantic parsing of clinical text
An annotated Treebank of 4,564 medication sentences based on a sublanguage grammar
PCFG effectively reduced the ambiguity when parsing medication sentences
Acknowledgments
Authors would like to thank to i2b2 organizers for providing dataset for research uses. We also thank Dr. Carol Friedman at Columbia University for her valuable comments to this paper. This study was partially supported by the grant from the US NIH: NCI R01CA141307 and NHGRI U01 HG004603.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shea S, Hripcsak G. Accelerating the use of electronic health records in physician practices. N Engl J Med. 362:192–5. doi: 10.1056/NEJMp0910140. [DOI] [PubMed] [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 3.Aronsky D, Fiszman M, Chapman WW, Haug PJ. Combining decision support methodologies to diagnose pneumonia. Proc AMIA Symp. 2001:12–6. [PMC free article] [PubMed] [Google Scholar]
- 4.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42:760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haug PJ, Christensen L, Gundersen M, Clemons B, Koehler S, Bauer K. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp. 1997:814–8. [PMC free article] [PubMed] [Google Scholar]
- 7.Heinze DT, Morsch ML, Holbrook J. Mining free-text medical records. Proc AMIA Symp. 2001:254–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc. 2008:404–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Denny JC, Miller RA, Waitman LR, Arrieta MA, Peterson JF. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform. 2009;78(Suppl 1):S34–42. doi: 10.1016/j.ijmedinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports. Stud Health Technol Inform. 2004;107:565–72. [PubMed] [Google Scholar]
- 12.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny JC, Smithers JD, Miller RA, Spickard A. “Understanding” Medical School Curriculum Content Using KnowledgeMap. Journal of the American Medical Informatics Association. 2003;10:351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 18.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42:839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–8. [PMC free article] [PubMed] [Google Scholar]
- 20.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Fiszman M, Ortiz E, Bray BE, Rindflesch TC. Semantic processing to support clinical guideline development. AMIA Annu Symp Proc. 2008:187–91. [PMC free article] [PubMed] [Google Scholar]
- 22.Sager N, Lyman M, Nhan NT, Tick LJ. Medical language processing: applications to patient data representation and automatic encoding. Methods Inf Med. 1995;34:140–6. [PubMed] [Google Scholar]
- 23.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris ZS. The structure of science information. Journal of Biomedical Informatics. 2002:35. doi: 10.1016/s1532-0464(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 25.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35:222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 26.Sager N, Friedman C, Chi E, Macleod C, Chen S, Johnson S. The analysis and processing of clinical narrative. MedInfo. 1986:1101–5. [Google Scholar]
- 27.Sager N, Friedman C, Lyman M. Medical language processing: computer management of narrative data. Reading, MA: Addison-Wesley; 1987. [Google Scholar]
- 28.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142–60. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman C, Cimino JJ, Johnson SB. A schema for representing medical language applied to clinical radiology. J Am Med Inform Assoc. 1994;1:233–48. doi: 10.1136/jamia.1994.95236155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins M. Three generative, lexicalised models for statistical parsing. Proceedings of the 35th Annual Meeting of the Association for Computational Linguistics and Eighth Conference of the European Chapter of the Association for Computational Linguistics; Madrid, Spain: Association for Computational Linguistics; 1997. [Google Scholar]
- 31.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics; Sapporo, Japan: Association for Computational Linguistics; 2003. [Google Scholar]
- 32.Bikel DM. On the parameter space of generative lexicalized statistical parsing models. 2004 [Google Scholar]
- 33.Charniak E, Johnson M. Coarse-to-fine n-best parsing and MaxEnt discriminative reranking. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics; Ann Arbor, Michigan: Association for Computational Linguistics; 2005. [Google Scholar]
- 34.Collins MJ. A new statistical parser based on bigram lexical dependencies. Proceedings of the 34th annual meeting on Association for Computational Linguistics; Santa Cruz, California: Association for Computational Linguistics; 1996. [Google Scholar]
- 35.Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, McCarty CA, Davis RL, Skaar T, Lamba J, Savova G. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 89:379–86. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]