Abstract
Background
Evidence for the associations of single nucleotide polymorphisms (SNPs) in the F7 gene and factor VII (FVII) levels and with risk of coronary heart disease (CHD) is inconsistent. We examined whether F7 tagging SNPs and haplotypes were associated with FVII levels, coagulation activation markers (CAMs) and CHD risk in two cohorts of UK men.
Methods
Genotypes for eight SNPs and baseline levels of FVIIc, FVIIag, and CAMs (including FVIIa) were determined in 2773 healthy men from the Second Northwick Park Heart Study (NPHS-II). A second cohort, Whitehall II study (WH-II, n=4055), was used for replication analysis of FVIIc levels and CHD-risk.
Results
In NPHS-II the minor alleles of three SNPs (rs555212, rs762635, and rs510317; haplotype H2) were associated with higher levels of FVIIag, FVIIc, and FVIIa, while the minor allele for two SNPs (I/D323, and rs6046; haplotype H5) were associated with lower levels. Adjusted for classical risk factors, H2 carriers had a CHD Hazard Ratio of 1.34 (CI 95%: 1.12–1.59; independent of FVIIc), while H5 carriers had a CHD-risk of 1.29 (CI 95%: 1.01–1.56; not independent of FVIIc) and significantly lower CAMs. Effects of haplotypes on FVIIc levels were replicated in WH-II, as was association of H5 with higher CHD-risk (pooled-estimate OR 1.16 [1.00–1.36], P=0.05), but surprisingly, H2 exhibited a reduced risk for CHD.
Conclusion
tSNPs in the F7 gene strongly influence FVII levels. The haplotype associated with low FVIIc level, with particularly reduced functional activity, was consistently associated with increased risk for CHD, while the haplotype associated with high FVIIc level was not.
Keywords: Factor VII, F7 gene, tagging SNPs, haplotypes, incident coronary heart disease
Introduction
Factor VII (FVII), a vitamin K-dependent glycoprotein secreted by the liver, plays an important role in the initiation of coagulation by tissue factor [1]. Following endothelial damage, exposed tissue factor binds and activates FVII [2–3]. Levels of FVII coagulant activity (FVIIc) determine the degree of thrombus formation after disruption of extensive atherosclerotic plaques following exposure to tissue factor [4]. Several [5–6], but not all [7–9] studies suggest that elevated levels of plasma FVIIc are also associated with increased risk of cardiovascular disease and venous thrombosis.
Plasma FVIIc levels are regulated by environmental factors, such as age, body mass index (BMI), dietary fat intake, plasma lipids, especially triglycerides (TG), and diabetes [10–13], and by genes, with heritability estimates from twin studies ranging from 57% to 63% of the variance attributable to a genetic component [14–15]. The F7 gene located at chromosome 13q34 includes nine exons [16] and several single nucleotide polymorphisms (SNPs) have been identified in the gene. Amongst the most studied variants, the minor alleles of the SNPs rs510335 (−401G/T), rs5742910 (−323 0 bp/10 bp insertion) in the promoter region of the gene and rs6046 (R353Q) in exon 9 of the gene, have been strongly and consistently associated with lower plasma FVIIc levels [17–19]. The R353Q variant has been shown previously to be in linkage disequilibrium with the I/D323 variant [20] and it has been suggested that the effects attributed to the R353Q may be due to the −323 insertion effect upon transcription [19, 21].
There are inconsistencies, however, concerning the association of F7 SNPs with the risk of coronary heart disease (CHD). Some studies have shown that these “FVII-lowering” SNPs are protective against myocardial infarction (MI) [4, 22–23] while others were not able to confirm the association [24–25]. A recent study of Indian Asians has also shown a positive association for R353Q and CHD [26]. The minor allele of the SNP rs510317 (−402G/A) in the promoter has been associated with both increased plasma FVIIc levels and an increased risk of MI in European men [4, 27]. Different findings on F7 SNPs and CHD may be due to the low statistical power of small studies, the presence of various confounders and from the possible impact of non-genotyped functional SNPs located elsewhere in the F7 gene.
Few studies have examined haplotypes in the F7 gene as risk factors for CHD. Sabater-Lleal et al. [28] identified 17 haplotypes from 43 SNPs that were at a frequency of >0.01. The 4 most frequent haplotypes encompassed half of the chromosomes studied and 12 haplotypes captured 80% of all chromosomes. One of the four haplotypes with highest frequency was associated with higher levels of FVII and another haplotype with lower levels of FVII, but their effect on risk for CHD was not assessed. Reiner et al. [29] have studied the effect of haplotypes on FVII level and CHD risk. A high expressing F7 haplotype was not associated with CHD-risk and a low expressing haplotype, also associated with lower BMI and lower HDL, was associated with a reduced risk of CHD. However, this association disappeared after adjustment for BMI and HDL.
The aim of the current study is 3-fold: (1) to evaluate the contribution of common tagging SNPs and haplotypes in the F7 gene to plasma FVII levels in a large group of healthy middle-aged men; (2) to examine whether SNPs and haplotypes associated with FVII levels are also associated with risk of CHD in this cohort and in a replication study; and (3), to obtain mechanistic insight by studying the association of these SNPs and haplotypes with downstream coagulation activation markers (CAMs) within the extrinsic coagulation pathway.
Methods
Study subjects and data collection
The prospective Second Northwick Park Heart Study (NPHS-II) commenced in 1989, and 3052 middle-aged men (49–64 years) were recruited from nine general medical practices in the UK. Participants were free of unstable angina, MI, evidence of silent infarcts, coronary surgery, anti-coagulant drugs (including aspirin), cerebrovascular disease, malignancy and any condition or disease preventing the attainment of written informed consent, or long-term follow-up. Information on lifestyle habits, height, weight, blood pressure was recorded at baseline and on subsequent prospective follow-up. Details of recruitment, blood processing and plasma measurements are described elsewhere [30]. Factor VII antigen (FVIIag), FVIIc (Brozovic method) and activated factor FVII (FVIIa) were determined as described previously [31–32] as were prothrombin F1+2 levels [33] and FIX activation peptide [2]. CHD end points up to15 years follow-up were as follows: 1. acute CHD events: sudden coronary death, fatal acute myocardial infarction, and nonfatal acute myocardial infarction (details of possible events were obtained through medical practices, hospitals, and coroners’ offices; the clinical history, ECGs, cardiac enzymes, and pathology were assessed by independent review according to World Health Organization criteria [34]; and normal limits for cardiac enzymes were those for the reporting laboratory); 2. a new major Q wave on the ECG after 5 years of follow-up (Minnesota codes 11, 12.1 to 12.7, and 12.8 plus 51 or 52) [35]; and 3. surgery for angina pectoris with CHD angiographically demonstrated. A sample of DNA was stored from 2773 men obtained at the time of recruitment, making it possible to study the effects of genotypes on incident heart disease. Interviews and repeat measurements were conducted annually for surviving participants.
Replication study: WH-II
The Whitehall II study (WH-II) recruited 10,308 participants (70% men) between 1985 and 1989 from 20 London-based Civil service departments [36–37]. Only men were used for current analysis, to make results comparable to NPHS-II. Blood samples for DNA were collected in 2002–2004 from more than 6000 participants [37]. Genetic risk analysis will therefore be assessed in survivors up to 17 years (n=4055). Blood samples were collected after either an 8-hour fast (participants presenting to the clinic in the morning) or at least 4 hours after a light fat-free breakfast (participants presenting in the afternoon) and stored at −70°C until analysis. Factor VII activity was measured in 1991–1993 by the Brozovic method [38]. CHD included first nonfatal myocardial infarction (MI) and first definite angina. Non-fatal MI was defined following MONICA criteria16 based on study electrocardiograms, hospital acute ECGs, and cardiac enzymes. Definite angina was defined on the basis of clinical records and nitrate medication use, excluding cases based solely on self-reported data without clinical verification and participants with definite angina at baseline. Prevalent data for CHD including both definite anginas, fatal and non-fatal MI at 17 years was used for the current analysis.
Genetic analysis in NPHS-II: tSNPs for the F7 gene were selected with Tagger [39] using the CEU panel of HapMap, applying an r2 threshold of 0.8 and a minor allele frequency threshold of 0.04. Five SNPs were genotyped using an Illumina GoldenGate candidate gene chip [40], F7: rs555212 (promoter), rs1475931 (intron 2), rs6046 (R353Q; Arg353Gln, exon 9), rs3093248, and rs3211719 (F10, intron 1). The tSNPs were optimized for the Illumina platform by preferentially selecting the SNP with the highest genotyping success rate in each block as a tag and re-evaluating the r2 in the sample. Additionally, 3 SNPs rs762635 (−670A>G, promoter), rs510317 (−402G>A, promoter), and I>D323 (−323A1/A2; A1-del/A2-ins, promoter) were included that had been previously genotyped by RFLP methods [41–42]. In WH-II study four SNPs were genotyped using the HumanCVD BeadChipI llumina [43], the remaining SNPs were imputed using phasing algorithms.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was assessed with chi-square test. Pairwise linkage-disequilibrium (LD) between the SNPs was calculated from the genotype data and measures as D′ and r2. Haplotype frequencies were calculated using the PHASE algorithm, [44–46]. Missing SNP genotypes were imputed by the phasing algorithm estimating the haplotype phase from genotype data using population genetic models, considered as the most accurate phasing method [47]. The levels of FVIIc and FIX activation peptide measurements were square root transformed and the levels of FVIIag, FVIIa, protthrombin F1+2 and TG measurements were logarithmically transformed to reduce skewness in their distribution. The t-test and chi-square test were used to compare a number of phenotypes between those with and those without a CHD event. General linear models were used to identify associations between the SNPs or haplotypes and plasma FVII levels. The common homozygote genotype for each SNP, (also the most frequent haplotype) was used as reference. A mixed model was used to analyse the effects of each of the SNPs or haplotypes on baseline and year 1–5 measures of plasma FVII. Since associations were formally examined for seven SNPs and five traits, some adjustment of the significance level is appropriate to account for multiple comparisons. However, there are high correlations between pairs of the phenotypes and some of the SNPs also showed a degree of linkage disequilibrium, therefore the Bonferroni correction would be too conservative [48–49]. In addition, type I errors are random and patterns in results that confirm previous reports, should be given more weight than isolated results with a single low P-value [48–49]. Also, correction for multiple comparisons largely increases the likelihood of type II errors and important differences are considered non-significant [48–49]. To take this into account, for individual tests of association, rather than applying a correction for multiple testing at global significance level of 5% (P<0.05) we defined statistical significance as less than 1% (P<0.01). We believe this to be appropriate because for example in the case of haplotypes there are 5 tests over 5 correlated measures with the P=0.01 cut off being close to the Bonferroni correction (0.05/5=0.01). Cox proportional hazards models were used to estimate hazard ratios (HR) and logistic regression analysis was used to estimate Odds Ratio (OR) and 95% confidence intervals (95% CI) for the associations of SNPs and haplotypes with risk of CHD events in NPHS-II study and WH-II study, respectively.
Results
The baseline characteristics of the NPHS-II and WH-II men who did or did not develop CHD are shown in supplementary Table S1. No significant differences were observed in FVIIc or FVIIag levels between those who did or did not subsequently have a CHD event, supplementary Table S1, while as previously reported [50], FVIIa levels were lower in those who later developed a CHD event in NPHS-II (no CHD 2.42 ng/mL (SD 1.49); CHD 2.05 ng/mL (SD 1.07), P=0.04). Overall, the mean FVIIc levels in NPHS-II were significantly higher than in the WH-II subjects (109±29% vs. 88±26%, P<0.001), but this was almost completely explained by the higher triglyceride levels in NPHS-II (2.08±1.26mmol/l vs. 1.57±1.25mmol/l, P<0.001). (See Supplementary Table 1).
Allele frequencies and pair-wise LD structure across the F7 gene
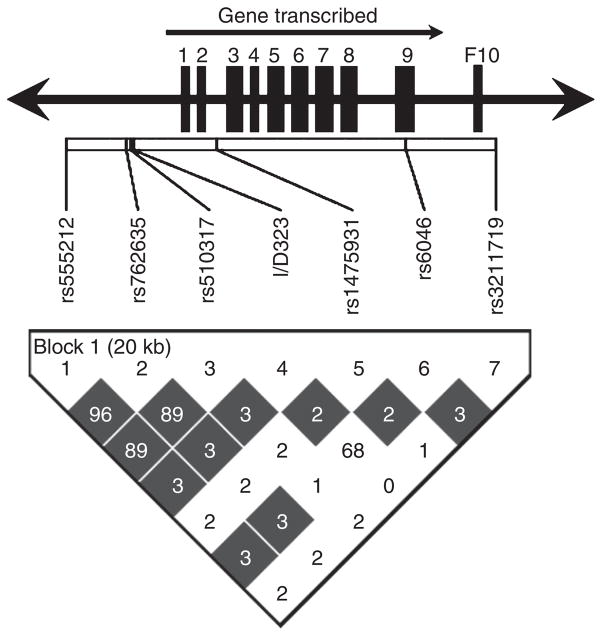
All polymorphisms genotyped were in Hardy-Weinberg equilibrium. The reported SNP rs3093248 was monomorphic so was not considered further. The LD structure, expressed as D′ and r2, is shown in Figure 1.
Figure 1.
Pair-wise linkage disequilibrium structure represent as a D′ (different color intensities) and r2 values (numbers) in 7 SNPs. The rs numbers and the relative physical distance between the SNPs are shown above (exons are the larger rectangular boxes). The color gradient indicates relative level of LD from black complete to white no LD.
Associations of F7 SNPs with FVIIag, FVIIc, and classical risk factors for CHD
In NPHS-II the minor alleles of three SNPs (rs555212, rs762635, and rs510317) were associated with higher FVIIag and FVIIc levels, while for two SNPs (I/D323, and rs6046) the minor alleles were associated with lower plasma FVIIc levels and FVIIag levels, Table 1. Similar results were seen in the WH-II study. No significant differences in BMI, diabetes status, or BP were seen associated with any of the F7 SNPs (data not shown).
Table 1.
The association between F7 genotypes and FVIIag, and FVIIc levels in NPHS-II study.
| SNPs | Genotype | n | FVIIag % (SE) | P-value | n | FVIIc % (SE) | P-value |
|---|---|---|---|---|---|---|---|
| rs555212 | AA | 1522 | 124 (0.86) | 1594 | 105 (0.69) | ||
| AG | 950 | 137 (1.09) | 1025 | 114 (0.87) | |||
| GG | 132 | 147 (2.92) | 5.37E-34 | 142 | 125 (2.33) | 4.46E-26 | |
|
| |||||||
| rs762635 | AA | 1530 | 124 (0.86) | 1602 | 105 (0.69) | ||
| AC | 947 | 137 (1.09) | 1022 | 114 (0.87) | |||
| CC | 127 | 147 (2.98) | 2.21E-33 | 137 | 124 (2.38) | 2.29E-25 | |
|
| |||||||
| rs510317 | GG | 1483 | 125 (0.88) | 1555 | 105 (0.71) | ||
| GA | 967 | 136 (1.09) | 1041 | 113 (0.87) | |||
| AA | 154 | 143 (2.72) | 2.99E-23 | 165 | 120 (2.18) | 1.81E-17 | |
|
| |||||||
| I/D 323* | A1A1 | 2084 | 134 (0.72) | 2221 | 114 (0.57) | ||
| A1A2 | 499 | 112 (1.48) | 517 | 91 (1.18) | |||
| A2A2 | 21 | 95 (7.21) | 4.59E-62 | 23 | 71 (5.57) | 1.11E-78 | |
|
| |||||||
| rs1475931 | TT | 1764 | 130 (0.82) | 1871 | 109 (0.65) | ||
| TG | 756 | 129 (1.25) | 801 | 110 (1.00) | |||
| GG | 84 | 131 (3.74) | 0.880 | 89 | 115 (3.00) | 0.085 | |
|
| |||||||
| rs6046 | GG | 2090 | 135 (0.71) | 2226 | 114 (0.56) | ||
| GA | 493 | 110 (1.47) | 510 | 89 (1.17) | |||
| AA | 21 | 93 (7.13) | 4.87E-79 | 25 | 71 (5.27) | 4.58E-98 | |
|
| |||||||
| rs3211719 | AA | 1625 | 130 (0.85) | 1726 | 109 (0.68) | ||
| AG | 850 | 129 (1.18) | 899 | 109 (0.95) | |||
| GG | 129 | 136 (3.02) | 0.059 | 136 | 115 (2.43) | 0.023 | |
A1-del, A2-ins; FVIIag, Factor VII antigen; FVIIc, factor VII coagulant activity.
Association between F7 haplotypes and FVIIag, or FVIIc levels
In NPHS-II haplotypes were inferred using seven SNPs (rs555212, rs762635, rs510317, I/D323, rs1475931, rs6046, rs3211719). Haplotypes with a relative frequency of >1.5% were used in the analysis, which included 93.8% of the inferred haplotypes. Six haplotypes were observed, and compared to the most common haplotype (H1, frequency 0.37), Table 2. Two haplotypes were independently associated with significant effects on plasma FVIIc, and FVIIag levels; Haplotype H2 (frequency 0.21) with 6.8% and 6.1% (P<0.01) higher levels of FVIIag, and FVIIc, respectively, and haplotype H5 (frequency 0.085) with 19.8% and 26.0% (P<0.01) lower levels of FVIIag, and FVIIc levels, respectively, Figure 2. FVIIc levels had also been measured in years 1–5. FVIIc increased over the five years, in all haplotypes, with H2 ranking highest at all time points and with H5 showing consistently lower plasma FVIIc levels over the period, supplementary Table S2.
Table 2.
Common haplotypes estimated for the F7 cluster and association with plasma FVIIag, FVIIc, FVIIc/FVIIag ratio, FVIIa, F 1+2, and FIXpep in NPHS-II and WH-II studies.
| NPHS-II | WH-II | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap. | rs555212 | rs762635 | rs510317 | I/D323* | rs1475931 | rs6046 | rs3211719 | Freq. (%) |
FVIIag % (SE) |
FVIIc % (SE) |
FVIIc /FVIIag Ratio |
FVIIa (ng/mL) |
F 1+2 (μmol/L) |
FIXpep (pmol/L) |
Freq. (%) |
FVIIc % (SE) |
| H1 | A | A | G | A1 | T | G | A | 36.51 | 130 (0.76) | 110 (0.61) | 0.87 (0.01) | 2.43 (0.05) | 0.75 (0.01) | 220 (3.60) | 38.36 | 88 (0.47) |
| H2 | G | C | A | A1 | T | G | A | 20.79 | 139 (1.01)‡ | 116 (0.80)‡ | 0.86 (0.01) | 2.58 (0.07)† | 0.79 (0.02)† | 217 (4.60) | 18.71 | 95 (0.67)‡ |
| H3 | A | A | G | A1 | T | G | G | 11.45 | 132 (1.35) | 110 (1.08) | 0.86 (0.01) | 2.35 (0.09) | 0.81 (0.02) | 220 (6.40) | 13.11 | 90 (0.79)‡ |
| H4 | A | A | G | A1 | G | G | A | 9.41 | 130 (1.48) | 111 (1.19) | 0.89 (0.01) | 2.54 (0.10) | 0.78 (0.03) | 205 (7.02) | 10.18 | 90 (0.91)‡ |
| H5 | A | A | G | A2 | T | A | A | 8.51 | 109(1.56)‡ | 87 (1.25)‡ | 0.84 (0.01)‡ | 1.60 (0.10)‡ | 0.77 (0.03) | 195 (7.07)‡ | 8.11 | 66 (1.01)‡ |
| H6 | A | A | G | A1 | G | G | G | 7.14 | 128 (1.71) | 110 (1.36) | 0.88 (0.01) | 2.40 (0.11) | 0.81 (0.03) | 208 (7.93) | 7.67 | 92 (1.05)‡ |
Hap., Haplotypes;
A1-del, A2-ins; FVIIag, Factor VII antigen; FVIIc, Factor VII coagulant activity; FVIIa Factor VII activated; F 1+2, Factor 1+2; FIX, Factor IX activation peptide; Freq., frequency;
P<0.05;
P<0.01.
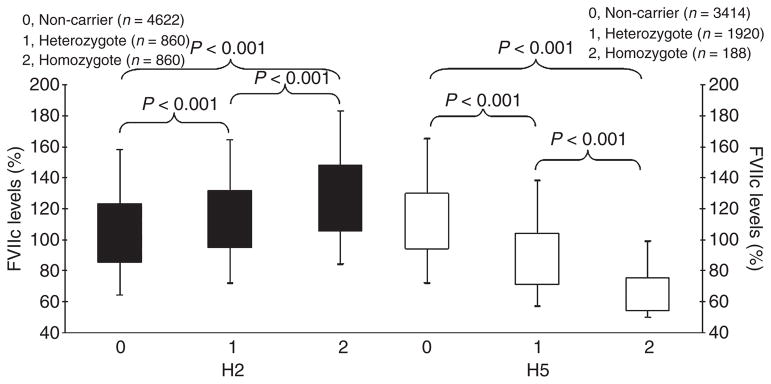
Figure 2.
The association between FVIIc levels and haplotypes H2/H5, in NPHS-II.
Subjects with two H5 alleles (0.7% of the sample) had FVIIag levels 29% lower and FVIIc levels 39% lower than non-carriers, with heterozygotes for the haplotype having intermediate FVIIc levels, (18% lower than non-carriers for FVIIag levels, 22% lower than non-carriers for FVIIc levels). This haplotype explained 8.5% (P<0.001) of sample variance in FVIIag levels and 12.1% (P<0.001) of sample variance in FVIIc levels. There was a small but significant reduction in FVIIc compared to FVIIag levels, for subjects carrying H5, shown by a statistically significant lower ratio of FVIIc/FVIIag (0.84, P<0.01), compared to the most frequent haplotype, H1, ratio 0.87. The ratios for H2, H3, H4 and H6 were not significantly different to H1.
In the WH-II study six haplotypes with a relative frequency of >1.7% were observed, which included 96.1% of the inferred haplotypes. Haplotype H2 and H5 were independently associated with significant effects upon plasma FVIIc levels: Haplotype H2 (frequency 0.19) with 12.7% (P<0.01) higher levels of FVIIc, and haplotype H5 (frequency 0.081) with 49.4% (P<0.01) lower levels of FVIIc levels, giving essentially the same results shown for NPHS-II.
To look for possible interactions, the pairwise effects on FVIIc levels of all SNPs showing significant effect in univariate analysis were examined. There was no statistically significant evidence for interaction between any pairs of SNPs in determining Factor VII coagulant activity in either study (data not shown), although there is limited power to detect such effects.
Association between F7 haplotypes and coagulation activation markers
In NPHS-II haplotypes H2 and H5 were associated with significant effects on FVIIa levels, which mirrored the effects seen in FVIIag and FVIIc, ie, higher levels of FVIIa in H2 carriers and lower levels of FVIIa in H5 carriers, Table 2. The higher levels of FVIIa associated with H2, were also associated with an increase in downstream activation of extrinsic coagulation, as determined by FIX activation peptide (FIXpep) or thrombin generation (prothrombin F1+2) levels. The lower level of FVIIa for haplotype H5, however, was associated with significantly lower levels of plasma FIXpep.
Associations of F7 SNPs and haplotypes with CHD
The univariate association between F7 genotypes and risk of a CHD-event in NPHS-II is shown in supplementary Table S3 and S4. There were no significant associations between individual SNPs and CHD. In addition, none of the inferred haplotypes were associated with CHD when compared separately to H1, Table 3. However, compared to non-carriers of H2, after combining individuals with one and two H2 alleles, there was a 26% excess risk of CHD after adjustment for age, BMI, diabetes, total cholesterol, smoking, and systolic blood pressure, and this effect was essentially unchanged after further adjustment for plasma levels of FVIIc (HR 1.34, CI 95%: 1.12–1.59). Similarly, after combining individuals with one and two alleles for haplotype H5 there was a 26% higher risk of CHD compared to the non-H5 carriers after adjustment for traditional CHD risk factors (HR 1.29 CI 95%: 1.01–1.56) although this effect was lost after adjustment for plasma levels of FVIIc.
Table 3.
Haplotype frequencies in CHD and non-CHD subjects and the association between F7 haplotypes and risk for CHD events in NPHS-II and WH-II studies
| Haplotype | Non-CHD % | CHD % | Adjusted modela HR (95% CI), P-value |
Adjusted model 1b HR (95% CI), P-value |
Adjusted model 2c HR (95% CI), P-value |
|---|---|---|---|---|---|
| NPHS-II | |||||
|
| |||||
| H1 | 36.6 | 35.9 | Ref. | Ref. | Ref. |
| H2 | 20.4 | 24.3 | 1.17 (0.94–1.46), 0.159 | 1.18 (0.94–1.47), 0.150 | 1.21 (0.97–1.52), 0.091 |
| H3 | 11.6 | 10.0 | 0.92 (0.68–1.25), 0.601 | 0.98 (0.73–1.33), 0.897 | 0.98 (0.73–1.33), 0.916 |
| H4 | 9.6 | 8.2 | 0.92 (0.66–1.27), 0.598 | 0.86 (0.62–1.20), 0.383 | 0.86 (0.61–1.19), 0.358 |
| H5 | 8.4 | 9.4 | 1.21 (0.89–1.64), 0.228 | 1.21 (0.89–1.65), 0.225 | 1.07 (0.78–1.47), 0.681 |
| H6 | 7.2 | 6.7 | 1.01 (0.71–1.43), 0.968 | 1.02 (0.71–1.45), 0.932 | 1.02 (0.71–1.45), 0.932 |
|
| |||||
| Non-carriers of H2 | 62.5 | 56.2 | Ref. | Ref. | Ref. |
| Carriers of H2 | 37.5 | 43.8 | 1.25 (1.06–1.48), 0.009 | 1.26 (1.06–1.49), 0.008 | 1.34 (1.12–1.59), 0.001 |
|
| |||||
| Non-carriers of H5 | 83.9 | 81.5 | Ref. | Ref. | Ref. |
| Carriers of H5 | 16.1 | 18.5 | 1.26 (1.01–1.56), 0.039 | 1.29 (1.01–1.56), 0.040 | 1.13 (0.89–1.44), 0.300 |
|
| |||||
| WH-II | |||||
|
| |||||
| H1 | 37.9 | 43.0 | Ref. | Ref. | Ref. |
|
| |||||
| H2 | 19.1 | 15.2 | 0.73 (0.58–0.91), 0.005 | 0.72 (0.56–0.92), 0.007 | 0.73 (0.57–0.94), 0.016 |
|
| |||||
| H3 | 13.2 | 12.4 | 0.85 (0.66–1.08), 0.180 | 0.84 (0.65–1.09), 0.191 | 0.89 (0.68–1.16), 0.392 |
|
| |||||
| H4 | 10.2 | 10.4 | 0.94 (0.73–1.23), 0.664 | 0.98 (0.75–1.29), 0.901 | 1.05 (0.79–1.38), 0.749 |
|
| |||||
| H5 | 8.1 | 8.2 | 0.95 (0.71–1.26), 0.704 | 0.95 (0.70–1.29), 0.766 | 0.87 (0.62–1.20), 0.387 |
|
| |||||
| H6 | 7.7 | 7.3 | 0.85 (0.63–1.15), 0.299 | 0.84 (0.60–1.17), 0.299 | 0.84 (0.60–1.19), 0.337 |
|
| |||||
| Non-carriers of H2 | 65.5 | 71.4 | Ref. | Ref. | Ref. |
|
| |||||
| Carriers of H2 | 34.5 | 28.7 | 0.77 (0.65–0.91), 0.003 | 0.75 (0.63–0.90), 0.002 | 0.77 (0.64–0.93), 0.008 |
|
| |||||
| Non-carriers of H5 | 84.8 | 84.4 | Ref. | Ref. | Ref. |
|
| |||||
| Carriers of H5 | 15.2 | 15.6 | 1.08 (0.88–1.33), 0.466 | 1.07 (0.86–1.34), 0.523 | 0.95 (0.74–1.22), 0.675 |
Percentages across the haplotype; HR, hazard ratio; OR, odds ratio (for WH-II); CI, confidence interval 95%; Ref., reference.
adjusted for age, clinic (only NPHS-II).
adjusted for age, clinic (only NPHS-II), BMI, diabetes, total cholesterol, smoking, and systolic blood pressure.
adjusted for age, clinic (only NPHS-II), BMI, diabetes, total cholesterol, smoking, systolic blood pressure, and FVIIc.
In WH-II study carriers of the minor allele for SNPs rs555121, rs762635, and rs510317 (showing strong LD) had a lower risk of a CHD event (OR=0.77 CI 95%: 0.60–0.98) compared with non-carriers after adjustment as above. This effect was lost after adjustment for plasma levels of FVIIc (OR=0.80 CI 95%: 0.62–1.03). SNPs I/D323 and rs6046 were associated with an increase in CHD-risk, but this effect did not reach statistical significance, P>0.49, supplementary Table 4. Haplotype H2 was associated with a significantly lower risk of a CHD event (OR 0.72 CI 95%: 0.56–0.92) compared with H1 after adjustment for age, BMI, diabetes, total cholesterol, smoking, and systolic blood pressure. Furthermore, the risk estimate did not change after further adjustment for FVIIc (HR 0.73, CI 95%: 0.57–0.94). Similarly, compared to non-carriers of H2, H2 allele carriers exhibited a 25% lower CHD-risk after adjustment for other CHD risk factors and this effect was also essentially unchanged after adjustment for plasma levels of FVIIc (OR=0.77, CI 95%: 0.64–0.93). H5 allele carriers tended towards increased CHD-risk only when compared to non-H5 carriers (after adjustment for age, BMI, diabetes, cholesterol, smoking, SBP and TG, 1.07 CI 95% 0.86–1.34).
Pooled Analysis: WH-II and NPHS-II association of F7 SNPs and haplotypes with CHD
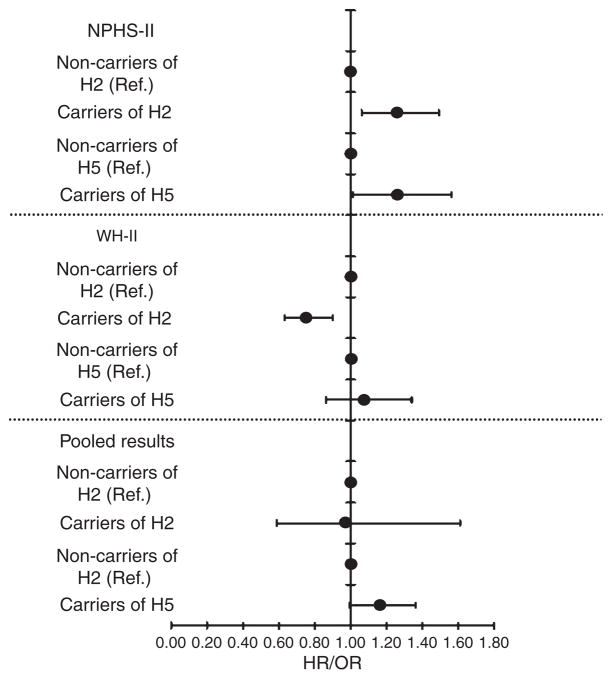
CHD-risk associated with H2 in NPHS-II and WH-II, were both statistically significant, but in different directions: in NPHS-II risk was higher (in all models, including a model with FVII levels), while in WH-II risk was lower, also in all models. By contrast, CHD-risk associated with H5 was significantly elevated in NPHS-II in a fully adjusted model (without inclusion of FVII levels) and was higher also in WH-II in a similar model, although not statistically significant, Figure 3. The pooled estimated of risk for H5 using the fully adjusted model (without FVII levels) was statistically significantly elevated (pooled-estimate HR 1.16 CI 95% 1.00–1.36, P=0.05).
Figure 3.
Association between haplotype H2, H5 and CHD in NPHS-II and WH-II studies.
HR, hazard ratio in NPHS-II study; OR, odds ratio in WH-II study. Adjusted for age, clinic, BMI, diabetes, total cholesterol, smoking, and systolic blood pressure, Lower panel shows pooled OR for both NPHS-II and WH-II.
Discussion
These two large cohort studies have confirmed the previous finding of genotypes and haplotypes in F7 having significant effects upon FVII levels. In accordance with other studies [17–19] one FVII raising haplotype, and one FVII lowering haplotype was identified. The effects identified in baseline samples were confirmed in years 1–5 for FVIIc levels in NPHS-II. The FVII lowering haplotype, H5, was associated with a small reduction in FVIIc/FVIIag ratio, which perhaps suggests that H5 gives rise to a protein with reduced function. The variant within the F7 promoter, I/D323, within H5, has been shown to be functional and to affect levels of transcription [19–21] but would not be expected to be associated with reduced activity of FVII protein. The amino acid substitution (R353Q) within H5 is a good contender for a reduced functional activity of the FVII protein.
When each of the F7 haplotypes were compared to the most frequent haplotype, H1, which contained the common allele at all sites, no significant associations with CHD could be identified, although there was a tendency for both H2 and H5 to exhibit a higher risk of CHD. In NPHS-II when carriers of H2 were compared to non-carriers (ie all other haplotypes), a similar higher CHD-risk was identified to that seen with individual genotypes within each haplotype (>25% increase in risk was identified) and these effects reached statistical significance. A similar trend for CHD-risk was also observed for H2 in the four-SNP model (data not shown). The CHD-risk association for H2 in NPHS-II was, however, completely reversed in WH-II, such that a reduced CHD-risk was associated with H2. It is possible that because DNA was not available in WH-II until year 17 of the study that there had been a survival bias against the H2 allele in WH-II. However, while there was a statistically significant difference between the allele frequencies for H2 in WH-II and NPHS-II, the frequency of H2 in WH-II was actually higher than that observed for NPHS-II, suggesting that more, rather than less of those with the H2 allele had survived in WH-II. It is possible that differences in CHD-risk between the studies were due to differences in demographics, WH-II having a more favorable risk profile with lower age, less smoking, lower BMI, lower blood pressure, (Supplementary Table S1). These finding require testing in further large prospective studies of heart disease.
It should be noted also that increased CHD-risk for H2 in NPHS-II remained in a model in which FVII levels were also included, suggesting that the risk shown in this study was not acting solely via the increased FVII levels. It is possible that the F7 allele is in LD with a risk allele outside the F7 gene, and further analysis to determine LD between the F7 gene and other SNPs in the surrounding region would be useful.
For H5 carriers, compared to non-carriers, a >25% increase in CHD-risk was identified in fully adjusted models, but the CHD-risk was no longer significant when FVIIc levels were included in the model, suggesting that risk is acting through the lower FVIIc levels identified for this haplotype. The findings of CHD-risk association were consistent in NPHS-II and WH-II, and a pooled estimate across the two studies was statistically significant. Interestingly, activated markers of coagulation downstream from FVII in NPHS-II were also significantly lower for this allele and the question arises as to what the pathological mechanism is for this lower, functionally less active FVII and lower levels of CAMs downstream from FVIIa. The findings point to reduced activation of the extrinsic coagulation pathway, resulting from a circulating FVII species with reduced coagulant activity. This observation is, however, counter-intuitive for an allele associated with increased risk of CHD. The processes of atherogenesis are complex, involving both plasma and vessel wall factors including proteins implicated in inflammation, dyslipidaemia, and thrombosis. However intra-plaque bleeding is also a recognized contributor to atherogenesis and it would be interesting to know, in future studies, whether this process is increased for the H5 haplotype. Interestingly there are reports of FVII deficiency associated with thrombotic events [51–52]. While the mechanism for FVII deficiency in thrombotic conditions has not been elucidated, those reported to date are functional deficiencies (normal or near normal antigen levels, with reduced FVIIc levels) and associated with amino acid substitutions. In the study of Marty et al. [52], 6 out of the 14 cases studied had the 353Q variant, associated with H5 in the current study. Reduced binding to tissue factor has been suggested as a contributory pathological mechanism in these cases [26]. However, it should be stated also that the observation may also be by chance and needs confirmation in further studies.
The combining of SNPs to form haplotypes and comparison of specific haplotypes to non-carriers of that haplotype, therefore, provided more statistically significant CHD-risk results than risk analysis for individual SNPs, or for assessment of risk in each haplotype, compared to the common haplotype. Reduced ability to detect CHD-risk when each haplotype was compared to the common haplotype is likely to be due to the lower numbers within the comparison group.
Study limitations
While the associations between F7 haplotypes and FVIIc levels are consistent across the studies and confirm previous reports, we have only studied UK Caucasian men, and effect sizes and associations may be different in women and in other ethnic groups. Since FVIIc was measured using the same method in both studies, the lower levels seen in the in WH-II men are likely to be due mainly to their younger age and lower triglyceride levels, although other unmeasured dietary or lifestyle factors may contribute. There are also differences in the definition of the CHD endpoint used in the two studies, with WH-II but not NPHS-II including definite angina, but it seems unlikely that the lower CHD risk associated with haplotype 5 can be confounded by this, and overall the prevalence of CHD was similar in both studies.
In summary we have identified a FVII raising haplotype (H2) that had significant, but opposite effects upon CHD-risk in two prospective studies, and a FVII lowering haplotype (H5), particularly associated with lower coagulant activity, that was associated with increased CHD-risk in two prospective studies. The elimination of significant CHD-risk associated with H5 when FVII levels were included in the model suggests that CHD-risk is acting via the lower (particularly coagulant) circulating FVII. Baseline FVII levels have been previously shown in cohort studies to be lower in those who developed a CHD-event [5–6], and in NPHS-II also, low FVIIa levels were previously shown to be associated with CHD-risk [50]. In combination, these findings suggest a non-linear association of FVII with CHD-risk with genotypes associated with both lower and potentially with higher levels of FVII (or FVIIa) being implicated in CHD.
Supplementary Material
Table S1: Baseline characteristic (Mean, SD) of the subjects who remained event free and who developed CHD in NPHS-II study and WH-II study.
Table S2: The association between F7 haplotypes and plasma FVIIc over the five years in NPHS-II study.
Table S3: SNP frequencies in CHD and non-CHD subjects and Cox proportional hazards models for the association between F7 SNPs and risk of CHD events in NPHS-II study.
Table S4: SNP frequencies in CHD and non-CHD subjects and logistic regression models for the association between F7 SNPs and risk of CHD events in WH-II study.
Acknowledgments
We acknowledge the contribution of the late Professor George Miller (1939–2006) who was the PI on the NPHS-II study. The British Heart Foundation support FD and SEH (PG2008/014). The NPHS-II study was supported by the Medical Research Council, the US National Institutes of Health (NHLBI 33014) and Du Pont Pharma. We also thank all the medical staff and patients who contributed to the NPHS-II study and the Office for National Statistics (NHS) Central Registry for provision of mortality data. We thank all participating civil service departments and their welfare, personnel, and establishment officers; the Occupational Health and Safety Agency; the Council of Civil Service Unions; all participating civil servants in the Whitehall II study; and all members of the Whitehall II study team. The Whitehall II study has been supported by grants from UK Medical Research Council (MRC), British Heart Foundation, Health and Safety Executive, Department of Health, National Heart Lung and Blood Institute (HL36310), National Institute on Aging (AG13196), Agency for Health Care Policy Research (HS06516) and MacArthur Foundation Research Network on Socio-economic Status and Health. M Kivimäki is supported by the Academy of Finland and a BUPA Foundation specialist research grant.
References
- 1.Rao LV, Rapaport SI. Activation of factor VII bound to tissue factor: a key early step in the tissue factor pathway of blood coagulation. Proc Natl Acad Sci U S A. 1988;85:6687–91. doi: 10.1073/pnas.85.18.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer KA, Kass BL, ten Cate H, Hawiger JJ, Rosenberg RD. Factor IX is activated in vivo by the tissue factor mechanism. Blood. 1990;76:731–6. [PubMed] [Google Scholar]
- 3.Osterud B, Rapaport SI. Activation of factor IX by the reaction product of tissue factor and factor VII: additional pathway for initiating blood coagulation. Proc Natl Acad Sci U S A. 1977;74:5260–4. doi: 10.1073/pnas.74.12.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozzini C, Girelli D, Bernardi F, Ferraresi P, Olivieri O, Pinotti M, Martinelli N, Manzato F, Friso S, Villa G, Pizzolo F, Beltrame F, Corrocher R. Influence of polymorphisms in the factor VII gene promoter on activated factor VII levels and on the risk of myocardial infarction in advanced coronary atherosclerosis. Thromb Haemost. 2004;92:541–9. doi: 10.1160/TH04-02-0130. [DOI] [PubMed] [Google Scholar]
- 5.Meade TW, Mellows S, Brozovic M, Miller GJ, Chakrabarti RR, North WR, Haines AP, Stirling Y, Imeson JD, Thompson SG. Haemostatic function and ischaemic heart disease: principal results of the Northwick Park Heart Study. Lancet. 1986;2:533–7. doi: 10.1016/s0140-6736(86)90111-x. [DOI] [PubMed] [Google Scholar]
- 6.Junker R, Heinrich J, Schulte H, van de Loo J, Assmann G. Coagulation factor VII and the risk of coronary heart disease in healthy men. Arterioscler Thromb Vasc Biol. 1997;17:1539–44. doi: 10.1161/01.atv.17.8.1539. [DOI] [PubMed] [Google Scholar]
- 7.Tracy RP, Bovill EG, Yanez D, Psaty BM, Fried LP, Heiss G, Lee M, Polak JF, Savage PJ. Fibrinogen and factor VIII, but not factor VII, are associated with measures of subclinical cardiovascular disease in the elderly. Results from The Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 1995;15:1269–79. doi: 10.1161/01.atv.15.9.1269. [DOI] [PubMed] [Google Scholar]
- 8.Ghaddar HM, Folsom AR, Aleksic N, Hearne LB, Chambless LE, Morrissey JH, Wu KK. Correlation of factor VIIa values with factor VII gene polymorphism, fasting and postprandial triglyceride levels, and subclinical carotid atherosclerosis. Circulation. 1998;98:2815–21. doi: 10.1161/01.cir.98.25.2815. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee AK, Pearson J, Gilliland EL, Goss D, Lewis JD, Stirling Y, Meade TW. A six year prospective study of fibrinogen and other risk factors associated with mortality in stable claudicants. Thromb Haemost. 1992;68:261–3. [PubMed] [Google Scholar]
- 10.Humphries SE, Lane A, Green FR, Cooper J, Miller GJ. Factor VII coagulant activity and antigen levels in healthy men are determined by interaction between factor VII genotype and plasma triglyceride concentration. Arterioscler Thromb. 1994;14:193–8. doi: 10.1161/01.atv.14.2.193. [DOI] [PubMed] [Google Scholar]
- 11.Simpson HC, Mann JI, Meade TW, Chakrabarti R, Stirling Y, Woolf L. Hypertriglyceridaemia and hypercoagulability. Lancet. 1983;1:786–90. doi: 10.1016/s0140-6736(83)91849-4. [DOI] [PubMed] [Google Scholar]
- 12.Feng D, Tofler GH, Larson MG, O’Donnell CJ, Lipinska I, Schmitz C, Sutherland PA, Johnstone MT, Muller JE, D’Agostino RB, Levy D, Lindpaintner K. Factor VII gene polymorphism, factor VII levels, and prevalent cardiovascular disease: the Framingham Heart Study. Arterioscler Thromb Vasc Biol. 2000;20:593–600. doi: 10.1161/01.atv.20.2.593. [DOI] [PubMed] [Google Scholar]
- 13.Mariani G, Bernardi F, Bertina R, Vicente VV, Prydz H, Samama M, Sandset PM, Di Nucci GD, Testa MG, Bendz B, Chiarotti F, Ciarla MV, Strom R. Serum phospholipids are the main environmental determinants of activated factor VII in the most common FVII genotype. European Union Concerted Action “Clotart”. Haematologica. 1999;84:620–6. [PubMed] [Google Scholar]
- 14.de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–5. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 15.Dubrey SW, Reaveley DR, Seed M, Lane DA, Ireland H, O’Donnell M, O’Connor B, Noble MI, Leslie RD. Risk factors for cardiovascular disease in IDDM. A study of identical twins. Diabetes. 1994;43:831–5. doi: 10.2337/diab.43.6.831. [DOI] [PubMed] [Google Scholar]
- 16.O’Hara PJ, Grant FJ, Haldeman BA, Gray CL, Insley MY, Hagen FS, Murray MJ. Nucleotide sequence of the gene coding for human factor VII, a vitamin K-dependent protein participating in blood coagulation. Proc Natl Acad Sci U S A. 1987;84:5158–62. doi: 10.1073/pnas.84.15.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kudaravalli R, Tidd T, Pinotti M, Ratti A, Santacroce R, Margaglione M, Dallapiccola B, Bernardi F, Fortina P, Devoto M, Pollak ES. Polymorphic changes in the 5′ flanking region of factor VII have a combined effect on promoter strength. Thromb Haemost. 2002;88:763–7. [PubMed] [Google Scholar]
- 18.Lane A, Green F, Scarabin PY, Nicaud V, Bara L, Humphries S, Evans A, Luc G, Cambou JP, Arveiler D, Cambien F. Factor VII Arg/Gln353 polymorphism determines factor VII coagulant activity in patients with myocardial infarction (MI) and control subjects in Belfast and in France but is not a strong indicator of MI risk in the ECTIM study. Atherosclerosis. 1996;119:119–27. doi: 10.1016/0021-9150(95)05638-6. [DOI] [PubMed] [Google Scholar]
- 19.Hunault M, Arbini AA, Lopaciuk S, Carew JA, Bauer KA. The Arg353Gln polymorphism reduces the level of coagulation factor VII. In vivo and in vitro studies. Arterioscler Thromb Vasc Biol. 1997;17:2825–9. doi: 10.1161/01.atv.17.11.2825. [DOI] [PubMed] [Google Scholar]
- 20.Girelli D, Russo C, Ferraresi P, Olivieri O, Pinotti M, Friso S, Manzato F, Mazzucco A, Bernardi F, Corrocher R. Polymorphisms in the factor VII gene and the risk of myocardial infarction in patients with coronary artery disease. N Engl J Med. 2000;343:774–80. doi: 10.1056/NEJM200009143431104. [DOI] [PubMed] [Google Scholar]
- 21.Pollak ES, Hung HL, Godin W, Overton GC, High KA. Functional characterization of the human factor VII 5′-flanking region. J Biol Chem. 1996;271:1738–47. doi: 10.1074/jbc.271.3.1738. [DOI] [PubMed] [Google Scholar]
- 22.Iacoviello L, Di Castelnuovo A, De Knijff P, D’Orazio A, Amore C, Arboretti R, Kluft C, Benedetta Donati M. Polymorphisms in the coagulation factor VII gene and the risk of myocardial infarction. N Engl J Med. 1998;338:79–85. doi: 10.1056/NEJM199801083380202. [DOI] [PubMed] [Google Scholar]
- 23.Nederhand RJ, de Maat MP, Jukema JW. Factor VII polymorphisms and myocardial infarction: what is special in Italians? REGRESS study group. Regression Growth Evaluation Statin Study. Thromb Haemost. 2001;85:746–7. [PubMed] [Google Scholar]
- 24.Doggen CJ, Manger Cats V, Bertina RM, Reitsma PH, Vandenbroucke JP, Rosendaal FR. A genetic propensity to high factor VII is not associated with the risk of myocardial infarction in men. Thromb Haemost. 1998;80:281–5. [PubMed] [Google Scholar]
- 25.Ardissino D, Mannucci PM, Merlini PA, Duca F, Fetiveau R, Tagliabue L, Tubaro M, Galvani M, Ottani F, Ferrario M, Corral J, Margaglione M. Prothrombotic genetic risk factors in young survivors of myocardial infarction. Blood. 1999;94:46–51. [PubMed] [Google Scholar]
- 26.Shanker J, Perumal G, Maitra A, Rao VS, Natesha BK, John S, Hebbagodi S, Kakkar VV. Genotype-phenotype relationship of F7 R353Q polymorphism and plasma factor VII coagulant activity in Asian Indian families predisposed to coronary artery disease. J Genet. 2009;88:291–7. doi: 10.1007/s12041-009-0042-x. [DOI] [PubMed] [Google Scholar]
- 27.Carew JA, Basso F, Miller GJ, Hawe E, Jackson AA, Humphries SE, Bauer KA. A functional haplotype in the 5′ flanking region of the factor VII gene is associated with an increased risk of coronary heart disease. J Thromb Haemost. 2003;1:2179–85. doi: 10.1046/j.1538-7836.2003.00424.x. [DOI] [PubMed] [Google Scholar]
- 28.Sabater-Lleal M, Martinez-Marchan E, Martinez-Sanchez E, Coll M, Vallve C, Mateo J, Souto JC, Fontcuberta J, Soria JM. Complexity of the genetic contribution to factor VII deficiency in two Spanish families: clinical and biological implications. Haematologica. 2003;88:906–13. [PubMed] [Google Scholar]
- 29.Reiner AP, Carlson CS, Rieder MJ, Siscovick DS, Liu K, Chandler WL, Green D, Schwartz SM, Nickerson DA. Coagulation factor VII gene haplotypes, obesity-related traits, and cardiovascular risk in young women. J Thromb Haemost. 2007;5:42–9. doi: 10.1111/j.1538-7836.2006.02279.x. [DOI] [PubMed] [Google Scholar]
- 30.Cooper JA, Miller GJ, Bauer KA, Morrissey JH, Meade TW, Howarth DJ, Barzegar S, Mitchell JP, Rosenberg RD. Comparison of novel hemostatic factors and conventional risk factors for prediction of coronary heart disease. Circulation. 2000;102:2816–22. doi: 10.1161/01.cir.102.23.2816. [DOI] [PubMed] [Google Scholar]
- 31.Miller GJ, Stirling Y, Esnouf MP, Heinrich J, van de Loo J, Kienast J, Wu KK, Morrissey JH, Meade TW, Martin JC, et al. Factor VII-deficient substrate plasmas depleted of protein C raise the sensitivity of the factor VII bio-assay to activated factor VII: an international study. Thromb Haemost. 1994;71:38–48. [PubMed] [Google Scholar]
- 32.Morrissey JH, Macik BG, Neuenschwander PF, Comp PC. Quantitation of activated factor VII levels in plasma using a tissue factor mutant selectively deficient in promoting factor VII activation. Blood. 1993;81:734–44. [PubMed] [Google Scholar]
- 33.Teitel JM, Bauer KA, Lau HK, Rosenberg RD. Studies of the prothrombin activation pathway utilizing radioimmunoassays for the F2/F1 + 2 fragment and thrombin--antithrombin complex. Blood. 1982;59:1086–97. [PubMed] [Google Scholar]
- 34.World Health Organization. Regional Office for Europe. Myocardial infarction community registers: results of a WHO international collaborative study coordinated by the Regional Office for Europe. Copenhagen: Regional Office for Europe, World Health Organization; 1976. [Google Scholar]
- 35.Prineas RJ, Crow RS, Blackburn HW. The Minnesota code manual of electrocardiographic findings: standards and procedures for measurement and classification. Boston, Mass: J. Wright; 1982. [Google Scholar]
- 36.Marmot M, Brunner E. Cohort Profile: the Whitehall II study. Int J Epidemiol. 2005;34:251–6. doi: 10.1093/ije/dyh372. [DOI] [PubMed] [Google Scholar]
- 37.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet. 2009;85:628–42. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brozovic M, Stirling Y, Harricks C, North WR, Meade TW. Factor VII in an industrial population. Br J Haematol. 1974;28:381–91. doi: 10.1111/j.1365-2141.1974.tb00819.x. [DOI] [PubMed] [Google Scholar]
- 39.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 40.Drenos F, Talmud PJ, Casas JP, Smeeth L, Palmen J, Humphries SE, Hingorani AD. Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum Mol Genet. 2009;18:2305–16. doi: 10.1093/hmg/ddp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas A, Lamlum H, Humphries S, Green F. Linkage disequilibrium across the fibrinogen locus as shown by five genetic polymorphisms, G/A-455 (HaeIII), C/T-148 (HindIII/AluI), T/G+1689 (AvaII), and BclI (beta-fibrinogen) and TaqI (alpha-fibrinogen), and their detection by PCR. Hum Mutat. 1994;3:79–81. doi: 10.1002/humu.1380030117. [DOI] [PubMed] [Google Scholar]
- 42.Baumann RE, Henschen AH. Linkage disequilibrium relationships among four polymorphisms within the human fibrinogen gene cluster. Hum Genet. 1994;94:165–70. doi: 10.1007/BF00202863. [DOI] [PubMed] [Google Scholar]
- 43.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham RR, Langefeld CD, Gaffney PM, Ortmann WA, Selby SA, Baechler EC, Shark KB, Ockenden TC, Rohlf KE, Moser KL, Brown WM, Gabriel SE, Messner RP, King RA, Horak P, Elder JT, Stuart PE, Rich SS, Behrens TW. Genetic linkage and transmission disequilibrium of marker haplotypes at chromosome 1q41 in human systemic lupus erythematosus. Arthritis Res. 2001;3:299–305. doi: 10.1186/ar319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin ZS, Munro HM, Abecasis GR, Donnelly P. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–50. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
- 49.Ken-Dror G, Talmud PJ, Humphries SE, Drenos F. APOE/C1/C4/C2 gene cluster genotypes, haplotypes and lipid levels in prospective Coronary Heart Disease Risk among UK healthy men. Mol Med. doi: 10.2119/molmed.2010.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller GJ, Ireland HA, Cooper JA, Bauer KA, Morrissey JH, Humphries SE, Esnouf MP. Relationship between markers of activated coagulation, their correlation with inflammation, and association with coronary heart disease (NPHSII) J Thromb Haemost. 2008;6:259–67. doi: 10.1111/j.1538-7836.2008.02819.x. [DOI] [PubMed] [Google Scholar]
- 51.Mariani G, Herrmann FH, Schulman S, Batorova A, Wulff K, Etro D, Dolce A, Auerswald G, Astermark J, Schved JF, Ingerslev J, Bernardi F. Thrombosis in inherited factor VII deficiency. J Thromb Haemost. 2003;1:2153–8. 395, pii. doi: 10.1046/j.1538-7836.2003.00395.x. [DOI] [PubMed] [Google Scholar]
- 52.Marty S, Barro C, Chatelain B, Fimbel B, Tribout B, Reynaud J, Schved JF, Giansily-Blaizot M. The paradoxical association between inherited factor VII deficiency and venous thrombosis. Haemophilia. 2008;14:564–70. doi: 10.1111/j.1365-2516.2007.01647.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Baseline characteristic (Mean, SD) of the subjects who remained event free and who developed CHD in NPHS-II study and WH-II study.
Table S2: The association between F7 haplotypes and plasma FVIIc over the five years in NPHS-II study.
Table S3: SNP frequencies in CHD and non-CHD subjects and Cox proportional hazards models for the association between F7 SNPs and risk of CHD events in NPHS-II study.
Table S4: SNP frequencies in CHD and non-CHD subjects and logistic regression models for the association between F7 SNPs and risk of CHD events in WH-II study.