Abstract
Human ether-á-go-go–related gene (hERG) potassium channels are critical for cardiac action potential repolarization. Cardiac hERG channels comprise two primary isoforms: hERG1a, which has a regulatory N-terminal Per-Arnt-Sim (PAS) domain, and hERG1b, which does not. Isolated, PAS-containing hERG1a N-terminal regions (NTRs) directly regulate NTR-deleted hERG1a channels; however, it is unclear whether hERG1b isoforms contain sufficient machinery to support regulation by hERG1a NTRs. To test this, we constructed a series of PAS domain–containing hERG1a NTRs (encoding amino acids 1–181, 1–228, 1–319, and 1–365). The NTRs were also predicted to form from truncation mutations that were linked to type 2 long QT syndrome (LQTS), a cardiac arrhythmia disorder associated with mutations in the hERG gene. All of the hERG1a NTRs markedly regulated heteromeric hERG1a/hERG1b channels and homomeric hERG1b channels by decreasing the magnitude of the current–voltage relationship and slowing the kinetics of channel closing (deactivation). In contrast, NTRs did not measurably regulate hERG1a channels. A short NTR (encoding amino acids 1–135) composed primarily of the PAS domain was sufficient to regulate hERG1b. These results suggest that isolated hERG1a NTRs directly interact with hERG1b subunits. Our results demonstrate that deactivation is faster in hERG1a/hERG1b channels compared to hERG1a channels because of fewer PAS domains, not because of an inhibitory effect of the unique hERG1b NTR. A decrease in outward current density of hERG1a/hERG1b channels by hERG1a NTRs may be a mechanism for LQTS.
INTRODUCTION
Human ether-á-go-go–related gene (hERG) potassium channels are members of the voltage-activated family of K+ channels, which contain six transmembrane domains and intracellular amino and carboxyl terminus domains (Warmke and Ganetzky, 1994). hERG subunits are the primary pore-forming units (Sanguinetti et al., 1995; Trudeau et al., 1995) of the rapid component of the delayed rectifier potassium current (IKr) in the heart (Noble and Tsien, 1969; Sanguinetti and Jurkiewicz, 1990, 1991). The physiological role of IKr is to help repolarize cardiac action potentials (Noble and Tsien, 1969; Sanguinetti and Jurkiewicz, 1990, 1991). Genetic mutations in two primary hERG1 subunits, hERG1a (Curran et al., 1995) and hERG1b (Sale et al., 2008), are linked to the long QT syndrome (LQTS), indicating the importance of both primary subunit isoforms in heart disease. Evidence suggests that mammalian ERG1a and ERG1b co-associate to form cardiac IKr (Lees-Miller et al., 1997; London et al., 1997; Jones et al., 2004; Sale et al., 2008) and also co-associate in the brain (Guasti et al., 2005). The two hERG isoforms are structurally different, as hERG1a channel subunits have a large intracellular N-terminal region (NTR; ∼390 amino acids in length) that contains a Per-Arnt-Sim (PAS) regulatory domain (Morais Cabral et al., 1998). In contrast, hERG1b subunits have a much shorter NTR (∼59 amino acids) and lack a PAS domain (Lees-Miller et al., 1997; London et al., 1997).
PAS domains are basic helix-loop-helix motifs found in a wide variety of proteins and are instrumental in a range of biological functions that include sensing environmental cues, regulating transcription, and mediating protein interactions (Jackson et al., 1986; Reddy et al., 1986; Hoffman et al., 1991; Nambu et al., 1991; Möglich et al., 2009). hERG PAS is a helix-loop-helix motif formed by amino acids 26–135 (Morais Cabral et al., 1998; Li et al., 2010; Muskett et al., 2011; Ng et al., 2011) and is capped by a short adjacent region composed of amino acids 1–26, of which residues 13–26 form a helix and residues 1–13 are unordered (Li et al., 2010; Muskett et al., 2011; Ng et al., 2011). Together, the PAS domain and the cap region (residues 1–135) are known as the “eag domain” (Morais Cabral et al., 1998). An intact eag domain is required for the slow time course of channel closing (deactivation) that is characteristic of hERG1a channels (Spector et al., 1996; Morais Cabral et al., 1998; Wang et al., 1998). The eag domain regulates gating by interacting directly with intracellular regions of hERG1a (Morais Cabral et al., 1998; Gustina and Trudeau, 2009), including the C-terminal cyclic nucleotide–binding domain (Gustina and Trudeau, 2011). Remarkably, the hERG eag domain retains its regulatory function when expressed as a fusion protein (Morais Cabral et al., 1998) or as a separate genetic fragment (Gustina and Trudeau, 2009). hERG1a channels with deletions of the eag domain exhibit approximately fivefold faster deactivation than wild-type channels (Spector et al., 1996; Morais Cabral et al., 1998; Wang et al., 1998). Likewise, naturally occurring hERG1b isoforms that lack eag domains have deactivation kinetics that are approximately fivefold faster than those of hERG1a (Lees-Miller et al., 1997; London et al., 1997).
Here, we asked whether hERG1b channels supported regulation by isolated hERG1a eag domains. To directly test this we constructed plasmids encoding a family of polypeptides that each contained the hERG1a eag domain plus additional regions of different lengths that corresponded to the proximal parts of the hERG1a NTR (Fig. 1, A and B). The lengths of the isolated polypeptides were also chosen because they were proposed to be formed from genetic mutations in the NTR that were linked to type 2 LQTS (Tester et al., 2005). Here, we report that all hERG1a NTRs functionally regulated gating in heteromeric hERG1a/hERG1b potassium channels and homomeric hERG1b channels, but not hERG1a channels. All NTRs markedly slowed deactivation kinetics of hERG1a/hERG1b channels and hERG1b channels, and reduced the magnitude of the I-V relationship. An NTR composed of just the eag domain (amino acids 1–135) was sufficient to regulate homomeric hERG1b channels. The NTRs effectively converted hERG1a/hERG1b or hERG1b channels into channels with properties similar to wild-type hERG1a channels. A reduction in the peak outward current in hERG1a/hERG1b channels by eag (and PAS) domain–containing NTRs may be a mechanism for LQTS.
Figure 1.
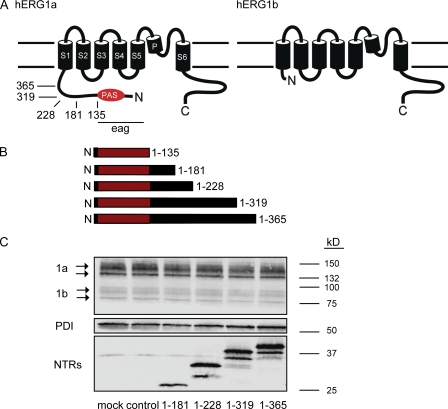
hERG1a NTRs form polypeptides. (A) Schematic of hERG1a channel subunit and hERG1b subunit. Arrows indicate positions of truncation mutants in the hERG1a NTR. (B) Schematic of hERG1a NTR polypeptides formed from truncation mutants. (C) Western blot of lysates from hERG1a/hERG1b stable cell line transfected with hERG1a NTR plasmids. (Top) Bands corresponding to hERG1a and hERG1b; (middle) bands corresponding to PDI, which is used as a loading control; and (bottom) bands corresponding to hERG1a NTR polypeptides as indicated. Lanes corresponding to control (Kir2.1) and mock (transfection reagent) experiments were also included, as indicated.
MATERIALS AND METHODS
Molecular biology and cell culture
We used a clonal stable cell line generated from human embryonic kidney 293 (HEK293) cells and hERG1a and hERG1b plasmids, as described previously (Jones et al., 2004). Cells were cultured at 37°C, 5% CO2 in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine. hERG1a and hERG1b subunit expression was maintained by supplementing DMEM with 1 µl/ml Zeocin and 500 µg/ml Geneticin. All hERG NTR plasmids were generated with PCR-based mutagenesis and had in-frame c-myc and 6x His epitope tags, or were genetically fused in-frame to enhanced cyan fluorescent protein (eCFP) at the C terminus. Clones for studies in mammalian cells were in the pcDNA3.1 vector, and clones for studies in oocytes were in the pGH19 vector. Transfections into hERG1a/hERG1b stable cell lines were performed using 1–2 µg hERG cDNAs and TransIT-LT1 transfection reagent (Mirus) according to the manufacturer’s protocol. Cells were incubated for 24–48 h before analysis by whole cell patch clamp and Western blot. Oocyte handling was performed as described previously (Gustina and Trudeau, 2009, 2011). As in previous studies with oocytes, hERG1a NTR plasmids were expressed at a 2:1 RNA ratio compared with hERG1a or hERG1b RNA (Gustina and Trudeau, 2009, 2011).
Electrophysiology
Whole cell patch-clamp recordings were performed from HEK293 cells that stably expressed hERG1a and hERG1b (see Fig. 2). Recordings were performed with a patch-clamp amplifier (EPC10; HEKA) and acquisition software (Patchmaster; HEKA). All recordings were performed at room temperature (22–24°C). Recording electrodes had initial resistances of 2–3 MΩ. The internal pipette solution contained 130 mM KCl, 1 mM MgCl2, 5 mM EGTA, 5 mM MgATP, and 10 mM HEPES, pH 7.2. The bath solution contained 137 mM NaCl, 4 KCl mM, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES, pH 7.4. A small endogenous K+ conductance in HEK293 cells was inhibited by 5 mM TEA. Two-electrode voltage-clamp recordings were performed as described previously (Gustina and Trudeau, 2009, 2011). All data were recorded at 1 kHz, and whole cell data were filtered (10 kHz; Bessel). To help identify HEK293 cells transfected with hERG1a NTR c-myc/6x His plasmids, cells were simultaneously transfected with 1 µg eCFP in preliminary experiments. We did not detect a measurable difference in the kinetics of hERG1a/hERG1b cells regulated by hERG1a 1–228-c-myc/6x His (τ deactivation [s] at −40 mV = 1.47 ± 0.18; n = 4) or hERG1a 1–228-eCFP (τ deactivation [s] = 1.58 ± 0.12; n = 6) at −40 mV, so to further facilitate identification of NTR-transfected cells, most experiments were performed with NTRs fused directly to eCFP at the C terminus. As a positive control, currents from untransfected stable hERG1a/hERG1b cells were recorded in parallel with currents from NTR-transfected stable hERG1a/hERG1b cells each time we split the stable cells. This was done to make sure that currents from hERG1a/hERG1b stable cells had characteristics of hERG1a/hERG1b channels (i.e., less rectification and faster deactivation kinetics compared with hERG1a). Conductance–voltage relationships were fit using a Boltzmann function, where y = 1/{1 + exp[(V1/2 − V)/k]}, and V is the membrane voltage, V 1/2 is the voltage at which half the channels are activated, and k is the slope. Current relaxations at negative potentials were fit with a single-exponential function (y = A1e−t/τ), where t is time and τ is the time constant for deactivation. Each point was the mean ± SEM. One-way ANOVA was used to determine statistical significance. The number of cells per experiment is represented by n.
Figure 2.
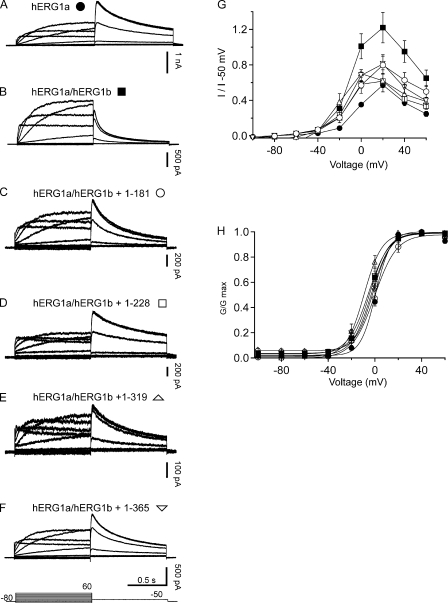
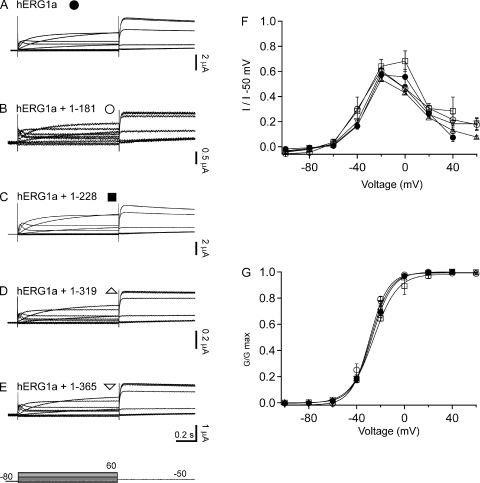
hERG1a NTR polypeptides regulate steady-state properties of hERG1a/hERG1b channels. Whole cell patch-clamp recordings of (A) hERG1a, (B) hERG1a/hERG1b channels, (C) hERG1a/hERG1b plus hERG1a 1–181, (D) hERG1a/hERG1b plus hERG1a 1–228, (E) hERG1a/hERG1b plus hERG1a 1–319, and (F) hERG1a/hERG1b plus hERG1a 1–365. The voltage protocol (shown at the bottom) consisted of steps from −100 to 60 mV for 2-s durations in 20-mV increments, followed by a step to −50 mV for 2 s. The holding potential was −80 mV. (G) Plot of I-V relationship for each channel in A–F normalized to peak tail current at −50 mV. (H) G-V relationship for channels in A–F. Bars in A–F, 0.5 s; n ≥ 6.
Western blot analysis
A stable cell line expressing hERG1a and hERG1b subunits was cultured in 60 × 15–mm cell culture dishes for 24 h and transfected with hERG1a NTR plasmids. 48 h after transfection, cells were washed with phosphate-buffered saline followed by 1 mL of lysis buffer (150 mM NaCl, 25 mM Tris-HCl, pH 7.4, 20 mM NaEDTA, and 10 mM NaEGTA, pH to 7.4 with NaOH). Cells were lysed with denaturing lysis buffer (lysis buffer plus 5 mM glucose, 1.0% Triton X-100, and protease inhibitors [10 µg/ml each]) at 4°C for 30 min with rotation. Cell debris was removed by centrifugation at 15,000 g for 15 min at 4°C. Protein concentrations were determined with Bradford assay reagent (Thermo Fisher Scientific). Lysates (20 µg of protein) were incubated with an equal volume of Laemmli sample buffer for 30 min at room temperature. Samples were loaded onto 4–15% SDS-PAGE gel (Bio-Rad Laboratories) and transferred onto a nitrocellulose membrane (Bio-Rad Laboratories). Membranes were blocked with 5% nonfat dry milk and 0.1% Tween 20 in Tris-buffered saline for 1 h. Membranes were separated into three sections: the first section (containing hERG1a or hERG1b) was incubated with a 1:20,000 dilution of goat anti–hERG-KA polyclonal antibody directed against the C terminus of hERG; the second section (containing protein disulfide isomerase [PDI]) was incubated with a 1:20,000 dilution of mouse anti-PDI (Abcam); and the third section (containing hERG1a NTR plasmids) was incubated with a 1:5,000 dilution of anti–c-myc antibody (Covance). Membranes were incubated overnight in primary antibody and for 1 h in a horseradish peroxidase–linked secondary antibody. Membrane sections were probed using an ECL detection kit (Thermo Fisher Scientific) and imaged using a ChemiDoc XRS imaging system (Bio-Rad Laboratories). Images were analyzed using Quantity One Software (version 4.5; Bio-Rad Laboratories).
Online supplemental material
Fig. S1 shows the regulatory effects of an NTR fragment composed of the hERG1a eag domain (amino acids 1–135) on hERG1a/hERG1b channels expressed in Xenopus laevis oocytes. The eag domain did not measurably change in the G-V relationship of hERG1a/hERG1b channels, but markedly slowed the time course of deactivation.
RESULTS
hERG1a NTRs formed polypeptides
To investigate the properties of hERG1a NTRs, we expressed plasmids encoding these regions in HEK293 cells stably expressing hERG1a and hERG1b (Fig. 1 C). Cell lysates were analyzed with SDS-PAGE and immunoblotted with an antibody directed to a c-myc epitope at the C terminus of hERG1a NTRs (see Materials and methods). We detected robust bands on Western blots (Fig. 1 C) corresponding to the predicted molecular weights for each of the hERG1a NTR polypeptides (1–181, 24 kD; 1–228, 29 kD; 1–319, 39 kD; 1–365, 44 kD). These results show that hERG1a NTRs were translated into polypeptides. Using an hERG1 antibody (hERG-KA) that recognized hERG1a and hERG1b, we detected a band at 150 kD, representing mature hERG1a, and at 135 kD, representing immature hERG1a. We also detected a band at 95 kD, representing mature hERG1b, and a band at 80 kD, representing immature hERG1b, in the stable cell line (Fig. 1 C). We did not detect a measurable change in the amount of total hERG1a or hERG1b protein, or a change in the ratio of mature to immature protein compared with control bands not expressed with NTRs. These results also suggest that hERG1a NTRs did not interfere with hERG1a or hERG1b channel synthesis or maturation.
Soluble hERG1a NTR polypeptides functionally regulated heteromeric hERG1a/hERG1b channels
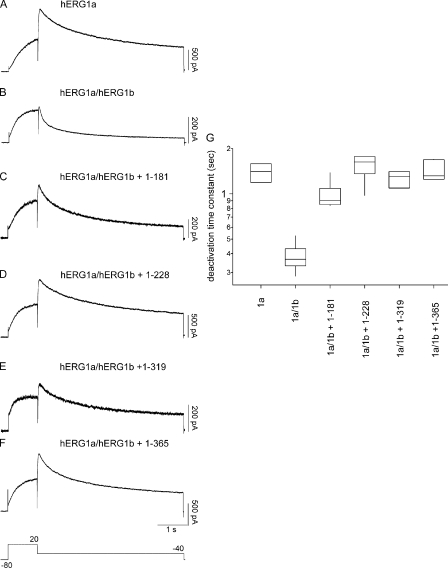
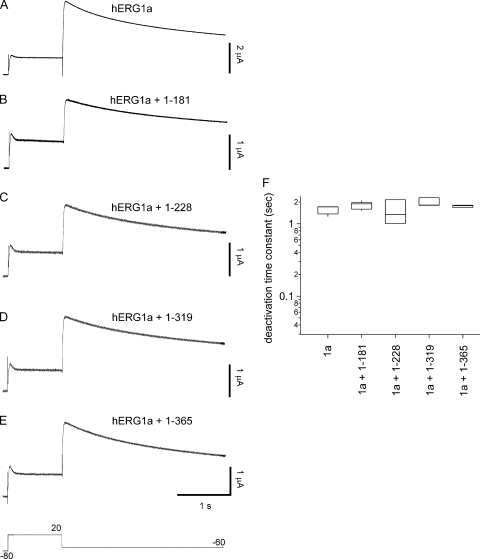
To examine a functional role for hERG1a NTRs, we performed whole cell patch-clamp recordings to measure ionic currents in an hERG1a/hERG1b stable cell line, as these currents closely recapitulate native IKr (London et al., 1997; Jones et al., 2004; Sale et al., 2008). We first recorded a family of currents from a stable cell line expressing heteromeric hERG1a/hERG1b channels as a positive control (Fig. 2). Compared with homomeric hERG1a channels (Fig. 2 A), hERG1a/hERG1b channels (Fig. 2 B) had a less rectifying I-V relationship (Fig. 2 G) but a similar G-V relationship (Fig. 2 H). hERG1a/hERG1b channels had a threefold faster time course of deactivation than that of hERG1a channels (Fig. 3, A, B, and G). We transfected the stable cell line with hERG1a NTR plasmids and performed whole cell patch-clamp recordings (Fig. 2, C–F). We found that, for cells expressing hERG1a NTRs, the I-V relationship was more rectifying than that of hERG1a/hERG1b channels and indistinguishable from that of hERG1a channels (Fig. 2 G). NTRs did not have a measurable effect on the G-V relationship of hERG1a/hERG1b channels (Fig. 2 H). We also found that the time course of deactivation of hERG1a/hERG1b currents in cells coexpressing hERG1a NTRs was markedly slower than that of hERG1a/hERG1b channels and more similar to that of hERG1a channels (Fig. 3, C–F and G). These results show that hERG1a NTRs changed the gating properties of hERG1a/hERG1b channels to more closely resemble those of hERG1a homomeric channels.
Figure 3.
hERG1a NTR polypeptides regulate deactivation time course of hERG1a/hERG1b channels. (A–F) Whole cell patch-clamp recording of currents from hERG1a/hERG1b and NTR polypeptides, as indicated. The voltage protocol was a step to 20 mV for 1 s, followed by a step to −40 mV for 5 s. The holding potential was −80 mV. (G) Box plot of deactivation time constant (τ) derived from single-exponential fits to current relaxations at −40 mV (after the pulse to 20 mV). Time courses of currents from A and C–F were significantly different from those in B (P < 0.01; ANOVA). Bars in A–F, 1 s; n ≥ 6.
Action potential voltage-clamp recordings show that hERG1a NTR polypeptides reduced peak hERG1a/hERG1b currents
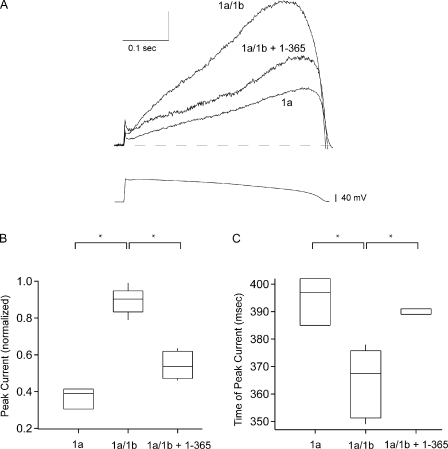
To determine the effect of hERG1a NTRs on peak hERG currents, we used a voltage pulse waveform that mimicked a ventricular action potential. As anticipated, in control experiments, the peak of the current for hERG1a and hERG1a/hERG1b channels occurred during the repolarization phase of the action potential clamp (Fig. 4 A). The peak hERG1a/hERG1b current, normalized to peak conductance (see Materials and methods), was larger than that of hERG1a (Fig. 4 B), and the time of the peak hERG1a/hERG1b current was earlier than that of hERG1a (Fig. 4 C), as reported previously at 34°C (Sale et al., 2008). Next, we chose to examine an hERG1a NTR (1–365), which had a representative effect on hERG1a/hERG1b kinetics (see Figs. 2 and 3). We found that 1–365 polypeptides reduced the peak current of hERG1a/hERG1b channels (Fig. 4, A and B) and increased the time at which the peak current occurred (Fig. 4 C). Thus, the hERG1a NTR regulated the shape of the hERG1a/hERG1b current and changed it to more closely resemble the shape of hERG1a currents.
Figure 4.
hERG1a/hERG1b currents generated by an action potential voltage clamp were reduced by hERG1a NTR polypeptides. (A) Currents elicited using an action potential voltage clamp (as indicated) from hERG1a, hERG1a/hERG1b, and hERG1a/hERG1b plus 1–365, as labeled. n ≥ 3 for each. For comparison, currents were normalized to the peak tail current at −50 mV after a pulse to 60 mV. Vertical scale bar is normalized units, and horizontal scale bar is time (0.1 s). (B) Box plot of magnitude of peak current. (C) Box plot of time after the onset of the voltage pulse at which peak current occurred (*, P < 0.05; ANOVA).
hERG1a NTR polypeptides regulated hERG1b currents
To distinguish whether NTRs differently regulated individual hERG subunits, we took advantage of the finding that hERG1b channels have small but measurable currents when expressed in Xenopus oocytes (Lees-Miller et al., 1997; London et al., 1997). We first examined whether hERG1a NTRs regulated hERG1a homomeric channels by performing two-electrode voltage-clamp recordings from oocytes (Fig. 5). We did not detect any measurable changes in the ionic currents (Fig. 5, A–E), the I-V relationship (Fig. 5 F), the G-V relationship (Fig. 5 G), or the time course of deactivation (Fig. 6) in hERG1a channels compared with hERG1a channels coexpressed with hERG1a NTRs. These results show that hERG1a NTRs did not measurably regulate wild-type hERG1a channels and suggest that hERG1a NTRs did not interact with hERG1a channels. This finding is consistent with our previous finding that isolated hERG1a eag domains did not interact with wild-type hERG1a channels (Gustina and Trudeau, 2009).
Figure 5.
hERG1a NTR polypeptides did not regulate steady-state properties of hERG1a channels. Two-electrode voltage-clamp recordings in response to voltage steps (shown at bottom) from Xenopus oocytes injected with (A) hERG1a, (B) hERG1a plus 1–181, (C) hERG1a plus 1–228, (D) hERG1a plus 1–319, and (E) hERG1a plus 1–365. The voltage was stepped from −100 to 60 mV for 1 s in increments of 20 mV, followed by a step to −50 mV for 750 msec. The holding potential was −80 mV. (F) Plot of I-V relationship. (G) G-V relationship for channels in A–E. Bars in A–E, 0.2 s; n ≥ 3.
Figure 6.
hERG1a NTR polypeptides did not measurably regulate deactivation gating of hERG1a channels. (A–E) Recording of hERG1a with NTR polypeptides, as indicated. (F) Box plot of deactivation time constant (τ) derived from single-exponential fits to current relaxations generated by a step to −60 mV for 3 s (after the 1-s pulse to 20 mV). Bars in A–E, 1 s; n ≥ 3.
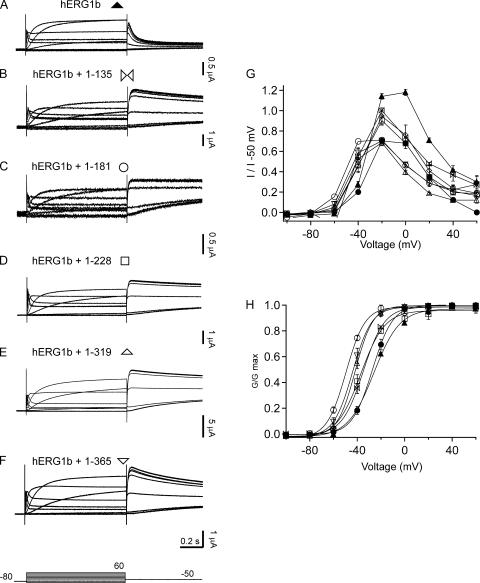
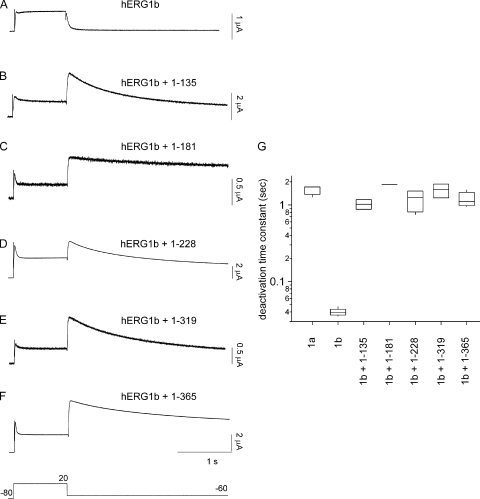
We next tested whether hERG1a NTRs specifically regulated hERG1b channels. We expressed hERG1b channels (Fig. 7 A) and hERG1b channels with hERG1a NTRs (Fig. 7, B–F). We found that when hERG1b was coexpressed with hERG1a NTRs, the I-V relationship was more rectifying and mimicked that of hERG1a (Fig. 7 G), but the G-V relationship (Fig. 7 H and Table I) was negatively shifted relative to either hERG1a or hERG1a/hERG1b channels (see Discussion). As anticipated, homomeric hERG1b channels had a very rapid time course of deactivation (Fig. 8, A and G). When hERG1b was coexpressed with hERG1a NTRs, the time course of deactivation was dramatically slowed (Fig. 8, B–G) and similar to that of hERG1a homomeric channels (Fig. 8 G; also see Fig. 6 A). We interpret these results to mean that the hERG1a NTRs regulated hERG1b channels.
Figure 7.
hERG1a NTR polypeptides regulate steady-state properites of homomeric hERG1b channels. Two-electrode voltage-clamp recordings in response to voltage steps (identical as in Fig. 5) from Xenopus oocytes injected with (A) hERG1b, (B) hERG1b plus 1–135, (C) hERG1b plus 1–181, (D) hERG1b plus 1–228, (E) hERG1b plus 1–319, and (F) hERG1b plus 1–365. (G) I-V relationships from channels in A–F. (H) G-V relationship for channels in A–F. Voltage protocol is the same as in Fig. 5. Bars in A–F, 0.2 s; n ≥ 3.
Table I.
Steady-state activation
| Channel | V1/2 | k | n |
| hERG1a | −27.2 ± 0.2 | 8.59 ± 0.15 | 4 |
| hERG1b | −26 ± 0.9 | 10.48 ± 0.8 | 3 |
| hERG1b + 1–135 | −34.84 ± 0.54 | 8.91 ± 0.47 | 3 |
| hERG1b + 1–181 | −49.48 ± 0.49 | 8.04 ± 0.35 | 3 |
| hERG1b + 1–228 | −37.09 ± 1.43 | 10.13 ± 1.31 | 4 |
| hERG1b + 1–319 | −41.69 ± 0.13 | 7.5 ± 0.14 | 5 |
| hERG1b + 1–365 | −43.88 ± 0.32 | 8.02 ± 0.3 | 5 |
V1/2, voltage at half-maximal activation; k, slope; n, number of oocytes.
Figure 8.
hERG1a NTR polypeptides regulate deactivation time course of hERG1b channels. (A–F) Two-electrode voltage-clamp recording of hERG1b channels and hERG1b channels with hERG1a NTR polypeptides, as indicated. (G) Box plot of deactivation time constant (τ) derived from single-exponential fits to current relaxations at −60 mV (after the pulse to 20 mV). Time courses of currents from B–F were significantly different from those in A (P < 0.01; ANOVA). Bars in A–F, 1 s. Voltage protocol is the same as in Fig. 6. n ≥ 3 for each.
The hERG1a NTRs in this study were of different lengths, but they all had in common an N-terminal eag domain. We found that the eag domain was sufficient for regulation of gating, as an NTR encoding just the eag domain markedly regulated hERG1b channels (Figs. 7, B, G, and H, and 8, B and G). We interpret this result to mean that the eag domain within each of the longer hERG1a NTRs was sufficient for regulation of deactivation gating in channels containing hERG1b subunits.
DISCUSSION
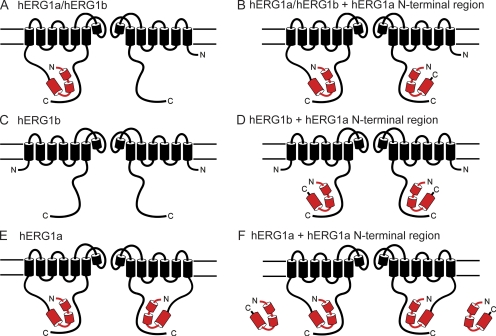
Here, we showed that hERG1a NTRs functionally regulated heteromeric hERG1a/hERG1b channels and homomeric hERG1b channels. The eag domain within the NTRs was sufficient for the regulatory function. Because the hERG1b subunit has a short NTR that does not contain an eag domain, we propose that hERG1b has an eag domain receptor site that is unoccupied by eag domains (Fig. 9, A and C). We propose a model where the hERG1a NTRs directly regulate hERG1b subunits in heteromeric hERG1a/hERG1b channels (Fig. 9 B) or homomeric hERG1b channels (Fig. 9 D) by interacting with unoccupied eag domain receptor sites. Because hERG1a NTRs did not measurably change the gating in homomeric hERG1a channels, we propose that the eag domain receptor site is occupied in hERG1a channels by the eag domain that is connected via the peptide bond and that, consequently, NTRs cannot interact with hERG1a (Fig. 9, E and F). As an alternative to this model, we considered if it was possible that the N-terminal eag domain of hERG1a channels interacted with hERG1b in heteromeric hERG1a/hERG1b channels, and that the hERG1a NTRs interacted with unoccupied eag domain receptor sites in hERG1a subunits. Indeed, there is evidence for functional intersubunit interactions in wild-type hERG1a channels between the eag domains and the C-terminal C-linker/cyclic nucleotide–binding domains (Gustina and Trudeau, 2011; Muskett et al., 2011). Either model of interaction is consistent with our finding that the hERG1a NTRs functionally converted hERG1a/hERG1b channels into hERG1a-like channels.
Figure 9.
Schematic for regulation of hERG channels by hERG1a NTR polypeptides. Schematic of (A) hERG1a/hERG1b, (B) hERG1a/hERG1b interaction with hERG1a NTR, (C) hERG1b, (D) hERG1b interaction with hERG1a NTR (E) hERG1a, and (F) lack of hERG1a interaction with hERG1a NTR.
Previously, it was proposed that the intermediate kinetics of heteromeric hERG1a/hERG1b channels could be a result of either a simple reduction in the number of eag domains because of heteromerization with hERG1b subunits (because hERG1b lacks eag and PAS domains), or a result of the regulatory effect of the eag domains being selectively inhibited by the short unique hERG1b NTR (Phartiyal et al., 2007; Sale et al., 2008). Our results address this issue by showing that hERG1a NTRs directly regulated heteromeric hERG1a/hERG1b channels and homomeric hERG1b channels, and effectively converted hERG1b subunit–containing channels into hERG1a-like channels. These results indicate that the most parsimonious explanation for why deactivation gating in hERG1a/hERG1b is faster than that of hERG1a channels but slower than that of hERG1b is not because of a specific inhibitory action of the unique hERG1b NTR on hERG1a eag domains, but rather because of a reduced number of eag domains in heteromeric hERG1a/hERG1b channels.
We found that hERG1a NTRs produced a left-shift in the G-V relationship of homomeric hERG1b channels expressed in oocytes. This was unanticipated because hERG1a NTRs did not produce a left-shift in the G-V of hERG1a/hERG1b channels expressed in HEK293 cells (Fig. 2 H) and because the midpoint of the G-V for hERG1a channels and hERG1b channels is similar (Fig. 7 H). What might be the basis of the NTR-induced left-shift in homomeric hERG1b channels? We ruled out technical differences between experiments with hERG1a/hERG1b channels in HEK293 cells (Fig. 2) and experiments with hERG1b in oocytes (Fig. 7) because in an additional experiment, an hERG NTR did not left-shift the G-V curve of hERG1a/hERG1b channels expressed in oocytes (Fig. S1). Perhaps for hERG1b channels that were regulated by NTRs, the unique combination of four hERG1b NTRs and (presumably) four hERG1a NTRs has a potentiating effect on steady-state activation. Future work will be required to sort out this mechanism.
We also observed that the left-shift in the hERG1b G-V relationship is greater for hERG1a 1–181 than for the other NTRs. We speculate that the region downstream of the PAS domain (3′ to the PAS domain) contained in hERG1a 1–181 has a potentiating effect on the G-V relationship and that the effect gets masked with the inclusion of longer sequences (for instance, hERG1a 1–365 potentiates much less) and/or the peptide bond in wild-type hERG1a channels.
In each of the lanes on Western blots that contained hERG1a NTRs, we observed a prominent lower molecular weight band. These smaller bands were most likely degradation products of each NTR (rather than partial transcripts) because they were of different sizes, they were not detected in control or mock lanes and they were recognized by the anti–c-myc antibody, the epitope for which is at the C-terminal end of the NTRs. We do not think that the degradation products interfered with or contributed to the regulation of the currents because the eag domain portion of the NTR was sufficient for regulation, and it is unlikely that the degradation products contained the eag domain.
Could regulation of hERG1a/hERG1b channels by hERG1a NTRs be a mechanism for type 2 LQTS? Earlier studies reported other hERG1a truncation mutants that were associated with type 2 LQTS. For instance, truncations in the C-terminal region of hERG channels resulted in dysregulation by 14–3-3 proteins (Choe et al., 2006) or functional channels with a reduced level of surface expression (Kupershmidt et al., 2002). A hERG1a frameshift mutation (Δ1261) produced a polypeptide that encoded the entire NTR and the S1 transmembrane domain and diminished hERG1a currents (Li et al., 1997) but had no effect on kinetics (Sanguinetti et al., 1996; Li et al., 1997). Smaller outward hERG currents are proarrhythmic in computational (Clancy and Rudy, 2001), animal (Arnaout et al., 2007; Brunner et al., 2008), and human stem cell models (Itzhaki et al., 2011) of type 2 LQTS. Furthermore, hERG1a currents are proarrhythmic in computational models, whereas hERG1a/hERG1b currents are not (Sale et al., 2008). Our results are most consistent with a mechanism where hERG1a NTRs dysregulate hERG1a/hERG1b channels, convert them into channels with hERG1a-like functional properties, and decrease outward repolarizing current. Alternatively, lessened protein stability or translation of hERG1a NTRs could account for disease. Indeed, hERG1a 1–181 polypeptides had relatively lower density on a Western blot, indicating less protein expression compared with the other NTRs, and this correlated with a less pronounced effect of hERG1a 1–181 on hERG1a/hERG1b channel deactivation properties. We also considered that NTR expression could be down-regulated by nonsense-mediated RNA degradation (Gong et al., 2007) but were unable to rule out an effect on RNA levels, as a control mini-gene (composed of exons 1–5 and introns 1–4 of the hERG1a NTR) did not make a detectable protein.
In summary, we have shown that isolated hERG1a NTRs changed the kinetics and steady-state properties of homomeric hERG1b and heteromeric hERG1a/hERG1b channels to more closely resemble those of hERG1a homomeric channels. We found that the eag domain was sufficient to regulate hERG1a/hERG1b and hERG1b channels, indicating that hERG1b subunits had sufficient machinery to support regulation by hERG1a N-terminal eag domain–containing polypeptides.
Acknowledgments
We thank A. Gustina, E. Gianulis, and C. Gallagher for helpful discussions and A. Gustina for collecting the data in the Fig. S1.
This work was supported by a National Institutes of Health grant (HL-083121 to M.C. Trudeau) and a gift from the Helen Pumphrey Denit Trust.
Author contributions: M.C. Trudeau and G.A. Robertson designed research; M.C. Trudeau, L.M. Leung, and E.R. Roti performed research and analyzed data; and M.C. Trudeau wrote the paper.
Christopher Miller served as editor.
Footnotes
Abbreviations used in this paper:
- HEK293
- human embryonic kidney 293
- hERG
- human ether-á-go-go–related gene
- LQTS
- long QT syndrome
- NTR
- N-terminal region
- PAS
- Per-Arnt-Sim
- PDI
- protein disulfide isomerase
References
- Arnaout R., Ferrer T., Huisken J., Spitzer K., Stainier D.Y., Tristani-Firouzi M., Chi N.C. 2007. Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. USA. 104:11316–11321 10.1073/pnas.0702724104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner M., Peng X., Liu G.X., Ren X.Q., Ziv O., Choi B.R., Mathur R., Hajjiri M., Odening K.E., Steinberg E., et al. 2008. Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J. Clin. Invest. 118:2246–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C.U., Schulze-Bahr E., Neu A., Xu J., Zhu Z.I., Sauter K., Bähring R., Priori S., Guicheney P., Mönnig G., et al. 2006. C-terminal HERG (LQT2) mutations disrupt IKr channel regulation through 14-3-3epsilon. Hum. Mol. Genet. 15:2888–2902 10.1093/hmg/ddl230 [DOI] [PubMed] [Google Scholar]
- Clancy C.E., Rudy Y. 2001. Cellular consequences of HERG mutations in the long QT syndrome: precursors to sudden cardiac death. Cardiovasc. Res. 50:301–313 10.1016/S0008-6363(00)00293-5 [DOI] [PubMed] [Google Scholar]
- Curran M.E., Splawski I., Timothy K.W., Vincent G.M., Green E.D., Keating M.T. 1995. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 80:795–803 10.1016/0092-8674(95)90358-5 [DOI] [PubMed] [Google Scholar]
- Gong Q., Zhang L., Vincent G.M., Horne B.D., Zhou Z. 2007. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long-QT syndrome. Circulation. 116:17–24 10.1161/CIRCULATIONAHA.107.708818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasti L., Cilia E., Crociani O., Hofmann G., Polvani S., Becchetti A., Wanke E., Tempia F., Arcangeli A. 2005. Expression pattern of the ether-a-go-go-related (ERG) family proteins in the adult mouse central nervous system: evidence for coassembly of different subunits. J. Comp. Neurol. 491:157–174 10.1002/cne.20721 [DOI] [PubMed] [Google Scholar]
- Gustina A.S., Trudeau M.C. 2009. A recombinant N-terminal domain fully restores deactivation gating in N-truncated and long QT syndrome mutant hERG potassium channels. Proc. Natl. Acad. Sci. USA. 106:13082–13087 10.1073/pnas.0900180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustina A.S., Trudeau M.C. 2011. hERG potassium channel gating is mediated by N- and C-terminal region interactions. J. Gen. Physiol. 137:315–325 10.1085/jgp.201010582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E.C., Reyes H., Chu F.F., Sander F., Conley L.H., Brooks B.A., Hankinson O. 1991. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 252:954–958 10.1126/science.1852076 [DOI] [PubMed] [Google Scholar]
- Itzhaki I., Maizels L., Huber I., Zwi-Dantsis L., Caspi O., Winterstern A., Feldman O., Gepstein A., Arbel G., Hammerman H., et al. 2011. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 471:225–229 10.1038/nature09747 [DOI] [PubMed] [Google Scholar]
- Jackson F.R., Bargiello T.A., Yun S.H., Young M.W. 1986. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 320:185–188 10.1038/320185a0 [DOI] [PubMed] [Google Scholar]
- Jones E.M., Roti Roti E.C., Wang J., Delfosse S.A., Robertson G.A. 2004. Cardiac IKr channels minimally comprise hERG 1a and 1b subunits. J. Biol. Chem. 279:44690–44694 10.1074/jbc.M408344200 [DOI] [PubMed] [Google Scholar]
- Kupershmidt S., Yang T., Chanthaphaychith S., Wang Z., Towbin J.A., Roden D.M. 2002. Defective human ether-à-go-go-related gene trafficking linked to an endoplasmic reticulum retention signal in the C terminus. J. Biol. Chem. 277:27442–27448 10.1074/jbc.M112375200 [DOI] [PubMed] [Google Scholar]
- Lees-Miller J.P., Kondo C., Wang L., Duff H.J. 1997. Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ. Res. 81:719–726 [DOI] [PubMed] [Google Scholar]
- Li Q., Gayen S., Chen A.S., Huang Q., Raida M., Kang C. 2010. NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker. Biochem. Biophys. Res. Commun. 403:126–132 10.1016/j.bbrc.2010.10.132 [DOI] [PubMed] [Google Scholar]
- Li X., Xu J., Li M. 1997. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J. Biol. Chem. 272:705–708 10.1074/jbc.272.2.705 [DOI] [PubMed] [Google Scholar]
- London B., Trudeau M.C., Newton K.P., Beyer A.K., Copeland N.G., Gilbert D.J., Jenkins N.A., Satler C.A., Robertson G.A. 1997. Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ. Res. 81:870–878 [DOI] [PubMed] [Google Scholar]
- Möglich A., Ayers R.A., Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 17:1282–1294 10.1016/j.str.2009.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral J.H., Lee A., Cohen S.L., Chait B.T., Li M., Mackinnon R. 1998. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 95:649–655 10.1016/S0092-8674(00)81635-9 [DOI] [PubMed] [Google Scholar]
- Muskett F.W., Thouta S., Thomson S.J., Bowen A., Stansfeld P.J., Mitcheson J.S. 2011. Mechanistic insight into human ether-à-go-go-related gene (hERG) K+ channel deactivation gating from the solution structure of the EAG domain. J. Biol. Chem. 286:6184–6191 10.1074/jbc.M110.199364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu J.R., Jr, Lewis J.O., Wharton K.A., Jr, Crews S.T. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 67:1157–1167 10.1016/0092-8674(91)90292-7 [DOI] [PubMed] [Google Scholar]
- Ng C.A., Hunter M.J., Perry M.D., Mobli M., Ke Y., Kuchel P.W., King G.F., Stock D., Vandenberg J.I. 2011. The N-terminal tail of hERG contains an amphipathic α-helix that regulates channel deactivation. PLoS ONE. 6:e16191 10.1371/journal.pone.0016191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D., Tsien R.W. 1969. Outward membrane currents activated in the plateau range of potentials in cardiac Purkinje fibres. J. Physiol. 200:205–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phartiyal P., Jones E.M., Robertson G.A. 2007. Heteromeric assembly of human ether-à-go-go-related gene (hERG) 1a/1b channels occurs cotranslationally via N-terminal interactions. J. Biol. Chem. 282:9874–9882 10.1074/jbc.M610875200 [DOI] [PubMed] [Google Scholar]
- Reddy P., Jacquier A.C., Abovich N., Petersen G., Rosbash M. 1986. The period clock locus of D. melanogaster codes for a proteoglycan. Cell. 46:53–61 10.1016/0092-8674(86)90859-7 [DOI] [PubMed] [Google Scholar]
- Sale H., Wang J., O’Hara T.J., Tester D.J., Phartiyal P., He J.Q., Rudy Y., Ackerman M.J., Robertson G.A. 2008. Physiological properties of hERG 1a/1b heteromeric currents and a hERG 1b-specific mutation associated with Long-QT syndrome. Circ. Res. 103:e81–e95 10.1161/CIRCRESAHA.108.185249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jurkiewicz N.K. 1990. Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. J. Gen. Physiol. 96:195–215 10.1085/jgp.96.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jurkiewicz N.K. 1991. Delayed rectifier outward K+ current is composed of two currents in guinea pig atrial cells. Am. J. Physiol. 260:H393–H399 [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jiang C., Curran M.E., Keating M.T. 1995. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 81:299–307 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Curran M.E., Spector P.S., Keating M.T. 1996. Spectrum of HERG K+-channel dysfunction in an inherited cardiac arrhythmia. Proc. Natl. Acad. Sci. USA. 93:2208–2212 10.1073/pnas.93.5.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector P.S., Curran M.E., Zou A., Keating M.T., Sanguinetti M.C. 1996. Fast inactivation causes rectification of the IKr channel. J. Gen. Physiol. 107:611–619 10.1085/jgp.107.5.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester D.J., Will M.L., Haglund C.M., Ackerman M.J. 2005. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2:507–517 10.1016/j.hrthm.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Trudeau M.C., Warmke J.W., Ganetzky B., Robertson G.A. 1995. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 269:92–95 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- Wang J., Trudeau M.C., Zappia A.M., Robertson G.A. 1998. Regulation of deactivation by an amino terminal domain in human ether-à-go-go–related gene potassium channels. J. Gen. Physiol. 112:637–647 10.1085/jgp.112.5.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke J.W., Ganetzky B. 1994. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. USA. 91:3438–3442 10.1073/pnas.91.8.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]