Abstract
The attachment of bioactive protein to surfaces underpins the development of biosensors and diagnostic microarrays. We present a surface treatment using plasma immersion ion implantation (PIII) to create stable covalent binding sites for the attachment of functional soya-bean peroxidase (SBP). Fourier transform infrared spectra of the surfaces show that protein is retained on the surface after boiling in sodium dodecyl sulphate and sodium hydroxide, which is indicative of covalent attachment. The activity of SBP on the treated surfaces remains high in comparison with SBP attached to control surfaces over the course of 11 days. Surface plasmon resonance was used to show that the surface coverage of the attached protein is close to a monolayer. We describe the potential of the PIII treatment method to be used as a one-step dry process to create surfaces for large-scale protein micro- or nanopatterning.
Keywords: plasma, surface modification, bioactive
1. Introduction
Recent research has shown advances in protein patterning methods, such as dip-pen nanolithography (Lee et al. 2002), imprint lithography (Hoff et al. 2004), soft lithographic techniques (Kane et al. 1999) and self-assembly (Agheli et al. 2006). The binding mechanisms responsible for the protein attachment can be classified into those using covalent chemical bonds and those which rely on physical adsorption. Almost all of the techniques resulting in covalent attachment use chemical linker molecules and a number of wet-chemistry steps to facilitate the attachment. Commonly used approaches rely on the biotin–avidin interaction (Hoff et al. 2004), thiol chemistry (Lee et al. 2002; Agheli et al. 2006) or silanization (Plueddemann 1991) to create covalent bonds between the protein and the surface. A suitable choice of the surface and linker attachment site on the protein can enhance the retention of bioactivity.
Another class of commonly used attachment methods (e.g. in soft lithography, dip-pen nanolithography, etc.) relies on physical adsorption via non-specific binding of target molecules and requires no wet-chemical processing. These are simple one-step processes but since the binding mechanism relies on a vast number of weak van der Waals or permanent dipole interactions between many sites on the protein and the surface, the efficacy and strength of binding on any given surface is protein dependent and highly variable. In some cases, serious loss of bioactivity through disruptions of the protein's conformation or weak attachment, easily disturbed in subsequent washing steps, can occur.
The research presented in this paper explores a new way to functionalize surfaces for covalent attachment of protein that relies only on a dry plasma treatment process with an inert gas. It is found that although this process does not require the use of chemical linker molecules and associated wet chemistry, it results in covalent attachment between the protein and the polymer surface as well as preservation of the bioactivity of the attached molecules which is greatly improved over that achievable by simple physical adsorption.
Plasma immersion ion implantation (PIII) is a technique in which a surface is implanted by ions from surrounding plasma via the application of a negative pulsed potential to the substrate by a conductive electrode (Conrad et al. 1987; Tendys et al. 1988). The capability of the PIII method to create sub-micrometre features has been demonstrated in the microelectronics industry (Lee et al. 2000). The technique therefore lends itself well to fine-scale protein patterning. Unlike conventional beamline ion implantation, PIII can be used to treat large areas in short time intervals and can handle substrates with three-dimensional features. Previous work (Kondyurin et al. 2006) on PIII of polystyrene surfaces with argon has focused on the structural changes introduced to the surface by the ion impacts. This work describes the attachment of the protein soya-bean peroxidase to PIII-treated surfaces.
A comparison of attachment to surfaces exposed to the argon plasma, but not to the PIII high-voltage bias pulses, shows that the PIII process is the key to retaining the protein's activity after attachment. We present evidence supporting a covalent interaction between the PIII-activated surface and the protein.
2. Surface modification procedure
Polystyrene (PS) sheets (Goodfellow, 0.25 mm thick, biaxially oriented) were cut into small samples of approximately 1×1 cm2 in size. These samples were then cleaned with methanol and transferred into the plasma treatment chamber. For surface plasmon resonance (SPR) measurements, the samples were PS films of a thickness of approximately 7.8±1.6 nm (N=4) as measured by ellipsometry, which were spin coated onto a 40 nm thick gold layer on glass slides. Spin-coating parameters were 0.1% by weight PS in toluene at 3000g. The uncoated surface of the glass slide was then coupled to a prism using immersion oil to match the refractive indices, while a flow cell was clamped to the PS-coated surface. The SPR experimental set-up is described in detail elsewhere (Barnes & Sambles 1986; Martin & Sambles 1990; Morrow et al. 2007).
The plasma source region in the PIII treatment system consists of a single-loop antenna of 16 cm in diameter wrapped around a borosilicate glass tube. The radio frequency power at 13.56 MHz was coupled to the antenna by a Comdel CPM-2000 matching network. An aluminium diffusion chamber located above the plasma source houses the sample holder. The outside of the aluminium chamber was surrounded by two pairs of copper coils to provide an axial magnetic field of approximately 5 mT (Gan et al. 2006). The base pressure in the vacuum chamber was 8×10−6 Torr. The plasma treatments were carried out in argon gas with a pressure of 2 mTorr prior to ignition and a post-ignition pressure of 2.2 mTorr. The forward power used in the process was 100 W. PIII was achieved using a high-voltage pulse bias of −20 kV and pulse duration of 25 μs at a frequency of 50 Hz. The pulse bias was applied to a conductive mesh in front of the substrate to accelerate the ions into the substrate. This technique, known as mesh-assisted PIII (Tian et al. 2005), is known to reduce the effects of surface charging by reflecting secondary electrons and to eliminate arcing by reducing the electric fields present at the surface of the polymer.
Two types of plasma treatment were applied using a 100 W, 2.2 mTorr inductively coupled radio-frequency discharge in argon. The first treatment was carried out with no bias applied to the substrate and the mesh, while the second one applied a pulsed bias (PIII) of −20 kV with respect to earth, for 25 μs at a frequency of 50 Hz, during the plasma treatment process. All protein attachment experiments were carried out on untreated control samples for comparison. In all cases involving a form of plasma treatment, the treatment time was 800 s, except for the surfaces prepared for SPR which were treated for only 40 s due to the small thickness of the film.
3. Buffers and solutions
Phosphate buffer (PB) was prepared with 10 mM NaH2PO4 and 10 mM Na2HPO4, and pH adjusted to pH 7.0. Standard phosphate-buffered saline (PBS) was prepared using PB containing 150 mM NaCl and adjusted to pH 7.4.
Seed coat soya-bean peroxidase (SBP) was from Sigma-Aldrich and was chosen because its activity on a surface is easily determined by the use of a colorimetric assay. In the assay, the reaction of an SBP substrate, 3,3′,5,5′-tetramethylbenzidine (TMB), is stopped with acid, forming a yellow reaction product, the optical density of which is read at 450 nm. Unlike horseradish peroxidase (HRP), SBP exists in only one isoform and generally has greater stability (Kamal & Behere 2002).
Lyophilized SBP was reconstituted into buffer. The extinction coefficient ε403=94.6 mM−1 cm−1 was then used to calculate the protein concentration (Kamal & Behere 2002). The protein was then diluted with buffer to the concentrations used in the experiments.
4. Experimental procedure
After the treatment, the polystyrene sheet samples for use in activity assays and the untreated controls were incubated overnight in a solution of buffer containing SBP added to a concentration of 50 μg ml−1 unless otherwise stated.
The samples were then transferred to a new container and washed six times with fresh buffer solution, resting on a rocker for a period of 20 min for each wash. The samples were then stored in a tube in fresh buffer until they were measured using the TMB assay. If the samples were to be stored for longer periods, the solution was replaced with fresh buffer daily.
The samples selected to be assayed on a given day were placed in small holders, which consisted of two metal layers with a 7 mm diameter hole in the centre of one layer surrounded by an O-ring to seal in the liquid. A solution of 75 μl TMB was allowed to react for 30 s, after which 50 μl of the reacted solution was removed and the reaction was stopped with acid prior to spectrophotometry at 450 nm. The absorbance measured is related to the amount of functional protein on the surface.
To determine relative estimates of the amount of protein (functional or not) left on the surface, infrared spectra were obtained using a Digilab FTS7000 Fourier transform infrared (FTIR) spectrometer. The spectra were taken in attenuated total reflectance (ATR) mode using a multiple bounce germanium crystal, at a resolution of 1 cm−1.
SPR was used to estimate the coverage of the surface-attached protein layer. In SPR, an evanescent wave probes the layer of protein as it adsorbs onto the surface. The effective index of refraction of the protein-coated surface can be obtained by calibrating the signal against that obtained from a solution of known refractive index in contact with the surface. We used milliQ water (n=1.33), a 40 g l−1 NaCl solution (n=1.340) and ethanol (n=1.35920) to perform the calibration; the angle shift measured through SPR was normalized to the index of refraction of each of the three test solutions. The effective index of refraction determined in this manner is independent of the index of refraction and the thickness of the PS film.
For the protein-binding experiment, the buffer solution containing protein was run through the flow cell across the surface. The experimentally measured effective index of refraction is an averaged index of refraction weighted with the depth-dependent evanescent field according to the formula (Jung et al. 1998)
| (4.1) |
where nexp is the experimentally measured effective index of refraction and dPD is the penetration depth of the evanescent electric field, given by
| (4.2) |
where λ is the wavelength of light used in the experiment (632.8 nm) and ϵM(ω) is the dielectric constant of the gold layer (−9.59 + 1.45i) (Raether 1988).
Modelling the protein as a confluent layer of unknown thickness T with water as the medium, the index of refraction n(z) as a function of distance from the surface is
| (4.3) |
Evaluating the integral in equation (4.1) for the model defined by equation (4.3) and solving the resulting equation for T leads to the expression
| (4.4) |
Comparing the calculated thickness with the dimensions of SBP indicates whether there is more or less than a monolayer of the protein present.
5. Results and discussion
5.1 Assessment of protein activity
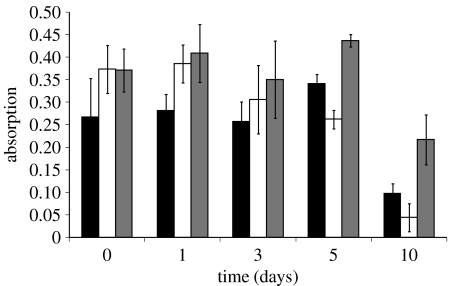
Figure 1 shows the results of a TMB activity assay on samples washed and stored over a 10-day period. Both the incubation and washing steps were done in phosphate buffer and the buffer was replaced daily. The initial (day 0) attachment of active protein is enhanced on the two plasma-treated surfaces. However, the surface treated using the PIII process shows much greater retention of active protein over the 10-day period compared with the untreated control and the surface treated without the PIII process.
Figure 1.
Bioactivity of surface-attached SBP as a function of time. Untreated (black), plasma-treated (white) and PIII (grey) surfaces with SBP attached were stored in PB for the duration of this experiment.
To test for any influence of the buffer choice, the experiment was repeated with incubation in PBS solution containing the protein. The last washing step was done in PB, as the salt is known to affect the TMB assay. The samples that were not to be assayed on a given day were stored in PBS until they were to be assayed.
In addition to the two previously described treatment methods, another batch of samples was treated with the argon PIII process followed by a 10 s exposure to oxygen plasma. The results are shown in figure 2. The functional attachment at day 0 was enhanced for all three plasma treatments compared with the untreated control. Once again, little functional protein was retained on the untreated and plasma surfaces over time while the surface treated using the PIII process retained about half the amount of functional protein up to day 11. The subsequent exposure to oxygen plasma appears to have reduced the effectiveness of the PIII plasma treatment in terms of functional attachment over time.
Figure 2.
Bioactivity of surface-attached SBP as a function of time. Untreated (black), plasma-treated (white), PIII (grey) and PIII/O2 plasma-treated (hatched) surfaces with SBP attached were stored in PBS for the duration of this experiment.
The results of figures 1 and 2 indicate that the surfaces exposed to the plasma as well as those exposed to the plasma with PIII treatment show an enhancement in initial binding. Only the PIII surface, however, has the ability to retain functional protein over longer periods. During the PIII treatment, ions penetrate many layers into the polystyrene, breaking and cross-linking the polymer chains at and below the surface (Kondyurin et al. 2006). We believe that the formation of a cross-linked or carbonized layer just below the surface, preventing reptation of the modified polymer chains into the bulk, could be the reason for the improved stability observed for the PIII-treated surfaces.
5.2 The nature of the attachment mechanism
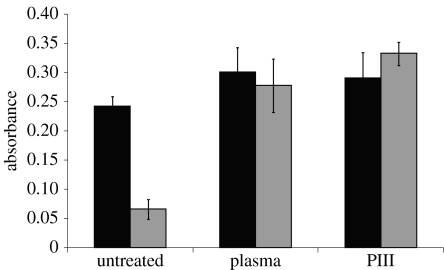
In order to differentiate between binding mechanisms on the treated surfaces versus the untreated controls, the surfaces were washed with detergent. The detergent Tween 20 is often used to block non-specific interactions (Shmanai 1999). Surfaces were soaked in protein-containing solutions overnight, and then washed with a solution of buffer containing 0.05% Tween 20. The results are shown in figure 3. The detergent removed most of the protein from the untreated surface, but not from either of the plasma-treated surfaces. This indicates that a strong interaction capable of resisting the detergent is responsible for at least some attachment on the plasma-treated surfaces.
Figure 3.
Effect of washing in Tween 20 on functional protein retained on the surface. The black bars are a measure of the functional protein attached prior to washing in Tween 20 detergent, while the grey bars are a measure of the functional protein attached after washing in Tween 20 detergent.
To further test the nature of attachment, the surfaces were boiled in a solution of 5% sodium dodecyl sulphate (SDS) for 10 min. The TMB assay was no longer effective, as SDS denatures the protein, so FTIR spectra of the surfaces were used to assess the quantity of protein remaining on the surfaces following the SDS treatment. While FTIR spectra of surfaces are often used to detect protein (Veiseh et al. 2002), the complexity of the spectrum of the underlying polystyrene made it difficult to see the peaks due to protein. To solve this problem, spectra of the surfaces were recorded both before and after incubation in protein, and then subtracted to give a ‘difference spectrum’. The resulting spectra, shown in figure 4, contain only peaks where the absorbance had changed from the original treated surface due to the attachment of protein. For both the treated and untreated surfaces, amide peaks between 1650 and 1660 cm−1 were present after incubation in protein solution, indicating the presence of protein. Nothing in the spectrum of the buffer used had an absorption in this band. After boiling in 5% SDS, there was no amide peak present for the untreated surface, while it was still present on the surface treated using PIII, although reduced in intensity compared with the initial peak. The peak had also shifted slightly higher in wavenumber to approximately 1675 cm−1, indicating unfolding of the protein as would be expected after boiling in SDS. SDS is therefore unable to detach all of the protein from the PIII-treated surface as indicated by the continued presence of a peak associated with the protein. This is consistent with protein attachment through a covalent bond. To confirm this result, the surface was boiled in a solution containing both 5% SDS and 1 M NaOH. The protein still remained bound on the surface.
Figure 4.
ATR–FTIR spectra of several surfaces were obtained. All spectra shown are after subtracting out the original spectrum of the surface taken before soaking in SBP. (a) Untreated surface after incubating in protein (black) and after boiling in SDS (grey). (b) Treated surface after incubating in protein (black) and after boiling in SDS (grey). (c) Various spectra after boiling the treated surface in different solutions: SDS (from (b)); 1 M HCl; 1 M HCl+5% SDS; and 1 M NaOH+5% SDS.
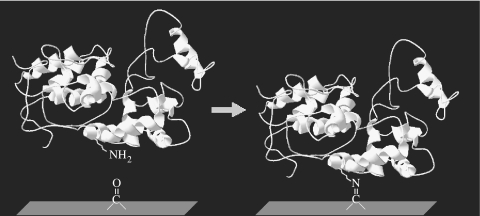
Previous infrared spectral analysis (Kondyurin et al. 2006) of PIII-treated polystyrene surfaces showed that upon atmospheric exposure, oxygen reacts with free radicals created by the treatment, generating carbonyl groups on the surface. It is known that carbonyls can form covalent bonds with amine groups through reactions such as Schiff base formation (Feeney et al. 1975). A reaction of this type could be responsible for the observed covalent attachment of SBP to the PIII-treated surfaces, as illustrated in figure 5. SBP has three exposed amino groups: two are in the form of lysines and one at the N-terminus. All three sites are on the opposite side of the protein from the active site, so attachment through these sites would not result in blocking the activity of SBP (Guex & Peitsch 1997).
Figure 5.
Schematic of a possible mechanism for covalent binding of protein to the surface (Guex & Peitsch 1997).
Another possibility is direct attachment of the protein to the long-lived free radicals created on the polystyrene surface in the plasma treatment process. Bombardment with energetic ions has been shown to break bonds in the polymer chains which leave highly reactive free radicals (Kuzuya et al. 1998). The free radicals in hydrocarbons created by ion bombardment rapidly convert to more stable unsaturated and aromatic groups. Free radicals conjugated with aromatic structures, such as graphitic rings, can be stable for long periods (Jahan et al. 1998), as is observed, for example, in benzylic free radicals.
To test the idea that amine groups on the protein are involved in the new binding mechanism associated with the treated surfaces, samples were soaked for 3 days in 0.2 M tris(hydroxymethyl)aminomethane (Tris) prior to exposure to SBP. The amine group of the Tris molecule would be expected to react with the active groups on the treated polymer surface, blocking these sites from subsequent interaction with the protein's amide groups. FTIR spectra were collected from the treated surface both before and after incubation in protein and then again after boiling in SDS. The spectra taken before incubation with protein were subtracted from those taken after incubation and after SDS exposure. The ‘difference spectra’ obtained in this way are shown in figure 6. The spectra show that most, if not all, of the protein was removed from the treated surface when boiled in SDS. This decrease in the amide peak is certainly much greater than the decrease from boiling the unblocked surface in SDS after incubation in protein solution. This result confirms that the covalent binding sites which bind the protein readily react with the C–NH2 or the C–OH functional groups found in Tris, indicating that they would be likely to react with these groups in the protein. However, this still leaves open the possibility of the surface binding other functional groups on the protein as well.
Figure 6.
ATR–FTIR spectra of the treated, Tris-blocked surface after incubating in protein (black) and after boiling in SDS (grey).
5.3 Estimation of protein coverage
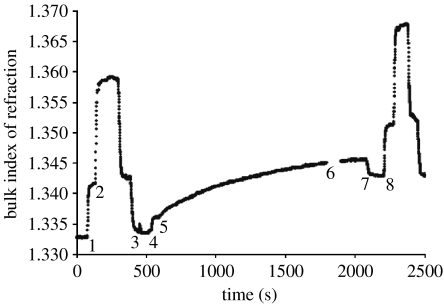
Figure 7 shows the effective index of refraction of the solution in contact with the treated surface over time, as measured using SPR. If we assume that SBP has a refractive index close to that of typical protein, i.e. nSBP=1.45–1.55, and that is the index of refraction of pure water, the measured nexp, where the protein adsorption had reached a steady state, is 1.343 (point 8). The thickness of the protein layer as calculated using equation (4.4) is then between 3.36 (nSBP=1.55) and 6.37 nm (nSBP=1.45). Since the dimensions of SBP are 6.1 and 3.1 nm (Guex & Peitsch 1997), these results indicate that there is approximately a monolayer of protein on the PIII-treated surface. A similar coverage was found by atomic force microscopy and ellipsometry for the enzyme HRP (Gan et al. 2007).
Figure 7.
Scaled surface plasmon resonance scan showing effective index of refraction, neff, at the polymer film surface as a function of time. Initially, milliQ water flows in the cell; at point 1, NaCl solution is introduced and at point 2, ethanol replaces the NaCl solution. These steps are reversed until at point 3 pure water is introduced again. Since the refractive indices of water, the NaCl solution and the ethanol are known, the vertical scale can be calibrated in terms of effective refractive index. At point 4, the buffer solution is introduced and at point 5, the solution of buffer and protein is introduced. At point 6, there is a gap in the stored data and at point 7, milliQ water is introduced. At point 8, a second scale calibration is made. The measured refractive index, nexp, is taken as the average index between points 7 and 8 and the difference in index between points 3–4 and 7–8 represents the change in refractive index due to protein adsorption.
6. Conclusion
Plasma treatment in argon gas on polystyrene with concurrent PIII produces a surface with enhanced binding capacity for functional SBP, as well as an enhanced ability to retain the protein function over time. The enhanced binding capacity seems to be at least in part due to the creation of functional groups which bind covalently to the protein. While covalent attachment is shown, future work must be done to isolate the dominant mechanism of covalent binding. This will lead to a greater understanding of the treatment and its possible applications.
Although we have as yet made no effort to optimize the treatment process with respect to treatment time and the PIII parameters, the improvements in active protein attachment are significant. Similar results have been found in the cases of horseradish peroxidase (Gan et al. 2007, in press; Ho et al. 2007) and catalase (Nosworthy et al. 2007) incubated with PIII-treated polyethylene, indicating that this treatment process has some generality of application with respect to the bound protein and the polymer surface. One of the studies with horseradish peroxidase showed that the treated surfaces, stored in ambient atmosphere, retain their ability to covalently bind protein for up to 1 year (Ho et al. 2007).
Advantages of using PIII to create functional sites for protein arrays and biosensors include the environmental friendliness and simplicity of the process, as well as its simple integration with currently existing methodologies for masking to create surface patterning. The process is completely dry, using only argon gas to create the functional sites, and no chemical linkers are needed to bind the protein. As such, it is low cost and environmentally friendly, producing minimal process waste, as compared with covalent attachment achieved using linker molecules which typically require wet-chemical processes.
An important aspect of creating protein arrays using masks to produce patterning will be minimizing background protein adhesion to the untreated areas. Our results show that a Tween 20 wash after incubation with protein gives a ratio of 1 : 5 (ratio of grey bars for untreated and PIII-treated surfaces in figure 3) in the functional protein remaining on the untreated and treated surfaces, respectively. One strategy to eliminate the background signal would be to treat the whole surface and mechanically place the protein on the desired sites, either through robotic placement, ink-jet-style protein printing (Schena et al. 1995) or dip-pen nanolithography. The whole surface could then be blocked with an agent such as Tween 20, which is ineffective in removing protein that is bound to PIII-treated surfaces.
Acknowledgments
The authors would like to recognize the generous support of the Australian Research Council, Australian Research Network for Advanced Materials and the Australian–American Fulbright Commission while conducting the research for this project. Many thanks to Rob Davies, who helped keep the PIII machine running, and to Keisuke Mizuno, Bee K. Gan, Alexey Kondyurin, Palli Thordarson and Neil Nosworthy for their help in various stages of the research. We also thank Scott Martin for his help with, and use of, the SPR equipment.
References
- Agheli H, Malmström J, Larsson E.M, Textor M, Sutherland D.S. Large area protein nanopatterning for biological applications. Nano Lett. 2006;6:1165–1171. doi: 10.1021/nl060403i. [DOI] [PubMed] [Google Scholar]
- Barnes W.L, Sambles J.R. Guided optical waves in Langmuir–Blodgett films of 22 tricosenoic acid. Surf. Sci. 1986;177:399–416. doi: 10.1016/0039-6028(86)90148-2. [DOI] [Google Scholar]
- Conrad J.R, Radtke J.L, Dodd R.A, Worzala F.J, Tran N.C. Plasma source ion-implantation technique for surface modification of materials. J. Appl. Phys. 1987;62:4591–4596. doi: 10.1063/1.339055. [DOI] [Google Scholar]
- Feeney R.E, Blankenhorn G, Dixon H.B. Carbonyl-amine reactions in protein chemistry. Adv. Prot. Chem. 1975;29:135–203. doi: 10.1016/s0065-3233(08)60412-x. [DOI] [PubMed] [Google Scholar]
- Gan B.K, Bilek M.M.M, Kondyurin A, Mizuno K, McKenzie D.R. Etching and structural changes in nitrogen plasma immersion ion implanted polystyrene films. Nucl. Instrum. Meth. B. 2006;247:254–260. doi: 10.1016/j.nimb.2006.01.063. [DOI] [Google Scholar]
- Gan B.K, Kondyurin A, Bilek M. Comparison of protein surface attachment on untreated and plasma immersion ion implantation treated polystyrene: protein islands and carpet. Langmuir. 2007;23:2741–2746. doi: 10.1021/la062722v. [DOI] [PubMed] [Google Scholar]
- Gan, B. K., Nosworthy, N. J., McKenzie, D. R., dos Remedios, C. G. & Bilek, M. M. M. In press. Plasma immersion ion implantation treatment of polyethylene for enhanced binding of active horseradish peroxidase. J. Biomed. Mater. Res. A ( 10.1002/jbm.a.31612) [DOI] [PubMed]
- Guex N, Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Ho J.P.Y, Nosworthy N.J, Bilek M.M.M, Gan B.K, McKenzie D.R, Chu P.K, dos Remedios C.G. Plasma-treated polyethylene surfaces for improved binding of active protein. Plasma Process. Polym. 2007;4:583–590. doi: 10.1002/ppap.200600182. [DOI] [Google Scholar]
- Hoff J.D, Cheng L, Meyhöfer E, Guo L.J, Hunt A.J. Nanoscale protein patterning by imprint lithography. Nano Lett. 2004;4:853–857. doi: 10.1021/nl049758x. [DOI] [Google Scholar]
- Jahan M.S, Thomas D.E, Banerjee K, Trieu H.H, Haggard W.O, Parr J.E. Effects of radiation–sterilization on medical implants. Radiat. Phys. Chem. 1998;51:593–594. doi: 10.1016/S0969-806X(97)00208-9. [DOI] [Google Scholar]
- Jung L, Campbell C.T, Chinowsky T.M, Mar M.N, Yee S.S. Quantitative interpretation of the response of surface plasmon resonance sensors to adsorbed films. Langmuir. 1998;14:5636–5648. doi: 10.1021/la971228b. [DOI] [Google Scholar]
- Kamal J.K.A, Behere D.V. Thermal and conformational stability of seed coat soybean peroxidase. Biochemistry. 2002;41:9034–9042. doi: 10.1021/bi025621e. [DOI] [PubMed] [Google Scholar]
- Kane R.S, Takayama S, Ostuni E, Ingber D.E, Whitesides G.M. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/S0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- Kondyurin A, Gan B.K, Bilek M.M.M, Mizuno K, McKenzie D.R. Etching and structural changes of polystyrene films during plasma immersion ion implantation with argon. Nucl. Instrum. Meth. B. 2006;251:413–418. doi: 10.1016/j.nimb.2006.06.027. [DOI] [Google Scholar]
- Kuzuya M, Yamashiro T, Kondo S, Sugito M, Mouri M. Plasma-induced surface radicals of low-density polyethylene studied by electron spin resonance. Macromolecules. 1998;31:3225–3229. doi: 10.1021/ma9709361. [DOI] [Google Scholar]
- Lee, K., Lee, B., Hoepfner, J., Economikos, L., Parks, C., Radens, C., Bemstein, J. & Kellennan, P. 2000 Plasma immersion ion implantation as an alternative deep trench buried-plate doping technology. In IEEE Proc. Conf. Ion Implantation Technology, 17–22 September, pp. 460–463. ( 10.1109/.2000.924187) [DOI]
- Lee K.B, Park S.J, Mirkin C.A, Smith J.C, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- Martin A.S, Sambles J.R. Guided wave study of Langmuir–Blodgett films of eicosanoic acid. Surf. Sci. 1990;225:390–396. doi: 10.1016/0039-6028(90)90459-L. [DOI] [Google Scholar]
- Morrow R, McKenzie D.R, Bilek M.M.M, MacDonald C.L, Stindt M, Anetsberger G, Martin A.S. Electric field effects on adsorption/desorption of proteins and colloidal particles on a gold film observed using surface plasmon resonance. Physica B. 2007;394:203–207. doi: 10.1016/j.physb.2006.12.054. [DOI] [Google Scholar]
- Nosworthy N.J, Ho J.P, Kondyurin A, McKenzie D.R, Bilek M.M.M. The attachment of catalase and poly-l-lysine to plasma immersion ion implantation treated polyethylene. Acta Biomater. 2007;3:695–704. doi: 10.1016/j.actbio.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Plueddemann E. 2nd edn. Plenum Press; New York, NY: 1991. Silane coupling agents. [Google Scholar]
- Raether H. Springer; Hamburg, Germany: 1988. Surface plasmons on smooth and rough surfaces and on gratings. [Google Scholar]
- Schena M, Shalon D, Davis R.W, Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Shmanai V.V. Blocking of non-specific sorption in ELISA on formylated polystyrene beads. J. Immunoassay. 1999;20:13–30. doi: 10.1080/01971529909349311. [DOI] [PubMed] [Google Scholar]
- Tendys J, Donnelly I.J, Kenny M.J, Pollock J.T.A. Plasma immersion ion implantation using plasmas generated by radio frequency techniques. Appl. Phys. Lett. 1988;53:2143–2145. doi: 10.1063/1.100299. [DOI] [Google Scholar]
- Tian X.B, Fu K.Y, Chu P.K, Yang S.Q. Plasma immersion ion implantation of insulating materials. Surf. Coat. Technol. 2005;196:162–166. doi: 10.1016/j.surfcoat.2004.08.166. [DOI] [Google Scholar]
- Veiseh M, Zareie M.H, Zhang M. Highly selective protein patterning on gold–silicon substrates for biosensor applications. Langmuir. 2002;18:6671–6678. doi: 10.1021/la025529j. [DOI] [Google Scholar]