Abstract
The surface microtexture of an orthopaedic device can regulate cellular adhesion, a process fundamental in the initiation of osteoinduction and osteogenesis. Advances in fabrication techniques have evolved to include the field of surface modification; in particular, nanotechnology has allowed for the development of experimental nanoscale substrates for investigation into cell nanofeature interactions. Here primary human osteoblasts (HOBs) were cultured on ordered nanoscale groove/ridge arrays fabricated by photolithography. Grooves were 330 nm deep and either 10, 25 or 100 μm in width. Adhesion subtypes in HOBs were quantified by immunofluorescent microscopy and cell–substrate interactions were investigated via immunocytochemistry with scanning electron microscopy. To further investigate the effects of these substrates on cellular function, 1.7 K gene microarray analysis was used to establish gene regulation profiles of mesenchymal stem cells cultured on these nanotopographies. Nanotopographies significantly affected the formation of focal complexes (FXs), focal adhesions (FAs) and supermature adhesions (SMAs). Planar control substrates induced widespread adhesion formation; 100 μm wide groove/ridge arrays did not significantly affect adhesion formation yet induced upregulation of genes involved in skeletal development and increased osteospecific function; 25 μm wide groove/ridge arrays were associated with a reduction in SMA and an increase in FX formation; and 10 μm wide groove/ridge arrays significantly reduced osteoblast adhesion and induced an interplay of up- and downregulation of gene expression. This study indicates that groove/ridge topographies are important modulators of both cellular adhesion and osteospecific function and, critically, that groove/ridge width is important in determining cellular response.
Keywords: osteoblasts, mesenchymal stem cells, focal adhesions, genomic regulation, nanotopography, nanobioscience
1. Introduction
The cell–biomaterial interface functions not only to define the boundary between tissue and implant, but also as a mediator of first stage protein interactions as well as later stage cell-specific adhesion and orientation. Functional modification of the cell–substrate interface is now recognized as a necessary process in orthopaedic design for successful integration and performance of a device in vivo, the paradigm of such being biomaterial-mediated regulation of osseoinduction and osteospecific function.
The phenomenon of cellular contact guidance on grooved topography has been well documented in literature (Den Braber et al. 1998; Karuri et al. 2004; Lenhert et al. 2005). Nanoscale grooves with dimensions similar to those found in vivo have been shown to cause contact guidance and alignment in studies involving human cell types, including corneal epithelial cells (Teixeira et al. 2003), fibroblasts (Dalby et al. 2003b) and osteoblasts (Lenhert et al. 2005).
Bone extracellular tissue is a natural composite biomaterial with a complex hierarchical structure, composed of a macromolecular network including proteins, glycoproteins and polysaccharides, into which mineral phase hydroxyapatite crystals are embedded, providing an environment rich in both biochemical and topographical cues. Macroscopically trabeculae form into struts that are typically orientated along lines of force resulting from mechanical loading. Cortical bone is composed of elongated, cylindrical osteons of approximately 10–500 μm in diameter (Rho et al. 1998). Each osteon is a cylinder consisting of multiple concentric lamellae of approximately 3–7 μm in thickness (Giraud-Guille 1988). Collagen fibres within each lamella are arranged into parallel bundles that lie obliquely relative to adjacent lamellae (Rubin et al. 2004); in addition, collagen molecules aggregate into highly ordered fibrils, composed of three protein chains, wound together in a triple helix of approximately 300 nm in length (Wagner & Weiner 1992), which also present a banded pattern at the ultrastructural level composed of 69 nm repeat banding units (Brodsky & Persikov 2005; Bozec et al. 2005).
Increased knowledge of this intricate extracellular environment, and how cells react within it, has resulted in the development of biomimetic materials to guide morphogenesis in tissue repair and tissue engineering, particularly in the field of topographical modification. Nanotechnology aims to create and use structures and systems in the size range of approximately 0.1–500 nm covering the atomic, molecular and macromolecular length scales.
Cell adhesion is an essential element of many physiological situations, including tissue maintenance, wound healing, development, angiogenesis and cell migration. Focal adhesions (FAs) were initially observed in 1964 (Curtis 1964), and later described by Abercrombie et al. (1971) as electron-dense regions along the ventral plasma membrane that make intimate contact with the substratum in cultured cells (Abercrombie et al. 1971). These structures play opposing roles in cell motility, aiding both in the generation of cellular strain to generate lamellopodium formation and polarized motility, and as anchoring complexes that resist detachment from the substratum and aid in cellular spreading. A highly ordered hierarchical recruitment of FA-associated proteins occurs during adhesion plaque assembly (Zamir & Geiger 2001). Vinculin is a plasma membrane-associated protein found in adhesion complexes and involved in the coupling of the actin-based microfilaments to the adhesion plaque (Zimerman et al. 2004). Vinculin is present in a number of adhesion subtypes and provides a definite detection system for FA sites by means of specific antibody detection.
However, the mechanical response of a cell to its surroundings and the onset of adhesion cannot be accurately described without considering the biochemical and genetic interplays that control signal transduction and protein production. FAs assemble into complex signalling systems where transmembrane integrin receptors transduce signals from the extracellular matrix (ECM) to regulate cellular function and gene expressions, such as cellular growth and differentiation. Work to date has focused on gaining a quantitative understanding of the interconnected mechanical and genetic networks that occur during cell mechanotransduction, specifically those processes that direct osseointegration, matrix deposition and bony tissue neogenesis. Alterations in gene and protein expression can occur within minutes of a cell adhering to a surface. Recently it has been suggested that adhesion complexes may transmit mechanical cues directly to the nucleus through cytoskeletal tension and that this mechanotransduction as well as adhesion kinetics could play a role in topographic induction of cellular differentiation (Dalby et al. 2006).
Here photolithography was used to produce three grooved topographies of identical groove depths with different widths (10, 25 and 100 μm repeat width, 330 nm groove depth and 2 cm length). Primary human osteoblasts (HOBs) derived from femoral head and condyle biopsies, and Stro-1 immunoselected human mesenchymal stem cells (MSCs) isolated from the marrow of a femoral diaphysis were used as cell models to investigate morphological and functional response, respectively, to grooved topographies. HOB adhesion complex formation was quantified and cytoskeletal morphology was investigated with scanning electron and fluorescence microscopy. Two extremes of groove width were used for the functional response study—100 and 10 μm groove arrays. This was then followed up with human 1718 gene cDNA microarrays to compare MSC reaction to the grooved topographies with response to a flat surface at 21 days of cell culture. Microarray allowed simultaneous gene detection in planar controls and grooved substrates. Data were then analysed by Ingenuity Pathways Analysis. Interesting changes were observed in the relative level of expression in genes involved in the areas of cell–cell signalling, cytoskeletal formation, ECM remodelling and adhesion-associated signalling.
2. Material and methods
2.1 Nanopatterning and die fabrication
Samples were made in a three-step process of photolithography, nickel die fabrication and embossing.
2.1.1 Photolithography
Silicon wafers were cleaned under acetone in an ultrasonic bath for 5 min. They were then rinsed thoroughly in reverse osmosis water (ROH2O) and blow-dried with an air gun. Next, they were spun with primer for 30 s at 4000g, then with S1818 photoresist for 30 s at 4000g and baked for 30 min at 90°C. The resulting layer was measured to be 1.8 μm thick. The photoresist layer was exposed to UV light through a chrome mask featuring an array of parallel slits on a Karl Suss MA6 mask aligner for 3.8 s. Then the resist layer was developed for 75 s in 50 : 50, Microposit developer: ROH2O.
The specific design pattern, in this case rectangular slits, was achieved by reactive ion etching using the exposed photoresist as a mask. The silicon substrate was etched in the silicon tetrachloride gas plasma of a Plasmalab System 100 machine (gas flow =18 sccm, pressure =9mTorr, rf power =250 W, DC bias =−300 V). The wafer was etched individually at 7 min at a nominal etch rate of 18 nm min−1. It was then stripped of resist in an acetone ultrasound bath for 5 min, followed by a 5 min soak in concentrated sulphuric acid/hydrogen peroxide mixture before rinsing thoroughly in ROH2O and drying in an air gun. The two subsequent topographies were 330 nm deep grooves that were 10, 25 or 100 μm in width.
2.1.2 Nickel shim fabrication
To facilitate polymeric replication of the topographical grooves, nickel shims were made directly from the patterned S1818 photoresist samples. A thin (50 nm) layer of Ni-V was sputter coated on both of the samples. That layer acted as an electrode in the subsequent electroplating process. The shims were plated to a thickness of approximately 300 nm (for more information about the procedure see Gadegaard et al. (2003)). The shims were finally trimmed to approximately 30×30 mm sizes using a metal guillotine. Imprinting of the nickel shims into poly(methyl-methacrylate) (pMMA) (Goodfellow, UK) was achieved using an Obducat nanoimprinter. Subsequent replicas possessed a nanoimprinted area of 1.5 cm2. Planar pMMA (average roughness of 1.17 nm over 10 mm2) was used as a control substrate.
2.2 HOB cell culture
The methodology has been described previously (Biggs et al. 2007). Briefly HOBs derived from a femoral head biopsy of an 84-year-old and a knee biopsy of a 74-year-old Caucasian woman (PromoCell, Heidelberg, Germany) were cultured in 75 cm2 culture flasks. HOBs were expanded to passage 4 following five weeks of culture and were subsequently trypsinized in 0.04% Trypsin/0.03% EDTA (Heidelberg, Germany) and seeded onto untreated experimental substrates. Cells were maintained in PromoCell osteoblast growth medium containing 10% foetal calf serum (FCS; Heidelberg, Germany) that was replaced every 2 days. Following 7 days of culture, the HOBs were maintained for 4 days without changing the medium, inducing a brief period of serum starvation. Osteoblast growth medium encompassing 10% FCS was subsequently introduced to the culture following 4 days of serum starvation and HOBs were allowed to metabolize for 17 hours, giving rise to a population of cells possessing a synchronized nuclear cycle. Following this period, the HOBs were cultured in osteoblast growth medium containing 10 μM 5-bromo-2-deoxyuridine (BrdU) for 3 hours.
MSCs were enriched using Stro-1 antibody and magnetic cell sorting as previously described (Howard et al. 2002; Mirmalek-Sani et al. 2006), from bone marrow samples obtained from haematologically normal patients undergoing routine hip replacement surgery. These cells represent the adherent bone marrow stromal cell fraction. MSCs were maintained in basal α-minimal essential medium (Invitrogen, Carlsbad, CA, USA) containing 10% FCS that was replaced every 2 days.
2.3 Immunocytochemistry for light microscopy
HOBs on test materials were fixed in 4% paraformaldehyde in phosphate-buffered solution (PBS), with 1% sucrose at 37°C for 15 min. Once fixed, the samples were washed with PBS. Samples were permeabilized with buffered 0.5% Triton X-100 (10.3 g sucrose, 0.292 g NaCl, 0.06 g MgCl2, 0.476 g (4-(2-hydroxyethyl)-1-piperazine-ethanesulphonic acid) (HEPES), 0.5 ml Triton X-100, in 100 ml water; pH 7.2) at 4°C for 5 min. Non-specific binding sites were blocked with 1% bovine serum albumin (BSA) in PBS at 37°C for 5 min and subsequently incubated for 2 hours with a 1 : 200 concentration of anti-vinculin (monoclonal anti-human raised in mouse (clone hVin-1; IgG1); Sigma, Poole, UK), followed by an incubation in 1 : 100 concentration of anti-BrdU/DNase solution for 1 hour (37°C) (RPN2001, Amersham Biosciences cell proliferation kit, Uppsala, Sweden). Simultaneously, rhodamine-conjugated phalloidin was added for the duration of this incubation (1 : 100, Molecular Probes, OR, USA). Non-specific charges (e.g. remaining aldehyde) were neutralized with 0.5% Tween 20/PBS (5×3 min) to minimize background labelling. A secondary biotin-conjugated antibody, (1 : 50 in 1% BSA/PBS, monoclonal horse anti-mouse (IgG), Vector Laboratories, UK) was added for 1 hour (37°C) followed by subsequent washing as mentioned above. Fluorescein isothiocyanate-conjugated streptavidin was added (1 : 50 in 1% BSA/PBS; Vector Laboratories, UK) at 4°C for 30 min, and given a final wash. Samples were mounted in Vectorshield mountant for fluorescence (Vector Laboratories, UK), then viewed with a Zeiss Axiovert 200 M microscope with a Zeiss Plan Neofluor 40× (0.75 NA) lens. Image manipulation in Adobe Photoshop was then used to superimpose the colour channels to show adhesion complexes (vinculin) in green, actin in red and S-phase nuclei in blue.
2.4 Immunocytochemistry for SEM
The immunolabelling method was a modified version of that first described by Richards et al. (2001). Briefly, the cells were rinsed for 2 min in 0.1M 1,4 piperazine bis(2-ethanosulphonic acid) (PIPES) buffer (pH 7.4) before being permeabilized with buffered 0.5% Triton X-100 (4×1 min; 10.3 g sucrose, 0.292 g NaCl, 0.06 g MgCl2, 0.476 g HEPES, 0.5 ml Triton X-100, in 100 ml water; pH 7.2) to remove the cell membrane. Cells were then stabilized in 4% paraformaldehyde with 1% sucrose in 0.1 M PIPES, pH 7.4, buffer, for 5 min and rinsed three times for 2 min in 0.1 M PIPES buffer to remove unreacted aldehyde. Non-specific binding sites were blocked with 1% BSA and 0.1% Tween 20 in 0.1 M PIPES buffer, pH7.4, for 15 min. The cells were then incubated with mouse anti-human vinculin (monoclonal anti-human raised in mouse (clone hVin-1; IgG1); Sigma, Poole, UK) diluted 1 : 200 in PIPES buffer +1% BSA+0.1% Tween 20 for 2 hours at 37°C. The cells were rinsed six times for 2 min in PIPES+1% BSA+0.1% Tween 20. Non-specific binding sites were blocked with 5% goat serum+1% BSA+0.1% Tween 20 in PIPES buffer for 15 min. The cells were labelled with goat anti-mouse 5 nm gold conjugate (BBI, Cardiff, UK) diluted 1 : 200 in PIPES buffer+1% BSA +0.1% Tween 20 overnight for 12 hours at 22°C. All samples were then rinsed six times for 2 min in PIPES buffer. The samples were fixed permanently in 2.5% glutaraldehyde for 5 min in PIPES buffer and rinsed three times for 2 min in PIPES buffer. Gold probes were silver enhanced with a silver developing solution for 7 min (BBI, Cardiff, UK). The samples were immediately rinsed two times in ultrapure water to remove any unreacted enhancer. Additional contrasting of the cell was accomplished by staining the cells with 1% osmium tetroxide in PIPES for 1 hour.
2.5 Scanning electron microscopy
The cells were dehydrated through an ethanol series (50, 60, 70, 80, 90, 96 and 100%) followed by a fluorisol/ethanol series (25, 50, 75 and 100%; fluorisol—1,1,2 trichloro,1,2,2, trifluoroethane). The samples were critical point dried (Polaron E3100, Quorum Technologies, UK), mounted on aluminium stubs and coated with a 12 nm layer of carbon using a Baltec CED030 carbon thread evaporator (Baltec, FL, USA). The samples (both with and without cells) were imaged using an Hitachi S-4700 field emission scanning electron microscope fitted with an Autrata yttrium aluminium garnet backscattered electron (BSE) scintillator type detector. The images were taken in both the secondary electron and BSE modes, with accelerating voltages between 2 and 10 kV; the BSE images were taken with an emission current of 50 μA, an aperture of 100 μm (apt1), working distances of 10–12 mm to allow low voltage imaging (Richards & Ap Gwynn 1995).
2.6 Image analysis
Cell images (n≥40 per substrate) were exported to Adobe Photoshop and adhesion complexes scored with a three-pixel wide straight line on a layer superimposed onto the background image creating an adhesion schematic (figure 1a–c). Analysis of individual adhesions (n≥60 per cell) was performed by Sigma Stat (SSI, San Jose, CA, USA) as previously described (Biggs et al. 2006; for more information see Biggs et al. (2007)). Criteria for adhesion classification were according to size restrictions described in current literature (Bershadsky et al. 1985; Bershadsky et al. 2006). Adhesion complexes measuring less than 1 μm in length were assigned as focal complexes (FXs), those from 1 to 5 μm in length were designated as FAs, while those measuring over 5 μm in length were accordingly classified as supermature adhesions (SMAs). Data were sorted into adhesion subtype and analysed using Kruskal–Wallis one-way analysis of variance on ranks, which compared like subtypes. Results of p<0.05 were considered significant (differences of p<0.05 denoted by asterisk, and hash symbol dependent on adhesion subtype).
Figure 1.
Adhesion quantification of S-phase HOBs and characterization of experimental substrates. (a–c) Adhesion-associated vinculin was immunolabelled, images were scored with a straight white line, and analysed by image analysis software. Substrates were 330 nm deep and had widths of (d) 100 μm, (e) 25 μm and (f) 10 μm.
2.7 RNA isolation and microarray analysis
RNA extraction from confluent layers of MSCs on experimental substrates (n=5) and microarray hybridization (n=5) was according to the methodology previously described (Dalby et al. 2005). Individual gene expression detection was carried out with 1.7 K distinct human transcripts spotted arrays obtained from the Ontario Cancer Institute Microarray Centre. The functional and canonical analyses were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). Changes in gene expression were associated with canonical pathways in the Ingenuity Pathways Knowledge Base. The significance of the association between the dataset and the canonical pathway was measured in two ways. (i) A ratio of the number of genes from the dataset that map to the pathway divided by the total number of genes that map to the canonical pathway is displayed. (ii) Fischer's exact t-test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone (for more information see Dalby et al. 2003b)
3. Results
3.1 Nanotopography and substrate characterization
The embossing process resulted in the fabrication of three groove/ridge topographies possessing identical groove depths and periodicity as verified by SEM (figure 1a–c). Grooves and ridges were 330 nm deep/high with widths of 10, 25 and 100 μm. Planar controls had an average roughness of 1.17 nm over 10 mm2.
3.2 HOB morphology and cytoskeletal organization
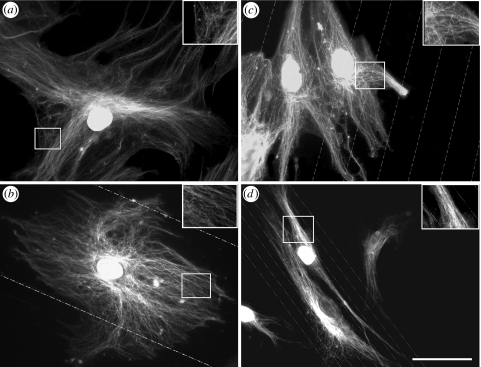
Nanogroove substrates modified S-phase HOB morphology and cytoskeletal organization relative to controls. HOBs cultured on all experimental substrates experienced increased cellular alignment, which was inversely proportional to groove/ridge width (figures 2–4). Actin cytoskeletal elements in S-phase HOBs on planar controls were prominent and organized into well-developed stress fibres, which typically spanned the entire cell body (figure 2a). Stress fibre networks in osteoblasts on 100 μm groove/ridge arrays appeared similar to cells on planar controls. Cells cultured on 25 μm groove/ridge arrays possessed ordered stress fibre organization and also demonstrated actin alignment in the groove direction. Actin fibres in HOBs on 10 μm groove/ridge arrays could be seen to run either perpendicular or parallel to the grooves, but stress fibres arranged both parallel and perpendicular to groove direction rarely occurred in a single cell (figure 2b–d). The HOBs on 10 μm groove/ridge topographies adopted a non-spread or a polarized elongated morphology, demonstrating contact guidance (figures 2–4d). Cells cultured on all experimental substrates but 10 μm groove/ridge arrays possessed a well-developed microtubule network (figure 3a–c). HOBs cultured on 10 μm grooves formed dense microtubule bundles that were orientated in the direction of cell polarity (figure 3d).
Figure 2.
Tri-fluorescent labelling of adhesion complexes and stress fibre organization in S-Phase HOBs on experimental substrates. (a) HOBs cultured on planar controls formed well-organized stress fibres and numerous SMAs. (b) Osteoblasts on 100 μm grooves possessed a well-developed actin cytoskeleton. Adhesions were large and numerous. (c) HOBs cultured on 25 μm grooves developed an organized actin cytoskeleton. Adhesion formation was reduced relative to planar controls. (d) HOBs cultured on 10 μm groove arrays disrupted cellular spreading. Adhesion formation was influenced by groove orientation. Bar=50 μm; red, actin; green, vinculin; blue, S-phase nuclei.
Figure 3.
Dual immunolabelling of the tubulin cytoskeleton and S-phase nuclei of HOBs on experimental substrates. (a,b) HOBs cultured on control and 100 μm groove substrates formed organized tubulin networks. Tubulin density decreased with increasing distance from the nucleus. (c) HOBs on 25 μm grooves demonstrated increased contact guidance. Cells were less spread and possessed a denser tubulin network. (d) Contact guidance was increased in cells on 10 μm groove substrates. S-phase HOBs formed a dense and polarized microtubule network. Bar=50 μm.
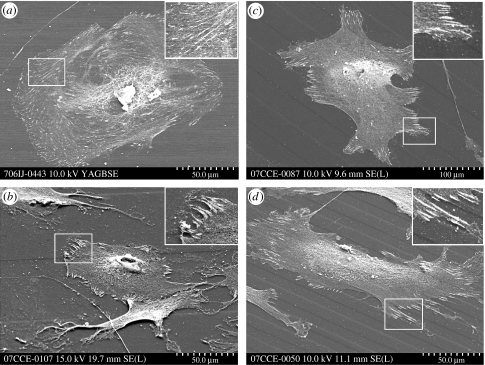
Figure 4.
Scanning electron immunolabelling of adhesion complexes in HOBs on experimental substrates. (a) HOBs cultured on flat pMMA formed numerous focal complexes. (b) Diffuse 100 μm grooves did not affect adhesion formation. (c) HOBs on 25 μm grooves formed adhesions predominantly on the raised ridge areas (indicated in insert). (d) HOBs cultured on 10 μm grooves influenced cellular and adhesion orientation, aligning both HOBs and adhesion complexes with groove direction.
3.3 Adhesion characterization
Quantification of adhesion distribution by vinculin immunolabelling revealed significant differences in adhesion formation in S-phase HOBs. The HOBs on planar controls formed numerous adhesions of all subtypes. SMAs were located predominantly at the perinuclear region and non-advancing areas of the cell periphery, while FAs and focal complexes were prominent at the region immediately preceding the leading edge and the extreme leading edge, respectively (figures 2a and 4a). Adhesion formation in HOBs on 100 μm groove/ridge substrates was similar to that of HOBs on planar controls (figures 2b and 4b); adhesion location and orientation were not significantly affected by this topography. Orientation and location of adhesions were significantly affected in HOBs on 25 and 10 μm groove/ridge arrays (figure 4c,d). Adhesion formation occurred predominantly on ridge structures on both substrates. Adhesion orientation was affected predominantly by 10 μm groove/ridge substrates; here adhesion orientation was largely either parallel or perpendicular to the groove angle (figures 2 and 4d).
3.4 Adhesion distribution and quantification
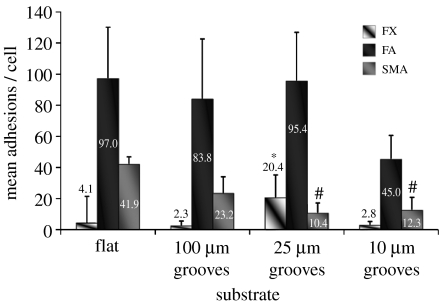
Significant differences in the number of generated adhesion subtypes relative to planar controls were observed on all topographies except the 100 μm groove/ridge arrays. Focal complexes were the least prominent adhesion subtype present in HOBs cultured on planar controls, yet were significantly upregulated in HOBs on 25 μm groove/ridge arrays (figures 5 and 6a).
Figure 5.
Quantification of adhesion complex formation in S-phase HOBs on experimental substrates. SMA formation was greatest in HOBs on planar control substrates, and significantly reduced in cells on 25 and 10 μm grooves. FA incidence was significantly decreased only in HOBs on 10 μm substrates. FX incidence was significantly increased in HOBs on 25 μm grooves. See table 1 for relevant statistics. Included are mean values. Results are mean+s.d.
Figure 6.
Adhesion complex distribution in S-phase HOBs on experimental substrates. (a) FXs measuring less than 1 μm in length were most abundant in HOBs on 25 μm grooves. Distribution of FXs approaching 2 μm in length was increased on flat and 100 μm groove substrates. All experimental substrates show a similar decline in adhesion complex distribution as FA lengths approach 4 μm. (b) SMAs are most pronounced in HOBs cultured on control substrates. SMA frequency declines on all substrates as length increases (diamonds, flat; triangles, 100 μm grooves; crosses, 25 μm grooves; squares, 10 μm grooves).
The HOBs on planar controls and 100 μm groove/ridge substrates formed predominantly FA subtypes. The HOBs cultured on planar controls formed on average 97 FAs per cell. This was reduced on both 100 and 25 μm groove/ridge topographies (83.8 and 95.4, respectively) and further reduced on 10 μm groove/ridge topographies (45). Distribution of FAs of greater than 2 μm declined in HOBS on all substrates except planar controls. This was most prominent in HOBs on 25 μm groove/ridge arrays (figures 5 and 6a).
Control substrates produced an average of approximately 41.9 SMAs per cell. SMA incidence was reduced on 100 μm groove/ridge arrays (23.2) and significantly reduced on 25 and 10 μm groove/ridge arrays (10.4 and 12.3, respectively). The frequency of SMAs of greater than 8 μm in length was less than 1 per cell in HOBs cultured on 25 and 10 μm groove/ridge arrays. Very large SMAs (greater than 10 μm) were observed only in HOBs cultured on planar control and 100 μm groove/ridge topographies (figures 5 and 6b).
3.5 Adhesion orientation
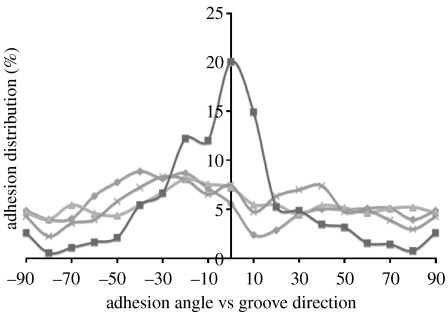
Planar controls, 100 and 25 μm groove/ridge topographies did not significantly affect adhesion orientation. Focal complexes, FAs and SMAs were randomly angled on planar and randomly aligned relative to groove direction on nanotopographies. Adhesion alignment in HOBs cultured on 10 μm groove/ridge arrays was significantly influenced. Here, 20% of all adhesions were parallel with respect to the groove angle (figure 7).
Figure 7.
Adhesion complex alignment on experimental substrates. Adhesion complex alignment was affected only by 10 μm groove substrates. The majority of adhesions on this topography deviated less than 30° from the groove direction. Approximately 20% of adhesions were aligned parallel with the grooves (diamonds, flat; triangles, 100 μm grooves; crosses, 25 μm grooves; squares, 10 μm grooves).
3.6 Microarray analysis
Because 10 μm groove/ridge arrays induced the most significant changes relative to planar controls in adhesion profile and the 100 μm groove/ridge arrays induced the least significant changes relative to control substrates in adhesion profile, they were selected for further microarray analysis with Stro+ progentior cells.
3.6.1 Canonical pathway analysis
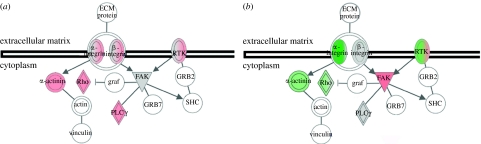
Analysis of microarray data revealed significant changes in canonical genetic pathways of MSCs cultured on nanogroove substrates (figure 8a). Changes in gene expression in MSCs cultured on nanogroove substrates relative to MSCs on planar controls were linked to several well-defined canonical pathways. Significant changes in p38 MAPK signalling were prominent on both substrates. A marked increase in PDGF signalling was observed in MSCs cultured on 100 μm groove/ridge arrays, signalling on 10 μm groove/ridge topographies however was reduced. Integrin signalling was upregulated in MSCs cultured on 100 μm groove/ridge arrays; however, integrin expression was significantly downregulated in MSCs cultured on 10 μm groove/ridge arrays (figure 9a,b). Significant upregulations were noted in VEGF signalling in cells on 10 μm groove/ridge arrays.
Figure 8.
Ingenuity analysis of Stro+progenitor cells on experimental substrates. (a) Canonical pathway analysis of Stro+progenitor cells on experimental substrates identified numerous well-defined signalling pathways that were most affected in cells cultured on groove/ridge substrates. Large changes in p38 MAPK signalling were induced by 100 μm grooves (black bars) while VEGF canonical pathways were most affected by 10 μm grooves (grey bars). Gene expression ratios between experimental and control substrates are indicated. (b) The functional analysis of a network of identified biological pathways that were most significant to the genes in the network affected by the groove/ridges. Progenitor cells cultured on both 10 and 100 μm groove nanopit arrays were associated with significant changes in gene expression and function; in particular, connective tissue development and cellular proliferation. A statistical threshold of significance p=0.05 indicates the probability that the association between the genes in the dataset canonical pathway is explained by chance alone.
Figure 9.
Integrin signalling in Stro+ progenitor cells on experimental substrates. (a) Expression of genes involved in integrin signalling pathways was upregulated in progenitor cells cultured on 100 μm grooves. (b) Downregulations were observed in similar genes in progenitor cells cultured on 10 μm grooves. Red, upregulated; green, downregulated; grey, no change.
3.6.2 Functional pathway analysis
Nanotopography significantly influenced genetic pathways linked to MSC function (figure 8b). Of most significance were large changes in connective tissue development in MSCs cultured on both grooved substrates. Trends indicate enhanced changes occurred in MSCs cultured on 10 μm relative to 100 μm groove/ridge arrays. Cellular movement, proliferation and protein synthesis were all significantly modified in MSCs cultured on 10 μm groove/ridge topographies. Cell morphology was significantly affected on both substrates, correlating with cytoskeletal changes (figures 2, 3 and 8b). Significant downregulations in cell proliferation were noted in cells cultured on 10 μm groove/ridge arrays. Upregulation of seven genes fundamental to skeletal system development was noted in HOBs on 100 μm groove/ridge arrays. Only five of these genes were upregulated in HOBs on 10 μm groove/ridge arrays (data not shown).
4. Discussion
Microtopography has been shown in previous studies to be an important mediator of cellular function, regulating cellular adhesion (Hamilton & Brunette 2007), spreading (Biggs et al. 2006; Gallant et al. 2007) and differentiation (Isa et al. 2006) as well as protein adsorption (Sela et al. 2007) and soft tissue response (Campbell & Von Recum 1989). While microscale topography significantly modulates cellular behaviour in vitro, due to the nanometre size of cellular components concerned with cell–biomaterial interactions, the surface structure of an implanted device influences the cell adhesion processes when the surface topography is within nanometre range (Dalby et al. 2003a; Cavalcanti-Adam et al. 2007; Lim et al. 2007). In order to elucidate the effects of topographical cues on primary HOB populations, we used biologically relevant nanoscale substrates to examine cell-substratum adhesion in osteoblasts and the functional response of Stro+ progenitor cells.
Mineralized tissues can be viewed as a dynamic system of self-regulating nanostructures containing grooves and ridges imparted by the collagen and mineral constituents. Previous studies have concluded that the interplay between both groove depth and width modulate cellular response and induce contact guidance (Sutherland et al. 2005). We hypothesized that topography with both nano- and microscale dimensions that closely approximate the size of in vivo topographical features found in mineralized tissues would better support osteoblast adhesion and MSC function. Loesberg et al. (2007) hypothesized that a cellular ‘point break’ is reached, with decreasing nanogroove width, where cells no longer display contact guidance; this cut-off point was found to be 100 nm in width and 35 nm in depth. A study by Hu et al. (2005) also on approximately 300 nm deep grooves concluded that the efficiency of smooth muscle cell alignment and elongation increases monotonically with the decrease of groove pitch, indicating that nanoscale structures produce more efficient alignment and elongation than micrometer-scale patterns. Similarly, we noted an increase in both cellular and adhesion alignment with decreasing groove pitch.
This study used photolithography to create anisotropic topographies of grooves and ridges possessing microscale widths and a constant nanoscale depth. Photolithography fabrication was followed by an embossing process to produce substrates with a constant surface chemistry that exhibited high fidelity from the master templates as well as high reproducibility. No satisfactory conclusions have been reached regarding the elastic properties of bone tissue; however, current literature places the elastic modulus of trabecular bone between 10 and 20 GPa, depending on the experimental procedure. To ensure a substrate stiffness comparable with that of trabecular bone, experimental substrates were fabricated in pMMA, an amorphous stiff polymer with an elastic modulus of approximately 3 GPa.
The rationale behind measuring cell adhesions in S-phase HOBs relates to variation in adhesion density measurements that occur as a cell progresses through the cell cycle, and that S-phase cells undergo extreme cell flattening relative to non-S-phase cells that display a more spherical morphology with less cell–substrate contact area (Hunter et al. 1995). Meredith et al. (2004) previously reported on the low sample variation with respect to adhesion density measurements in S-phase fibroblasts, facilitating accurate adhesion quantification. In this study, changes observed in HOBs cultured on nanogroove substrates were directly linked to the topographical modification, rather than to morphological changes that occur with HOB progression through the cell cycle.
In this study, we characterized vinculin-positive SMAs in primary HOBs on nanogroove and planar substrates. These structures appeared similar to elongated FAs and were prominent in well-spread osteoblasts such as those found on control and 100 μm groove/ridge substrates. We hypothesized that the mechanosensory function of FAs facilitates increased intracellular tension on these substrates and anisotropic protein recruitment at the adhesion plaque resulting in the generation of elongated fibrillar-like adhesions. These SMAs, although morphologically fibrillar, retain the molecular composition of classic FAs including vinculin and FA kinase (FAK; Zamir et al. 2000; Cukierman et al. 2001). It is probable that the generation of these SMAs and the conservation of FA-associated proteins, most notably FAK, induces increased upregulation in many major signalling pathways.
The S-phase HOBs cultured on planar controls formed predominantly mature FAs and SMAs, indicative of a well-adhered, well-spread phenotype. The HOBs cultured on experimental substrates demonstrated varying degrees of contact guidance and adhesion formation dependent on groove width. Adhesion complex formation was most perturbed in HOBs cultured on 10 μm groove/ridge arrays. Here, frequency of both FAs and SMAs was reduced. The increased number of focal complexes in HOBs on 25 μm groove/ridge substrates indicates a possible increase in cell motility and filopodia formation, a phenomenon that has been shown to occur on nanoscale topographies previously (Diehl et al. 2005). SMAs were significantly reduced in S-phase HOBs on 25 and 10 μm groove/ridge topographies, indicating a reduction in cell adhesion. It has been reported that aligned and elongated fibroblasts on micro- and nanoscale grooves secrete elevated levels of ECM as compared with non-aligned cells (Chou et al. 1995; Zhu et al. 2005). In this study, a correlation between cellular alignment and increased connective tissue synthesis was noted, supporting the hypothesis that cells dynamically interact with underlying topography and alteration of the ECM is reflected in cell function and behaviour (Adams 2002).
Immuno-SEM revealed adhesion formation of all subtypes to occur predominantly on the top of the ridge structures, a phenomenon previously reported by Teixeira et al. (2003). Adhesion to the ridge surface was prevalent in HOBs cultured on 10 and 25 μm groove/ridge arrays (figure 4c) but was absent in HOBs on 100 μm groove/ridge topographies, as increased inter-ridge area facilitated increased osteoblast spreading on both ridge and groove areas (figure 4b). Groove regions facilitated FA and SMA formation in the direction of groove propagation, aligning both osteoblasts and peripheral adhesions with groove orientation (figure 4d). Integrin-mediated adhesion aligning also occurred in the absence of cellular alignment, a phenomenon previously observed only on microgroove topographies (Richards et al. 1997). It has been suggested that the geometrical dimensions of the FA plaques force propagation on a grooved surface to be orientated parallel to these ridges (Ohara & Buck 1979). FAs show highly anisotropic growth dynamics in which additional proteins are mostly accumulated in the direction of the force exerted by the cytoskeleton (Besser & Safran 2006). Cellular and cytoskeletal alignment has generally been found to be more pronounced on patterns with ridge widths between 1 and 5 μm than on grooves and ridges with larger lateral dimensions (Matsuzaka et al. 2000; Teixeira et al. 2003; Karuri et al. 2004) such as those used in this study.
Integrin-mediated adhesion plaques act as sites for signal transduction, linking the transcriptional machinery of a cell to its outside environment. Indirect mechanotransductive pathways act to propagate mechanical forces from the ECM to the cell nucleus, regulating cellular response through a dynamic network of protein interaction. It is hypothesized that via direct linkage of the cytoskeletal intermediate filaments to the nucleoskeletal intermediate filaments (lamins) and the association of lamins with the interphase chromosomes, direct linkage from cellular adhesions to DNA is achieved and genomic regulation may thus be controlled by cell adhesion (Dalby 2005; Dalby et al. 2006). Indeed, a study by Jean et al. (2004) concluded that nuclear deformation appeared to be in direct and immediate response to alterations in cell adhesion area, hence it seems probable that direct mechanotranscriptive pathways play a major role in modulating cellular function in progenitor cells cultured on nanogrooved topographies.
Nanogrooves induced changes in osteoblast cytoskeletal organization, cellular morphology and, primarily, adhesion formation, which were correlated to significant alterations in osteoprogenitor gene expression (figure 9). HOBs cultured on 10 μm groove/ridge arrays demonstrated a downregulation in Rho expression (figure 9a), typical in cells undergoing increased cell migration, when focal complexes and lamellapodia are more prominent than FAs and stress fibres (Bershadsky et al. 2003). Conversely, this protein was upregulated in Stro+ progenitors cultured on 100 μm groove/ridge arrays (figure 9b), typical of well-spread, adhered cells. One of the main downstream targets of Rho is mDia (Riveline et al. 2001), a protein that may regulate microtubule polymerization (Schwarz & Bischofs 2005).
Expression of α-actinin was increased in progenitor cells cultured on 100 μm and reduced in cells cultured on 10 μm groove/ridge topographies, a major stress fibre-associated protein that plays a key role in bundling and maintaining the stability of the stress fibres (Byers et al. 1984; Burridge et al. 1988). In addition to functioning as an organizer of stress fibres, α-actinin also directly connects the stress fibres to FAs via the structural protein vinculin (Katoh et al. 2007). A downregulation in α-actinin expression will result in decreased tension-dependent adhesion growth, cellular tension and subsequent adhesion-mediated cellular signalling in cells cultured on 10 μm groove/ridge arrays. It appears that nanogroove substrates act as mediators of cellular function through both the regulation of adhesion formation and subsequent signalling pathways activated/deactivated by cellular elongation.
5. Conclusion
Dynamic regulation of FA structure and the integrin–cytoskeleton associations modulating cellular function play a central role in balancing adhesive and migratory stimuli in the cell (Cohen et al. 2004). Nanogroove feature dimension influences the generation of adhesion complex subtypes in S-phase HOBs as well as modulates the functional response in Stro+ progenitor populations. Increased groove width is associated with reduced contact guidance and increased adhesion formation and osteospecific function. Conversely, decreased nanogroove widths were associated with increased contact guidance, reduced adhesion formation and an increase in angiospecific function. These findings are intriguing as they indicate that a critical frequency of nanofeatures is necessary to modulate cellular adhesion and subsequent cellular function, and may provide valuable insight for the fabrication of osteoinductive biomaterials with significant clinical implications.
Table 1.
Statistically significant data pertaining to adhesion subtype quantification. (Kruskal–Wallis one-way analysis of variance was used to estimate the equality of population medians among substrate groups. The largest rank differences were seen between SMA formation on planar and 10 μm groove groups. Data group comparisons indicate a significance of p<0.05.)
| comparison | difference of ranks |
|---|---|
| flat SMA versus 10 μm grooves SMA | 117.018 |
| flat SMA versus 25 μm grooves SMA | 125.878 |
| 25 μm groove FX versus 10 μm groove FX | 114.578 |
| 25 μm groove FX versus 100 μm groove FX | 121.862 |
| 25 μm groove FX versus flat FX | 107.298 |
Acknowledgments
This work was supported by a grant 04-D81 from the AO Research Fund, Switzerland. M.J.D. is a BBSRC David Phillips Fellow and is supported through this fellowship. We would like to acknowledge the work of the Sir Henry Wellcome Functional Genomics Facility and thank the following people: Prof. Adam Curtis and Dr Mathis Riehle for their interesting discussions and support, John Pedersen (SDC Dandisc A/S, Denmark) for the organizing and preparation of nickel shims, Bill Monaghan and Mary Robertson for embossing into pMMA and materials characterization. We also thank Dr Rahul Tare, Bone & Joint Research Group, University of Southampton, for the provision of immunoselected enriched MSC populations and Mrs Pamela Furlong for technical assistance with SEM sample preparation.
References
- Abercrombie M, Heaysman J.E, Pegrum S.M. The locomotion of fibroblasts in culture. IV. Electron microscopy of the leading lamella. Exp. Cell Res. 1971;67:359–367. doi: 10.1016/0014-4827(71)90420-4. [DOI] [PubMed] [Google Scholar]
- Adams J.C. Molecular organisation of cell–matrix contacts: essential multiprotein assemblies in cell and tissue function. Expert Rev. Mol. Med. 2002;4:1–24. doi: 10.1017/S1462399402004039. [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D, Tint I.S, Neyfakh A.A, Jr, Vasiliev J.M. Focal contacts of normal and RSV-transformed quail cells. Hypothesis of the transformation-induced deficient maturation of focal contacts. Exp. Cell Res. 1985;158:433–444. doi: 10.1016/0014-4827(85)90467-7. [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D, Balaban N.Q, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Biol. 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Besser A, Safran S.A. Force-induced adsorption and anisotropic growth of focal adhesions. Biophys. J. 2006;90:3469–3484. doi: 10.1529/biophysj.105.074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs M.J, Richards R.G, Gadegaard N, Wilkinson C.D, Dalby M.J. Regulation of implant surface cell adhesion: characterization and quantification of S-phase primary osteoblast adhesions on biomimetic nanoscale substrates. J. Orthop. Res. 2006;25:273–282. doi: 10.1002/jor.20319. [DOI] [PubMed] [Google Scholar]
- Biggs M.J, Richards R.G, Gadegaard N, Wilkinson C.D, Dalby M.J. The effects of nanoscale pits on primary human osteoblast adhesion formation and cellular spreading. J. Mater. Sci. Mater. Med. 2007;18:399–404. doi: 10.1007/s10856-006-0705-6. [DOI] [PubMed] [Google Scholar]
- Bozec L, De Groot J, Odlyha M, Nicholls B, Horton M.A. Mineralised tissues as nanomaterials: analysis by atomic force microscopy. IEE Proc. Nanobiotechnol. 2005;152:183–186. doi: 10.1049/ip-nbt:20050004. [DOI] [PubMed] [Google Scholar]
- Brodsky B, Persikov A.V. Molecular structure of the collagen triple helix. Adv. Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu. Rev. Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- Byers H.R, White G.E, Fujiwara K. Organization and function of stress fibers in cells in vitro and in situ. A review. Cell Muscle Motil. 1984;5:83–137. doi: 10.1007/978-1-4684-4592-3_2. [DOI] [PubMed] [Google Scholar]
- Campbell C.E, Von Recum A.F. Microtopography and soft tissue response. J. Invest. Surg. 1989;2:51–74. doi: 10.3109/08941938909016503. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam E.A, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz J.P. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou L, Firth J.D, Uitto V.J, Brunette D.M. Substratum surface topography alters cell shape and regulates fibronectin mRNA level, mRNA stability, secretion and assembly in human fibroblasts. J. Cell Sci. 1995;108(Pt 4):1563–1573. doi: 10.1242/jcs.108.4.1563. [DOI] [PubMed] [Google Scholar]
- Cohen M, Joester D, Geiger B, Addadi L. Spatial and temporal sequence of events in cell adhesion: from molecular recognition to focal adhesion assembly. Chembiochem. 2004;5:1393–1399. doi: 10.1002/cbic.200400162. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens D.R, Yamada K.M. Taking cell–matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Curtis A.S. The mechanism of adhesion of cells to glass. A study by interference reflection microscopy. J. Cell Biol. 1964;20:199–215. doi: 10.1083/jcb.20.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M.J. Topographically induced direct cell mechanotransduction. Med. Eng. Phys. 2005;27:730–742. doi: 10.1016/j.medengphy.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Dalby M.J, Riehle M.O, Johnstone H.J, Affrossman S, Curtis A.S. Nonadhesive nanotopography: fibroblast response to poly(n-butyl methacrylate)-poly(styrene) demixed surface features. J. Biomed. Mater. Res. A. 2003a;67:1025–1032. doi: 10.1002/jbm.a.10139. [DOI] [PubMed] [Google Scholar]
- Dalby M.J, Riehle M.O, Yarwood S.J, Wilkinson C.D, Curtis A.S. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp. Cell Res. 2003b;284:274–282. doi: 10.1016/S0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- Dalby M.J, Riehle M.O, Sutherland D.S, Agheli H, Curtis A.S. Morphological and microarray analysis of human fibroblasts cultured on nanocolumns produced by colloidal lithography. Eur. Cell Mater. 2005;9:1–8. doi: 10.22203/ecm.v009a01. [DOI] [PubMed] [Google Scholar]
- Dalby M.J, Biggs M.J, Gadegaard N, Kalna G, Wilkinson C.D, Curtis A.S. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell Biochem. 2006;100:326–338. doi: 10.1002/jcb.21058. [DOI] [PubMed] [Google Scholar]
- Den Braber E.T, De Ruijter J.E, Ginsel L.A, Von Recum A.F, Jansen J.A. Orientation of ECM protein deposition, fibroblast cytoskeleton, and attachment complex components on silicone microgrooved surfaces. J. Biomed. Mater. Res. 1998;40:291–300. doi: 10.1002/(SICI)1097-4636(199805)40:2%3C291::AID-JBM14%3E3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Diehl K.A, Foley J.D, Nealey P.F, Murphy C.J. Nanoscale topography modulates corneal epithelial cell migration. J. Biomed. Mater. Res. A. 2005;75:603–611. doi: 10.1002/jbm.a.30467. [DOI] [PubMed] [Google Scholar]
- Gadegaard N, Mosler M, Larsen M.B. Biomimetic polymer nanostructures by injection molding. Macromol. Mater. Eng. 2003;288:76–83. doi: 10.1002/mame.200290037. [DOI] [Google Scholar]
- Gallant N.D, Charest J.L, King W.P, Garcia A.J. Micro- and nano-patterned substrates to manipulate cell adhesion. J. Nanosci. Nanotechnol. 2007;7:803–807. doi: 10.1166/jnn.2007.507. [DOI] [PubMed] [Google Scholar]
- Giraud-Guille M.M. Twisted plywood architecture of collagen fibrils in human compact bone osteons. Calcif. Tissue Int. 1988;42:167–180. doi: 10.1007/BF02556330. [DOI] [PubMed] [Google Scholar]
- Hamilton D.W, Brunette D.M. The effect of substratum topography on osteoblast adhesion mediated signal transduction and phosphorylation. Biomaterials. 2007;28:1806–1819. doi: 10.1016/j.biomaterials.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Howard D, Partridge K, Yang X, Clarke N.M, Okubo Y, Bessho K, Howdle S.M, Shakesheff K.M, Oreffo R.O. Immunoselection and adenoviral genetic modulation of human osteoprogenitors: in vivo bone formation on PLA scaffold. Biochem. Biophys. Res. Commun. 2002;299:208–215. doi: 10.1016/S0006-291X(02)02561-5. [DOI] [PubMed] [Google Scholar]
- Hu W, Yim E.K.F, Reano R.M, Leong K.W, Pang S.W. Effects of nanoimprinted patterns in tissue-culture polystyrene on cell behavior. J. Vasc. Sci. Technol. B. 2005;23:2984–2989. doi: 10.1116/1.2121729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Archer C.W, Walker P.S, Blunn G.W. Attachment and proliferation of osteoblasts and fibroblasts on biomaterials for orthopaedic use. Biomaterials. 1995;16:287–295. doi: 10.1016/0142-9612(95)93256-D. [DOI] [PubMed] [Google Scholar]
- Isa Z.M, Schneider G.B, Zaharias R, Seabold D, Stanford C.M. Effects of fluoride-modified titanium surfaces on osteoblast proliferation and gene expression. Int. J. Oral Maxillofac. Implants. 2006;21:203–211. [PubMed] [Google Scholar]
- Jean R.P, Gray D.S, Spector A.A, Chen C.S. Characterization of the nuclear deformation caused by changes in endothelial cell shape. J. Biomech. Eng. 2004;126:552–558. doi: 10.1115/1.1800559. [DOI] [PubMed] [Google Scholar]
- Karuri N.W, Liliensiek S, Teixeira A.I, Abrams G, Campbell S, Nealey P.F, Murphy C.J. Biological length scale topography enhances cell-substratum adhesion of human corneal epithelial cells. J. Cell Sci. 2004;117:3153–3164. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kano Y, Ookawara S. Rho-kinase dependent organization of stress fibers and focal adhesions in cultured fibroblasts. Genes Cells. 2007;12:623–638. doi: 10.1111/j.1365-2443.2007.01073.x. [DOI] [PubMed] [Google Scholar]
- Lenhert S, Meier M.B, Meyer U, Chi L, Wiesmann H.P. Osteoblast alignment, elongation and migration on grooved polystyrene surfaces patterned by Langmuir–Blodgett lithography. Biomaterials. 2005;26:563–570. doi: 10.1016/j.biomaterials.2004.02.068. [DOI] [PubMed] [Google Scholar]
- Lim J.Y, Dreiss A.D, Zhou Z, Hansen J.C, Siedlecki C.A, Hengstebeck R.W, Cheng J, Winograd N, Donahue H.J. The regulation of integrin-mediated osteoblast focal adhesion and focal adhesion kinase expression by nanoscale topography. Biomaterials. 2007;28:1787–1797. doi: 10.1016/j.biomaterials.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Loesberg W.A, Te Riet J, Van Delft F.C, Schon P, Figdor C.G, Speller S, Van Loon J.J, Walboomers X.F, Jansen J.A. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials. 2007;28:3944–3951. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Matsuzaka K, Walboomers F, De Ruijter A, Jansen J.A. Effect of microgrooved poly-l-lactic (PLA) surfaces on proliferation, cytoskeletal organization, and mineralized matrix formation of rat bone marrow cells. Clin. Oral Implants Res. 2000;11:325–333. doi: 10.1034/j.1600-0501.2000.011004325.x. [DOI] [PubMed] [Google Scholar]
- Meredith D.O, Owen G.R, Ap Gwynn I, Richards R.G. Variation in cell-substratum adhesion in relation to cell cycle phases. Exp. Cell Res. 2004;293:58–67. doi: 10.1016/j.yexcr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Mirmalek-Sani S.H, Tare R.S, Morgan S.M, Roach H.I, Wilson D.I, Hanley N.A, Oreffo R.O. Characterization and multipotentiality of human fetal femur-derived cells: implications for skeletal tissue regeneration. Stem Cells. 2006;24:1042–1053. doi: 10.1634/stemcells.2005-0368. [DOI] [PubMed] [Google Scholar]
- Ohara P.T, Buck R.C. Contact guidance in vitro. A light, transmission, and scanning electron microscopic study. Exp. Cell Res. 1979;121:235–249. doi: 10.1016/0014-4827(79)90002-8. [DOI] [PubMed] [Google Scholar]
- Rho J.Y, Kuhn-Spearing L, Zioupos P. Mechanical properties and the hierarchical structure of bone. Med. Eng. Phys. 1998;20:92–102. doi: 10.1016/S1350-4533(98)00007-1. [DOI] [PubMed] [Google Scholar]
- Richards R.G, Ap Gwynn I. Backscattered electron imaging of the undersurface of resin-embedded cells by field emission scanning electron microscopy. J. Microsc. 1995;177:43–52. doi: 10.1111/j.1365-2818.1995.tb03532.x. [DOI] [PubMed] [Google Scholar]
- Richards R.G, Rahn B.A, Ap Gwynn I. A quantitative method of measuring cell adhesion areas. Review. Cells Mater. 1997;7:15–30. [Google Scholar]
- Richards R.G, Stiffanic M, Owen G.R, Riehle M, Ap Gwynn I, Curtis A.S. Immunogold labelling of fibroblast focal adhesion sites visualised in fixed material using scanning electron microscopy, and living, using internal reflection microscopy. Cell Biol. Int. 2001;25:1237–1249. doi: 10.1006/cbir.2001.0807. [DOI] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban N.Q, Schwarz U.S, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky A.D. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin M.A, Rubin J, Jasiuk I. SEM and TEM study of the hierarchical structure of C57BL/6J and C3H/HeJ mice trabecular bone. Bone. 2004;35:11–20. doi: 10.1016/j.bone.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Schwarz U.S, Bischofs I.B. Physical determinants of cell organization in soft media. Med. Eng. Phys. 2005;27:763–772. doi: 10.1016/j.medengphy.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Sela M.N, Badihi L, Rosen G, Steinberg D, Kohavi D. Adsorption of human plasma proteins to modified titanium surfaces. Clin. Oral Implants Res. 2007;18:630–638. doi: 10.1111/j.1600-0501.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- Sutherland J, Denyer M, Britland S. Contact guidance in human dermal fibroblasts is modulated by population pressure. J. Anat. 2005;206:581–587. doi: 10.1111/j.1469-7580.2005.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A.I, Abrams G.A, Bertics P.J, Murphy C.J, Nealey P.F. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H.D, Weiner S. On the relationship between the microstructure of bone and its mechanical stiffness. J. Biomech. 1992;25:1311–1320. doi: 10.1016/0021-9290(92)90286-A. [DOI] [PubMed] [Google Scholar]
- Zamir E, Geiger B. Molecular complexity and dynamics of cell–matrix adhesions. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- Zamir E, et al. Dynamics and segregation of cell–matrix adhesions in cultured fibroblasts. Nat. Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- Zhu B, Lu Q, Yin J, Hu J, Wang Z. Alignment of osteoblast-like cells and cell-produced collagen matrix induced by nanogrooves. Tissue Eng. 2005;11:825–834. doi: 10.1089/ten.2005.11.825. [DOI] [PubMed] [Google Scholar]
- Zimerman B, Volberg T, Geiger B. Early molecular events in the assembly of the focal adhesion–stress fiber complex during fibroblast spreading. Cell Motil. Cytoskeleton. 2004;58:143–159. doi: 10.1002/cm.20005. [DOI] [PubMed] [Google Scholar]