Abstract
Neurological injury is a frequent cause of morbidity and mortality from general anesthesia and related surgical procedures that could be alleviated by development of effective, easy to administer and safe preconditioning treatments. We seek to define the neural immune signaling responsible for cold-preconditioning as means to identify novel targets for therapeutics development to protect brain before injury onset. Low-level pro-inflammatory mediator signaling changes over time are essential for cold-preconditioning neuroprotection. This signaling is consistent with the basic tenets of physiological conditioning hormesis, which require that irritative stimuli reach a threshold magnitude with sufficient time for adaptation to the stimuli for protection to become evident.
Accordingly, delineation of the immune signaling involved in cold-preconditioning neuroprotection requires that biological systems and experimental manipulations plus technical capacities are highly reproducible and sensitive. Our approach is to use hippocampal slice cultures as an in vitro model that closely reflects their in vivo counterparts with multi-synaptic neural networks influenced by mature and quiescent macroglia / microglia. This glial state is particularly important for microglia since they are the principal source of cytokines, which are operative in the femtomolar range. Also, slice cultures can be maintained in vitro for several weeks, which is sufficient time to evoke activating stimuli and assess adaptive responses. Finally, environmental conditions can be accurately controlled using slice cultures so that cytokine signaling of cold-preconditioning can be measured, mimicked, and modulated to dissect the critical node aspects. Cytokine signaling system analyses require the use of sensitive and reproducible multiplexed techniques. We use quantitative PCR for TNF-α to screen for microglial activation followed by quantitative real-time qPCR array screening to assess tissue-wide cytokine changes. The latter is a most sensitive and reproducible means to measure multiple cytokine system signaling changes simultaneously. Significant changes are confirmed with targeted qPCR and then protein detection. We probe for tissue-based cytokine protein changes using multiplexed microsphere flow cytometric assays using Luminex technology. Cell-specific cytokine production is determined with double-label immunohistochemistry. Taken together, this brain tissue preparation and style of use, coupled to the suggested investigative strategies, may be an optimal approach for identifying potential targets for the development of novel therapeutics that could mimic the advantages of cold-preconditioning.
Keywords: Neuroscience, Issue 43, innate immunity, hormesis, microglia, hippocampus, slice culture, immunohistochemistry, neural-immune, gene expression, real-time PCR
Protocol
Sterile and aseptic techniques are critical to the preparation, maintenance, and use of slice cultures for extended periods. Furthermore, the rationale for our use of slice cultures only after 18 days in vitro is based on evidence that indicates excitatory and inhibitory synaptic transmission becomes most mature, and glia (astrocytes and microglia) become quiescent and consistent with their in vivo counterparts (Figure 1).
1. Preparation & Maintenance of Hippocampal Slice Cultures
The same day of culturing, equilibrate sterile inserts in 1.1 mL growth media consisting of 50 mL Basal Medium Eagle, 25 mL Earle's Balanced Salt Solution, 23 mL Horse Serum, 0.5 mL Glutamax (200 mM stock), 0.1 mL gentamicin (10 mg/mL stock), 0.4 mL Fungizone (250 μg/mL), 1.45 mL D-glucose (45%; 42 mM total).
Keep six-well trays in an incubator at 36°C, 5% carbon dioxide and 95% humidity.
For maximal sterility, culturing is ideally done in a HEPA filtered positive pressure culture room with the air also purified via self-contained ultra-violet light air-cleansing fans, and while wearing a clean lab coat and sterile gloves stretched over the lab coat sleeves.
Prepare slice cultures in a BSL-1 laminar fume hood that includes all needed materials (i.e., McIlwain tissue chopper, related Teflon inserts, razor slicing blade, dissection cold (3-4°C) plate, surgical tools, and dissection Petri dishes) using a Wild (M8) stereomicroscope.
- Allow the cooling plate (run from a nearby water bath) to reach 3-4°C temperature for at least 30 min while at the same time exposing dissection materials to ultraviolet light to further establish sterility of the dissection area and tools.
- The laminar flow hood is periodically tested to ensure the ultraviolet light has sufficient power at the tabletop level with a shortwave ultraviolet light measuring meter.
Use rat pups (P8-P9 and ~23 g/ea. from litters culled to 10 at birth) anesthetized with 100% carbon dioxide in a small animal box on an aseptic bench behind the fume hood and cleansed by dipping in 100% ethanol within the fume hood, where all subsequent steps are performed.
Decapitate pups in one half of a 100 mm Petri dish and place the head in the second half, using a fresh dish per pup.
Dissect out the brain and place it into the bottom half of a 60 mm Petri dish containing 10 mL sterile cold (3-4°C) Gey's Balanced Salt Solution supplemented with D-glucose to 6.5 mg/mL (i.e., 7.5 mL of 45% d-glucose per 500 mL bottle of Gey's.
Dissect out the hippocampus from each hemisphere and place them onto a Teflon disk for the McIlwain chopper. Spread media away from the brain tissue using an iris spatula to prevent cut brain sections from adhering to the cutting blade.
Section the hippocampi perpendicular to their long axis using the McIlwain chopper, with section thickness set at 350-400 μm.
Briskly wash freshly cut slices off of the Teflon inserts and into the top half of a 60 mm Petri dish containing cold (3-4°C) Gey's Balanced Salt Solution with 6.5 mg/mL D-glucose using a one mL pipettor.
Inspect slices under the stereomicroscope for an intact dentate gyrus and pyramidal cell layer.
Gently place the largest slices onto an insert (three per insert and 12 slices/pup) and maintain under normal incubation conditions in an incubator cleaned every six months and calibrated every three months with an infrared carbon dioxide analyzer and thermometer accurate to one decimal place.
Refresh growth media in culture dishes every 3-4 days in vitro using a BSL-2 hood and sterile technique.
After 7 days in vitro, replace media with 1.1 mL serum-free media (SFM) consisting of 97 mL Neurobasal, 2 mL B27, 0.5 mL Glutamax (200 mM), 0.1 mL ascorbic acid (0.5 M), and 0.68 mL D-Glucose (45%; 42 mM total).
2. Pre-screen Vitality of Slice Cultures
Aliquot Sytox Green stock (10 μL) and store at -20°C.
Dilute Sytox stock to 500 nM working concentration in serum-free growth media (i.e., 10 μL in 100 mL media). Store Sytox media at 4°C and use for up to one week.
Place 1.1 mL of Sytox per well in 6-well culture dishes and warm to 36°C at 5% carbon dioxide for a sufficient period (i.e., as evidenced by lack of condensate on culture dishes).
Meanwhile, allow an ultraviolet lamp (i.e., a fluorescent light source with an FITC filter) powered via a stabilized power supply to ensure uniform light intensity to warm to temperature for at least 20 min.
Place slice cultures (18 days in vitro) in Sytox-media for 10 min and then back to SFM to screen for irreversible CA1 pyramidal layer injury.
View cultures on an aseptic surface of an inverted microscope, examined under 5x magnification.
Accept cultures with fewer than 30 Sytox positive cells in the CA1 area.
3. Cold-Preconditioning
Place 1.1 mL of SFM in 6-well culture dishes. Allow cooled media (e.g., 30°C) to equilibrate in an incubator (5% carbon dioxide and 95% humidity) for at least 20 min before use. Absence of condensate on culture dishes indicates that adequate time for equilibration has occurred.
Transfer slice culture inserts to cooled media (1.1 mL/well) tray kept at 30°C and incubate for 90 min using sterile technique in a BSL-2 fume hood.
Place slice cultures back into normal SFM under normal incubation conditions for 24 hours, again using sterile technique via a BSL-2 fume hood.
4. Excitotoxic Injury
- Begin slice culture experimental use prescreen by exposing to Sytox media for 20 min.
- Acquire individual slice images with slices maintained at an orientation similar to that used for prescreen (background images). This is done by placing a mark to the left and two marks to the right with marker pen at "10" and "two" o'clock on each insert. This maneuver facilitates using the same area of interest shape for each culture set of images.
At the same time, turn on the ultraviolet lamp source (under regulated power supply), and CCD camera for a minimum of 20 min so they warm to temperature.
- Then, calibrate the CCD camera with a fluoroscein standard.
- "Clear" the CCD by exposing to light for 50 cycles unless an automatically calibrating CCD camera is used. We have established a program within MetaMorph to clear our Cool Snap camera used for injury quantification.
- Place 10 μL of fluoroscein in 90 mL PBS (10 mM phosphate buffer, 150 mM NaCl at 7.3 pH) and vortex.
- Pipette 10 μL of fluoroscein mixture onto a 100 μm deep hemacytometer.
- Adjust full image intensity to 1000/4096.
Collect background images of slice cultures to verify that cold-preconditioning did not cause injury (i.e., discard cultures with ≥ 250 CA1 area of interest intensity).
- Prepare trays with NMDA media.
- Prepare 10 mM stock solution of NMDA at least weekly.
- For excitotoxic injury, dilute NMDA to 20 50 μM in SFM and equilibrate to normal incubation conditions for at least 20 min.
Using the BSL-2 hood and sterile technique, place inserts into the NMDA-media for 60 min under normal incubation conditions.
Rinse NMDA off of inserts by dipping each insert three times in three separate 60 mm dishes each containing 10 mL Neurobasal warmed to 36°C. Do not use more than three inserts for each set of three 60 mm wash dishes.
Return cultures to normal incubation conditions.
- Collect injury images 24 hours after exposure to NMDA.
- Calibrate camera with fluoroscein standard as described above.
- Place cultures in Sytox media for 20 min.
- Quantitate Injury:
- Using MetaMorph software, draw an AOI around CA1 region.
- Measure intensity of selected region for "injury."
- Copy and paste AOI from "injury" to "background" image.
- Enter values from "injury" and "background" into Excel.
- Quantitate "numerator-background" for Control and Cold-preconditioned cultures.
- Using statistical software (e.g., SigmaStat), run appropriate statistical tests to compare injury of experimental group(s) versus a control group, which should always be included with each experimental run.
- Note: If injury levels are low in one-day post-injury images, carry-out experiment to two three days. Protection in cultures may be masked due to a lack of susceptibility to injury (Figure 2).
- Note: The issue of "protection" prompted us to move to measuring injury levels and not ratios for all comparisons (Figure 3).
5. Co-treatments Applied with Cold-preconditioning
An important advantage of slice cultures is that environmental conditions can be accurately controlled. This means that cytokine signaling from cold-preconditioning can be measured, mimicked, and modulated to dissect the critical node aspects.
Recombinant (e.g., agonist) proteins and neutralizing (e.g., antibody or soluble receptor) proteins can be used to mimic and modulate, respectively, cytokine signaling.
In general, these proteins are reconstituted and stored as aliquots at -20°C for use within six months to ensure adequate bioactivity.
- Here, we describe exemplary use of TNF-α signaling abrogation using soluble TNF receptor 1 (sTNFR1).
- Reconstitute sTNFR1 in 0.1% bovine serum albumin in PBS to a stock of 50 μg/mL, aliquot in 20-50 μL amounts, and store at -20°C.
- For use, dilute sTNFR1 to 200 ng/mL in slice cultures growth media warmed to 36°C and place slice cultures in sTNFR1-media for 20 min prior to cold-preconditioning.
- Note: To reduce variance, it is best to dilute 200 ng/mL sTNFR1 in a large volume of growth media (i.e., 1:250 dilution from 50 μg/mL sTNFR1 stock in 10 mL of growth media requires 40 μL sTNFR1) vs. adding sTNFR1 directly to each well in dish (4.4 μL per 1.1 mL media).
- Dilute sTNFR1 to 200 ng/mL in media and place 1.1 mL in each insert well. Allow media to equilibrate to 30°C for at least 20 min before exposing slice cultures to cold-preconditioning for 90 min as described above.
- Place slice cultures back at 36°C with sTNFR1 present in media.
- 24 hours later, expose cultures to Sytox media for 20 min as previously described and collect background images to verify that cold-preconditioning did not injure cultures.
Quantitate data as described above.
Refresh media with components every 3-4 days in vitro.
6. Immediate and Delayed Excitotoxic Injury after Cold-preconditioning
- Immediate injury with cold-preconditioning.
- Perform cold-preconditioning as described above.
- After cold-preconditioning, return cultures to normal incubation for 20 min.
- Then, expose cultures to 20-50 μM NMDA injury for one hour.
- Rinse cultures three times as described above and return to normal incubation.
- After 24 hours, acquire injury photos as described above.
- Delayed effects of cold-preconditioning.
- Perform cold-preconditioning as described above.
- Expose cultures to NMDA injury 24 hours later as described above.
- Continue to acquire injury images for up to three days after initial cold-preconditioning to monitor protection.
7. RNA Isolation
The following gene expression procedures are scaled for a single slice culture.
Transfer slice cultures from growth media and normal incubation to 3 mL RNAlater in 6-well culture dishes to stabilize RNA. Store at 4°C for up to three days until processed as described below.
Gently lift the slice culture off the insert with a fine tip paint brush and place in 1 mL of cold (4°C) PBS in a 1.5 mL (DNase, RNase, and DNA free) microcentrifuge tube.
Centrifuge samples for 30 sec and remove PBS supernatant.
Store samples at -80°C.
To isolate RNA from slice cultures (~250 ng/ea.), thaw samples on ice.
- Isolate RNA using Qiagen MicroRNeasy kit via the following procedures that are described in further detail and adapted from the RNeasy Micro Handbook (Qiagen).
- Place 350 μL Buffer RLT (containing 10 μL β-mercaptoethanol per mL) into each microcentrifuge tube containing one slice culture.
- Homogenize samples by vortexing for 30 sec. Triturate tissue, if necessary (i.e., tissue pellet still visible) using a disposable pestle until completely homogenized in Buffer RLT.
- Place 350 μL of 70% ethanol into the lysate mix and pipette to mix.
- Transfer homogenate to a spin column and place in a collection tube (both spin column and collection tube are provided by Qiagen).
- Centrifuge sample for 15 sec at max speed. Retain the column.
- Wash the spin column by adding 350 μL Buffer RW1 plus centrifuge for 15 sec at max speed. Discard buffer.
- Dilute 10 μL of DNase I stock in 70 μL Buffer RDD to yield 80 μL total volume. Pipette gently to mix do not vortex. Prepare 80 μL of diluted DNase stock per sample. Incubate at room temperature for 30 min.
- Add 350 μL Buffer RW1 to sample spin column and centrifuge for 15 sec. Discard collection tube.
- Using a new collection tube, add 500 μL Buffer RPE to sample and centrifuge for 15 sec and discard buffer.
- Place 500 μL of 80% ethanol on sample and centrifuge for 2 min. Discard the collection tube.
- Dry the spin columns by centrifuging for 5 min at max speed with tube caps open, discard collection tubes, and transfer spin column to a 1.5 mL collection tube supplied by the kit.
- To collect isolated RNA, place 14 μL RNase-free water on the center of the spin column membrane. Centrifuge for one min and collect the fluid.
- Add 2 μL of RNasin (diluted at 1 U/ μL in TE buffer) and store at -80°C. TE buffer consists of 10 mM Tris (tris[hydroxymethyl]aminomethane), 1mM EDTA (ethylenediaminetetraacetic acid disodium salt hydrate) at 8.0 pH.
8. RNA Quantification
Note: RNasin does not interfere with the RiboGreen assay.
- Dilute RiboGreen 1:200 in RNase-free TE buffer as follows.
- TE (mL): 1; Ribogreen (μL): 5;
- TE (mL): 4; Ribogreen (μL): 20;
- TE (mL): 6; Ribogreen (μL): 30.
- RNA standards are prepared as follows.
- Yeast tRNA is used as the RNA standard. tRNA is stored (-20°C) as 1 mg/mL in TE buffer.
- Dilute the 1 mg/mL stock 1:100 in TE buffer, using (DNase, RNase, and DNA free) 1.5 mL microcentrifuge tubes to produce a 1 μg/mL working standard.
- Prepare the standard curve directly in a 96-well fluorescent assay plate by adding the TE buffer diluent first, then adding the appropriate amount of the 1 μg/mL standard into the TE buffer already in the plate wells as shown in the adjacent table. Make duplicate wells for each standard as follows.
- Vol. of std. (μL): 100; Vol. of TE (μL): 0; RNA in well (ng): 100;
- Vol. of std. (μL): 80; Vol. of TE (μL): 20; RNA in well (ng): 80;
- Vol. of std. (μL): 60; Vol. of TE (μL): 40; RNA in well (ng): 60;
- Vol. of std. (μL): 40; Vol. of TE (μL): 60; RNA in well (ng): 40;
- Vol. of std. (μL): 20; Vol. of TE (μL): 80; RNA in well (ng): 20;
- Vol. of std. (μL): 0; Vol. of TE (μL): 0; RNA in well (ng): 0.
- The assay is prepared as follows.
- Add 99 μL of TE buffer to each of the sample wells of the 96 well plate. The most efficient way to do this is to use a multichannel pipettor.
- Add 1 μL of RNA to the experimental sample wells.
- Add 100 μL of the diluted RiboGreen to each well using the multichannel pipettor and triturate with one or two strokes of the pipettor.
- Read the plate on a fluorescent plate reader at 480 nm excitation and 520 nm emission.
- Construct a standard curve using Microsoft Excel with measured fluorescence intensities of RNA standards.
- Calculate RNA concentrations in samples using the resulting linear equation from the standards curve.
9. SYBR Green Quantitative PCR
- PCR procedures are best done in a clean area reserved specifically for work with RNA (Figure 4).
- Decontaminate the area and related lab equipment from potential DNA contamination and RNase activity before use with ultraviolet light, 10% bleach or RNase Away.
- Wear gloves throughout all procedures.
- Use RNase free water for all procedures as provided with the kit. Do not use DEPC-(diethylpyrocarbonate) treated water.
- Close all PCR-related tubes immediately after use to retard aerosol contamination of foreign DNA.
Develop and characterize primers for the mRNA target(s) of interest. These methods are detailed in the Supplementary Methods to Hulse et al., Journal of Neuroscience, 2008 13.
- cDNA is produced from 30-200 ng total RNA via reverse transcription using iScript.
- The choice of starting RNA quantity is determined by the total amount of RNA available with the goal of performing RT-PCR analysis on a portion of the sample (i.e., 10%-20% of the total).
- The remaining larger portion of RNA is stored for future analysis.
- The iScript kit utilizes a blend of random hexamers and oligo-dT primers, and a modified MMLV reverse transcriptase in a total volume of 20 μL.
- Reverse transcription proceeds for 25°C for 30 min followed by 42°C for 50 min, and the reverse transcriptase is subsequently denatured at 85°C for 5 min.
- Dilute cDNA in TE buffer to a final concentration of 0.4 ng/μL relative to starting RNA quantity.
- SYBR Green PCR based strategy is used to amplify cDNA.
- PCR reactions are monitored in real-time by monitoring SYBR Green incorporation.
- The 50 μL reactions contain 3 mM MgCl2, 200 μM dNTPs, 15 pmol each of forward and reverse primers, 1 μM fluorescein, 1x SYBR Green dye, 10 μL (4 ng) slice culture cDNA, and 1.25 U Platinum Taq polymerase in reaction buffer consisting of 50 mM KCl and 10 mM Tris, 8.3 pH.
- PCR is performed using the iCycler thermocycler that can collect data in real time.
- Cycling parameters consist of 15 sec denaturation (95°C) followed by 30 sec annealing/extension (60°C) repeated 45 times.
- Optical measurements are taken during the annealing/extension step and Ct values were determined using the iCycler system software.
- cDNA for cytokine targets of interest and β-actin are used to construct standard curves.
- Copy number is determined from the molecular weight and mass of the plasmid cDNA.
- Standard curves of copy number/mass are constructed and included in each assay.
- Ct values for each cDNA dilution are used to determine the copy number v. Ct curve.
- Cytokine and β-actin copy number levels are determined for samples using standard curves.
10. Quantitative PCR for Microarrays
Quantitative real-time qPCR array screening is a highly sensitive and reproducible means to probe for low-level inflammatory mediator expression changes.
The RT2 Profiler PCR array from SABiosciences is used for inflammatory mediator gene expression changes, with the following steps described in further detail by the manufacturer.
The assay uses 0.1-1.0 μg RNA. We use local slice areas (e.g., CA1 which provides ~250 ng total RNA from two pooled samples) or whole single slices that contain a similar amount of total RNA.
- Sample and control RNAs are reverse transcribed to cDNA using a first strand kit (e.g., #C-03).
- The resultant cDNA is mixed with SYBR Green-based PCR mix (#PA-011) and 25 μL aliquotted into each of the 96 wells of the PCR array plate (measuring 84 unique experimental genes and 16 housekeeping genes).
- One array plate is prepared for the experimental sample and a second plate is prepared for the control.
- Note: The SYBR Green master mix provided by the manufacturer is thermal cycler-specific.
Thermal cycling is done using an iCycler with a 40 cycle protocol consisting of a denaturation step of 15 sec at 95°C followed by an extension step of one min at 60°C. Optical data is collected during the extension step. Thermal cycling is followed by melt curve analysis scanning the 55-95°C temperature range in 0.5°C increments.
The relative expression of genes in treated and control plates was determined using the 2-ΔΔCt method using the provided Excel based software and expressed as a fold increase or decrease relative to controls.
Genes with two-fold increases or decreases in expression are considered for further evaluation.
- Genes identified from PCR array screening (e.g., using #PARN-011A for cold-preconditioning) are further confirmed using qPCR.
- PCR arrays identify which genes are regulated in response to stimuli.
- In the example of cold-preconditioning, we identified the IL-11 gene as positively regulated in response to cold-preconditioning.
- The next step in the analysis is to confirm that the newly identified gene is regulated in slice cultures using the qPCR assay detailed above.
11. Multiplexed Microsphere Flow Cytometric Proteomic Assay
Expose slice cultures to cold-preconditioning as described above.
- Harvest slice cultures for total protein assay.
- Gently lift slices off the insert using a fine-tip brush and place in 1 mL cold (4°C) PBS.
- Centrifuge tubes and remove PBS off slice cultures. Place on dry ice until harvesting is finished.
- Store tubes at -80°C.
- Homogenize slice culture samples for performing a total protein assay.
- Prepare cell lysis buffer for homogenization.
- Following the manufacturer's instructions (Bio-Rad), add protease inhibitors to 5 mL of lysis buffer and set aside on ice.
- Place 100 μL of lysis buffer on slice cultures and agitate slices by pipetting up and down five times with a 100 μL pipette tip cut open to 1 mm.
- Shake samples on plate shaker at 4°C for 20 min.
- Centrifuge samples at 13,000 rpm for 15 min at 4°C.
- Transfer supernatant to a clean tube and set aside on ice.
- Perform total protein assay.
- Prepare BSA (bovine serum albumin) protein standards.
- Reconstitute stock BSA with ultrapure water according to manufacturer's instructions.
- Prepare protein standards ranging 0-1000 μg/mL, diluted in lysis buffer.
- Calculate the volume of working reagent needed according to kit instructions.
- For example, (eight standards + 18 unknown samples) x (two replicates) x (200 μL working reagent required per sample) = 10.4 mL of total volume of working reagent required to load onto a 96 well microplate (e.g., microsphere flow cytometric assay, see below).
- When mixing Reagent A with Reagent B, some turbidity may occur, but should disappear shortly.
- Load 10 μL of standards and samples onto microplate.
- Load 0.2 mL of working reagent into wells.
- Cover microplate with sealing tape and shake on plate shaker for 30 sec.
- Incubate microplate at 37°C for 30 min.
- Cool microplate to room temperature (about 10 min) before reading on a plate reader at 595 nm wavelength.
- Construct a standard curve using Microsoft Excel with measured absorbances of BSA standards.
- Calculate protein concentrations in samples using the resulting linear equation from the standards curve.
- Dilute all samples to the lowest concentration measured with lysis buffer and store at -80°C.
Perform single-plex and multiplex cytokine tissue (or fluid) assays as outlined in complete detail in references #12 and 16.
12. Immunohistochemistry
- Fix cultures for 24 hours with PLP fixative consisting of 10 mL 16% paraformaldehyde, 1.096 g lysine, 0.42 g sodium phosphate, and 0.17 g sodium periodate, filled to a total volume of 80 mL with ultrapure water (fixative is 6.2 pH).
- We find that PLP fixative is a gentle fixative that allows detection of low-level immunostaining that otherwise would not be evident using other fixatives.
- Cultures are fixed for 24 hours and then transferred with a fine brush to PBS containing sodium azide (100 mg/L).
- Bright field immunostaining of floating whole slice cultures is accomplished as follows with all steps coupled to shaking at ~ 50 rpm.
- Wash slice cultures in 3 mL PBS three times for 10 min.
- Quench slice cultures in 0.3% H2O2.
- Dilute 30% H2O2 stock solution in PBS.
- Wash slice cultures in PBS three times, 10 min each.
- Block slice cultures for one hour at room temperature in 3 mL blocking solution in a six-well plate consisting of 10 mL goat serum, 0.75 mL Triton X-100, and 89.25 mL PBS.
- Serum for the blocking solution should come from the same species in which the secondary antibody was raised.
- Incubate slice cultures overnight in primary antibody diluted in blocking solution at 4°C.
- The maximum volume of primary antibody solution for small glass Pyrex dish wells should be 0.3 mL higher volumes tend to run over the edge of the plate.
- Place the dish in a humidified closed container. We do not cover the dish with adherent film since sections might be spun onto the overlying film and lost for further workup.
- Adjust shaking speed of plate shaker to just high enough to circulate slices in the antibody solution.
- Remove slices from glass plate and wash in PBS three times for 10 min per wash.
- Incubate slices in secondary antibody at room temperature for one hour while shaking.
- Use small glass Pyrex dishes to load 0.3 mL secondary antibody solution.
- Wash three times in PBS, 10 min per wash.
- Visualize staining with 3'-diaminobenzidine (DAB).
- Dissolve 30 mg DAB in 3 mL dimethyl sulfoxide.
- Filter DAB using two #1 filter papers and wash with 54 mL PBS.
- Immediately before use, add 20 μL of 30% H2O2.
- Incubate cultures in DAB solution for 5-7 min at room temperature.
- Note: DAB is a carcinogen and must be disposed of properly. Dispose of all DAB solutions in a marked container stored in a fume hood, and place all dishes in bleach solution to deactivate DAB. All DAB solutions should ultimately be discarded via the Institutional Safety Office.
- Wash slice cultures in PBS three times for 10 min each.
- Wet mount slice cultures to gelatin (or silane) coated slides with 100 μL dish soap dissolved in distilled water. Dry overnight at room temperature.
- Avoid using slide warmers excessive heat may cause cracking in cultures mounted to slides.
- Dehydrate slides through a series of graded ethanol (50, 75, 95, 95, 100, 100 %) for 30 sec each.
- Clear slides with xylene four times for ten min each.
- Using a glass Pasteur pipette, place 0.1 mL of mounting media on top of slide and gently coverslip to avoid air bubbles forming. Dry overnight at room temperature.
- Fluorescent immunostaining.
- Section slice cultures to 20 μm thick using a cryostat.
- Adjust cryostat settings to -16°C for internal temperature, and -12°C for object temperature.
- Place a small amount of water on metal chuck using a Pasteur pipette and freeze on dry ice.
- Apply a thin disk layer of Tissue-Tek media on top of frozen chuck.
- Allow chuck to freeze and equilibrate to cryostat chamber temperature.
- Section Tissue-Tek media to create a flattened surface suitable for laying slice cultures flat.
- Note the orientation of the chuck while sectioning this provides a consistent angle for sectioning that will prevent mounting media from falling off the chuck.
- When frozen, take chuck out and place in a holder. Using a metal spatula and a fine-tip paint brush, slide one slice culture onto spatula and transfer over to the middle of the flattened layer of Tissue-Tek media on the chuck.
- Place a thin layer of Tissue-Tek media over mounted slice culture and freeze at chamber temperature for at least 10 min.
- With the "trim" button activated, section the top layers of Tissue-Tek media until slice culture is visible.
- Switch from "trim" to "section" preset at 20 μm.
- Pick-up 20 μm sections with gelatin-coated slides that are room temperature and dry slides overnight.
- Tissue-Tek media will be visible on slides, but dissolves in PBS washes.
- Immunostaining.
- Wash slides in PBS three times for 10 min each.
- Quench in 0.3% H2O2 for 15 min at room temperature, shaking.
- Wash slides in PBS three times for 10 min each.
- Using SFX signal enhancer, apply a few drops of Component A directly on top of sections on slide and incubate for 30 min at room temperature in a humidified chamber.
- Incubation with Component A replaces the blocking step with 10% goat serum solution performed in bright field immunohistochemistry.
- Incubate slides in primary antibody diluted in 0.75% Triton X-100 in PBS for two hours at 37°C.
- Apply primary antibody solution directly to top of slides and incubate in humidified chamber to prevent evaporation.
- Do not include serum in any of the antibody solutions only use PBS and Triton X-100.
- Wash slides in PBS three times for 10 min each.
- Incubate in secondary antibody for one hour at room temperature and protect slides from light.
- Centrifuge all fluorescent secondary antibodies for 20 min to prevent any aggregates from getting into the antibody solution.
- Prepare antibody dilutions using 0.75% Triton X-100 in PBS.
- Wash slides in PBS three times for 10 min each.
- Dip slides once in distilled water to wash off salts from PBS.
- Dry slides overnight at room temperature, covered away from light.
- Coverslip slides with ProLong Antifade media.
- Thaw Component A in microwave for 5-10 sec and add approximately 1 mL to a vial of Component B. Mix together using a Pastuer pipette, being careful not to introduce air bubbles into solution.
- Centrifuge for 5 min at max speed to remove bubbles.
- Apply a thin layer of ProLong media to top of slide using a Pastuer pipette, careful to avoid creating any air bubbles.
- Gently place cover slip on top of slide and dry overnight, covered away from light.
- Double-label fluorescent Immunostaining (Figure 5).
- Perform fluorescent Immunostaining as described above, except dilute both primary antibodies in the same incubation solution.
- Dilute all secondary antibodies in the same solution and incubate as described above.
- Serial incubation periods using two antibodies resulted in diminished immunostaining.
13. Representative Results
 Figure 1. Appearance of hippocampal slice cultures and microglia. The left hand image shows a typical mature (i.e., 21 day in vitro) hippocampal slice culture stain with NeuN (green) to illustrate the cytoarchitecture of the principal neurons. Pyramidal neurons are shown in areas CA1 and CA3 and dentate gyrus (DG) neurons to the left. Scale bar is 250 μm. The right hand image is derived from the CA1 area and shown at higher power to illustrate the branched quality of quiescent microglia within mature slice cultures. Cells were marked with the microglial surface marker, cd11b. Scale bar is 50 μm.
Figure 1. Appearance of hippocampal slice cultures and microglia. The left hand image shows a typical mature (i.e., 21 day in vitro) hippocampal slice culture stain with NeuN (green) to illustrate the cytoarchitecture of the principal neurons. Pyramidal neurons are shown in areas CA1 and CA3 and dentate gyrus (DG) neurons to the left. Scale bar is 250 μm. The right hand image is derived from the CA1 area and shown at higher power to illustrate the branched quality of quiescent microglia within mature slice cultures. Cells were marked with the microglial surface marker, cd11b. Scale bar is 50 μm.
 Figure 2. Cold-preconditioning neuroprotection in hippocampal slice cultures. Cultures are incubated with Sytox Green, a fluorescent marked dead cell marker. The top row shows sham control cultures and the bottom row shows slices exposed to 28°C for 90 min. Left hand, pre-screen images show no CA1 injury. Relative injury color calibration scale is shown in the left upper image. Middle row images show relative slice culture injury 24 hours after exposure to 20 μM NMDA. Notice that sham control injury is greater than that of cultures exposed to cold-preconditioning (CP). Traditionally, cultures are then exposed to 20 μM NMDA overnight to maximize CA1 neuronal injury levels and relative injury of CP v. sham, noted as a ratio of injury/maximal injury. However, exposure to maximal injury stimuli may not be sufficient to overcome neuroprotection from preconditioning. Accordingly, use of ratios of injury/ maximal injury may not accurately reflect neuroprotection from preconditioning. This is evident in the right hand images that show CP maximal levels are less than those of the sham controls.
Figure 2. Cold-preconditioning neuroprotection in hippocampal slice cultures. Cultures are incubated with Sytox Green, a fluorescent marked dead cell marker. The top row shows sham control cultures and the bottom row shows slices exposed to 28°C for 90 min. Left hand, pre-screen images show no CA1 injury. Relative injury color calibration scale is shown in the left upper image. Middle row images show relative slice culture injury 24 hours after exposure to 20 μM NMDA. Notice that sham control injury is greater than that of cultures exposed to cold-preconditioning (CP). Traditionally, cultures are then exposed to 20 μM NMDA overnight to maximize CA1 neuronal injury levels and relative injury of CP v. sham, noted as a ratio of injury/maximal injury. However, exposure to maximal injury stimuli may not be sufficient to overcome neuroprotection from preconditioning. Accordingly, use of ratios of injury/ maximal injury may not accurately reflect neuroprotection from preconditioning. This is evident in the right hand images that show CP maximal levels are less than those of the sham controls.
 Figure 3. Schematic illustrating the utility of using initial, first day measurements to quantitate injury levels in cold-preconditioning experiments. As noted above, we found that use of ratios (injury/maximal injury) could obscure neuroprotection from cold-pre-conditioning. This can be seen from the schematic to the left where sham injury is shown in red and that after cold-preconditioning in blue. One day after NMDA exposure cold-preconditioning shows a relative "3" level of injury v. sham ("5") control, consistent with 40% neuroprotection. However, if a traditional format using ratios (i.e., injury/maximal injury) is used, no protection is evident [i.e., (5/10)=50% for sham v. (3/6)=50% for cold-preconditioning].
Figure 3. Schematic illustrating the utility of using initial, first day measurements to quantitate injury levels in cold-preconditioning experiments. As noted above, we found that use of ratios (injury/maximal injury) could obscure neuroprotection from cold-pre-conditioning. This can be seen from the schematic to the left where sham injury is shown in red and that after cold-preconditioning in blue. One day after NMDA exposure cold-preconditioning shows a relative "3" level of injury v. sham ("5") control, consistent with 40% neuroprotection. However, if a traditional format using ratios (i.e., injury/maximal injury) is used, no protection is evident [i.e., (5/10)=50% for sham v. (3/6)=50% for cold-preconditioning].
 Figure 4. Typial qPCR result are shown. Upper curve RNA copy number v. threshold cycle for PCR amplification showing controls (blue) and experimental samples (red). Lower image shows typical amplification profiles for four samples (black, green, yellow and purple). Notice the latter Ct threshold cycles (marked by orange line) occur at 26.0, 29.5, 31.0, and 32.5.
Figure 4. Typial qPCR result are shown. Upper curve RNA copy number v. threshold cycle for PCR amplification showing controls (blue) and experimental samples (red). Lower image shows typical amplification profiles for four samples (black, green, yellow and purple). Notice the latter Ct threshold cycles (marked by orange line) occur at 26.0, 29.5, 31.0, and 32.5.
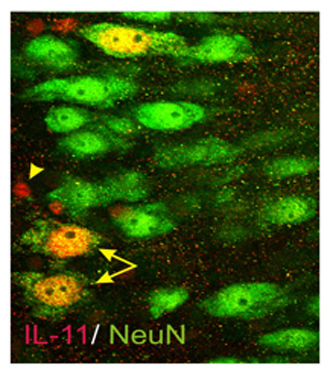 Figure 5. Double-label immunostaining used to confirm qPCR and qPCR array results. A slice culture was processed for IL-11 (red) and NeuN (green; to mark neurons) to probe for the cellular expression loci of IL-11. Notice that some pyramidal neurons (arrows) show IL-11 and NeuN staining (yellow) while a few smaller cells (arrows), presumed to be astrocytes, show only increased IL-11 staining (red).
Figure 5. Double-label immunostaining used to confirm qPCR and qPCR array results. A slice culture was processed for IL-11 (red) and NeuN (green; to mark neurons) to probe for the cellular expression loci of IL-11. Notice that some pyramidal neurons (arrows) show IL-11 and NeuN staining (yellow) while a few smaller cells (arrows), presumed to be astrocytes, show only increased IL-11 staining (red).
Discussion
Two fundamentally important concepts important to delineation of the cytokine signaling system involved in cold-preconditioning neuroprotection are illustrated in Figures 6 and 7. First, cytokines are extremely low concentration signaling molecules in normal brain. Nonetheless, physiological cytokine concentration changes have an immense potential to alter brain structure and function (i.e., phenotype) because of their ability to alter gene expression (Figure 6). Furthermore, cytokines are highly redundant and pleiotropic in that multiple cytokines can have similar effects and a single cytokine can have variable effects (Figure 7). Thus, to accurately establish the innate cytokine-bases for neuroprotection from cold-preconditioning (or other physiological preconditioning stimuli), composite analysis of related signaling variables must be determined. This is accomplished via multiplexed assay strategies. This will establish the cytokine "signature" of cold-preconditioning neuroprotection.
 Figure 6. Power of brain cytokine signaling. The illustrations convey the immense signaling power of physiological concentrations of brain cytokines compared to the concentrations of other well-recognized counterparts. Concentration is represented as the inverse of distance, beginning with a single cat whisker as the reference point. Sodium (10-1M illustrated by a grain of pepper) and potassium (10-3M illustrated by a tomato) are present in the interstitial brain space at levels of around 150 and 3 mM, respectively, and have well-recognized roles in neural cell electrophysiological function. Similarly, pH (i.e., ~10-7M levels of hydrogen ions and illustrated as 780 meters along Chicago's lakefront) and calcium (i.e., ~10-8M levels shown as 7.8 km seen from a satellite Google image along Chicago's lakefront from McCormick Place to Promontory Point in Hyde Park near the University of Chicago). In addition, neurotransmitters released to interstitial space over these concentrations affect local brain region activity. In contrast, cytokines (shown as the distance from Earth to Mars) can alter brain function at concentrations more than ten million times less.
Figure 6. Power of brain cytokine signaling. The illustrations convey the immense signaling power of physiological concentrations of brain cytokines compared to the concentrations of other well-recognized counterparts. Concentration is represented as the inverse of distance, beginning with a single cat whisker as the reference point. Sodium (10-1M illustrated by a grain of pepper) and potassium (10-3M illustrated by a tomato) are present in the interstitial brain space at levels of around 150 and 3 mM, respectively, and have well-recognized roles in neural cell electrophysiological function. Similarly, pH (i.e., ~10-7M levels of hydrogen ions and illustrated as 780 meters along Chicago's lakefront) and calcium (i.e., ~10-8M levels shown as 7.8 km seen from a satellite Google image along Chicago's lakefront from McCormick Place to Promontory Point in Hyde Park near the University of Chicago). In addition, neurotransmitters released to interstitial space over these concentrations affect local brain region activity. In contrast, cytokines (shown as the distance from Earth to Mars) can alter brain function at concentrations more than ten million times less.
 Figure 7. Interactive signaling of innate cytokine pathways. Cytokines are highly redundant and pleiotropic in that multiple cytokines can have similar effects and a single cytokine can have variable effects. Such diversity stems from complex interactive signaling that occurs at the level of ligands, receptors, and phosphoproteins. Further complexity stems from the fact that brain consists of different cell types, with each capable of cell-specific innate cytokine, receptor, and related phosphoprotein changes. For illustrative purposes here, only innate cytokine signaling pathways (derived from immune cell studies) are shown. For simplicity, the brain is drawn as a single cell (white line) showing the potential interactions for innate cytokines (IL-1α and IL-1β (referred to here as IL-1α/β), TNF-α, IL-6, IFN-γ and IL-10), receptors (IL-1R1, TNFR1, IL-6/gp130, IFNγR, and IL-10R), and phosphoproteins (i.e., kinases ERK1/2, P38 (P38-MAPK) and JNK) and transcription factors (ATF-2, NFκB, and STAT3). For example, TNF-α signals via TNFR1 to JNK, p38-MAPK and ERK1/2, which triggers gene expression via ATF-2. TNF-α also alters gene expression directly through NFκB activation. Together, activation of these transcription factors evoke increased (arrow) and decreased (blunt end) expression of cytokines and their receptors as indicated. Importantly, these pathways show that changes in one cytokine (e.g., TNF-α) influence production of other (e.g., IL-1β) cytokines. Thus, to establish accurately the cytokine-bases for neuroprotection from cold-preconditioning, composite analysis of related signaling variables must be determined. This will establish the innate cytokine "signature" of cold-preconditioning. (Image was compiled from data of reference #25.)
Figure 7. Interactive signaling of innate cytokine pathways. Cytokines are highly redundant and pleiotropic in that multiple cytokines can have similar effects and a single cytokine can have variable effects. Such diversity stems from complex interactive signaling that occurs at the level of ligands, receptors, and phosphoproteins. Further complexity stems from the fact that brain consists of different cell types, with each capable of cell-specific innate cytokine, receptor, and related phosphoprotein changes. For illustrative purposes here, only innate cytokine signaling pathways (derived from immune cell studies) are shown. For simplicity, the brain is drawn as a single cell (white line) showing the potential interactions for innate cytokines (IL-1α and IL-1β (referred to here as IL-1α/β), TNF-α, IL-6, IFN-γ and IL-10), receptors (IL-1R1, TNFR1, IL-6/gp130, IFNγR, and IL-10R), and phosphoproteins (i.e., kinases ERK1/2, P38 (P38-MAPK) and JNK) and transcription factors (ATF-2, NFκB, and STAT3). For example, TNF-α signals via TNFR1 to JNK, p38-MAPK and ERK1/2, which triggers gene expression via ATF-2. TNF-α also alters gene expression directly through NFκB activation. Together, activation of these transcription factors evoke increased (arrow) and decreased (blunt end) expression of cytokines and their receptors as indicated. Importantly, these pathways show that changes in one cytokine (e.g., TNF-α) influence production of other (e.g., IL-1β) cytokines. Thus, to establish accurately the cytokine-bases for neuroprotection from cold-preconditioning, composite analysis of related signaling variables must be determined. This will establish the innate cytokine "signature" of cold-preconditioning. (Image was compiled from data of reference #25.)
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was funded by grants from the National Institute of Neurological Disorders and Stroke (NS-19108), the Migraine Research Foundation, and the White Foundation to Richard P. Kraig. Ms. Marcia P. Kraig assisted in preparation of culture media and maintenance of slice cultures. We thank Yelena Grinberg for her comments and revisions on a final version of this article.
References
- Aptowicz CO, Kunkler PE, Kraig RP. Homeostatic plasticity in hippocampal slice cultures involves changes in voltage-gated Na+ channel expression. Brain Res. 2004;998:155–163. doi: 10.1016/j.brainres.2003.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie EC. Control of synaptic strength by glial TNFalpha. Science. 2002;295:2282–2285. doi: 10.1126/science.1067859. [DOI] [PubMed] [Google Scholar]
- Breder CD, Dewitt D, Kraig RP. Characterization of inducible cyclooxygenase in rat brain. J Comp Neurol. 1995;355:296–315. doi: 10.1002/cne.903550208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano AO, Breder CD, Kraig RP. Long-term elevation of cyclooxygenase-2, but not lipoxygenase, in regions synaptically distant from spreading depression. J Comp Neurol. 1996;376:447–462. doi: 10.1002/(SICI)1096-9861(19961216)376:3<447::AID-CNE7>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano AO, Kraig RP. Eicosanoids and nitric oxide influence induction of reactive gliosis from spreading depression in microglia but not astrocytes. J Comp Neurol. 1996;369:93–108. doi: 10.1002/(SICI)1096-9861(19960520)369:1<93::AID-CNE7>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano AO, Kraig RP. Prostaglandin E2 and 4-aminopyridine prevent the lipopolysaccharide-induced outwardly rectifying potassium current and interleukin-1beta production in cultured rat microglia. J Neurochem. 1998;70:2357–2368. doi: 10.1046/j.1471-4159.1998.70062357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano AO, Kraig RP. Prostaglandin E receptor subtypes in cultured rat microglia and their role in reducing lipopolysaccharide-induced interleukin-1beta production. J Neurochem. 1999;72:565–575. doi: 10.1046/j.1471-4159.1999.0720565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese EJ. Drug therapies for stroke and traumatic brain injury often display U-shaped dose responses: occurrence, mechanisms, and clinical implications. Crit Rev Toxicol. 2008;38:557–577. doi: 10.1080/10408440802014287. [DOI] [PubMed] [Google Scholar]
- Calabrese EJ. Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose-response framework. Toxicol Appl Pharmacol. 2007;222:122–128. doi: 10.1016/j.taap.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA. Hormesis: the dose-response revolution. Annu Rev Pharmacol Toxicol. 2003;43:175–197. doi: 10.1146/annurev.pharmtox.43.100901.140223. [DOI] [PubMed] [Google Scholar]
- Hansel NN. Analysis of CD4+ T-cell gene expression in allergic subjects using two different microarray platforms. Allergy. 2008;63:366–369. doi: 10.1111/j.1398-9995.2007.01540.x. [DOI] [PubMed] [Google Scholar]
- Hulse RE, Kunkler PE, Fedynyshyn JP, Kraig RP. Optimization of multiplexed bead-based cytokine immunoassays for rat serum and brain tissue. J Neurosci Methods. 2004;136:87–98. doi: 10.1016/j.jneumeth.2003.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse RE, Swenson WG, Kunkler PE, White DM, Kraig RP. Monomeric IgG is neuroprotective via enhancing microglial recycling endocytosis and TNF-alpha. J Neurosci. 2008;28:12199–12211. doi: 10.1523/JNEUROSCI.3856-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse RE, Winterfield J, Kunkler PE, Kraig RP. Astrocytic clasmatodendrosis in hippocampal organ culture. Glia. 2001;33:169–179. doi: 10.1002/1098-1136(200102)33:2<169::aid-glia1016>3.0.co;2-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraig RP. TNF-alpha and Microglial Hormetic Involvement in Neurological Health and Migraine. International Dose-Respose Society. 2010. [DOI] [PMC free article] [PubMed]
- Kunkler PE, Hulse RE, Kraig RP. Multiplexed cytokine protein expression profiles from spreading depression in hippocampal organotypic cultures. J Cereb Blood Flow Metab. 2004;24:829–839. doi: 10.1097/01.WCB.0000126566.34753.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkler PE, Hulse RE, Schmitt MW, Nicholson C, Kraig RP. Optical current source density analysis in hippocampal organotypic culture shows that spreading depression occurs with uniquely reversing currents. J Neurosci. 2005;25:3952–3961. doi: 10.1523/JNEUROSCI.0491-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkler PE, Kraig RP. Reactive astrocytosis from excitotoxic injury in hippocampal organ culture parallels that seen in vivo. J Cereb Blood Flow Metab. 1997;17:26–43. doi: 10.1097/00004647-199701000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkler PE, Kraig RP. Calcium waves precede electrophysiological changes of spreading depression in hippocampal organ cultures. J Neurosci. 1998;18:3416–3425. doi: 10.1523/JNEUROSCI.18-09-03416.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkler PE, Kraig RP. P/Q Ca2+ channel blockade stops spreading depression and related pyramidal neuronal Ca2+ rise in hippocampal organ culture. Hippocampus. 2004;14:356–367. doi: 10.1002/hipo.10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen KL. Microglia protect neurons against ischemia by synthesis of tumor necrosis factor. J Neurosci. 2009;29:1319–1330. doi: 10.1523/JNEUROSCI.5505-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK. Regulator of G-protein signaling 10 promotes dopaminergic neuron survival via regulation of the microglial inflammatory response. J Neurosci. 2008;28:8517–8528. doi: 10.1523/JNEUROSCI.1806-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Hormesis and disease resistance: activation of cellular stress response pathways. Hum Exp Toxicol. 2008;27:155–162. doi: 10.1177/0960327107083417. [DOI] [PubMed] [Google Scholar]
- Ning B. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen. 2008;13:194–201. doi: 10.1177/1087057108315513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim J, Feldman M. In: in Cytokine Reference. Oppenheim JJ, Feldman M, editors. Vol. 1. New York: Academic; 2001. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45–e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D, Beattie EC, Seo JY, Malenka RC. Differential regulation of AMPA receptor and GABA receptor trafficking by tumor necrosis factor-alpha. J Neurosci. 2005;25:3219–3228. doi: 10.1523/JNEUROSCI.4486-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-alpha. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- Streit WJ. Microglia as neuroprotective, immunocompetent cells of the CNS. Glia. 2002;40:133–139. doi: 10.1002/glia.10154. [DOI] [PubMed] [Google Scholar]
- Wilde GJ, Pringle AK, Sundstrom LE, Mann DA, Iannotti F. Attenuation and augmentation of ischaemia-related neuronal death by tumour necrosis factor-alpha in vitro. Eur J Neurosci. 2000;12:3863–3870. doi: 10.1046/j.1460-9568.2000.00273.x. [DOI] [PubMed] [Google Scholar]


