Abstract
The biological basis for the development of the cerebro-cerebellar structures required for posture and gait in humans is poorly understood. We investigated a large consanguineous family from Turkey exhibiting an extremely rare phenotype associated with quadrupedal locomotion, mental retardation, and cerebro-cerebellar hypoplasia, linked to a 7.1-Mb region of homozygosity on chromosome 17p13.1–13.3. Diffusion weighted imaging and fiber tractography of the patients' brains revealed morphological abnormalities in the cerebellum and corpus callosum, in particular atrophy of superior, middle, and inferior peduncles of the cerebellum. Structural magnetic resonance imaging showed additional morphometric abnormalities in several cortical areas, including the corpus callosum, precentral gyrus, and Brodmann areas BA6, BA44, and BA45. Targeted sequencing of the entire homozygous region in three affected individuals and two obligate carriers uncovered a private missense mutation, WDR81 p.P856L, which cosegregated with the condition in the extended family. The mutation lies in a highly conserved region of WDR81, flanked by an N-terminal BEACH domain and C-terminal WD40 beta-propeller domains. WDR81 is predicted to be a transmembrane protein. It is highly expressed in the cerebellum and corpus callosum, in particular in the Purkinje cell layer of the cerebellum. WDR81 represents the third gene, after VLDLR and CA8, implicated in quadrupedal locomotion in humans.
Developmental abnormalities of the cerebellum are a rare and genetically heterogeneous group of disorders characterized by loss of balance and coordination. Identification of the genes responsible for these disorders provides mechanistic insights into the regulation of neuronal development, differentiation, morphogenesis, migration, and organization (Fogel and Perlman 2007). These genes can be identified by exploiting targeted genomic sequencing in combination with linkage analysis and homozygosity mapping (Ropers 2007; Bilguvar et al. 2010). We applied this approach to the analysis of cerebellar hypoplasia and quadrupedal locomotion in an extended consanguineous family from southern Turkey.
Multiple families have been reported with cerebellar ataxia, mental retardation, and disequilibrium syndrome (CAMRQ) (Tan 2006; Turkmen et al. 2006, 2009; Moheb et al. 2008; Ozcelik et al. 2008; Kolb et al. 2010). All the reported CAMRQ families are consanguineous with recessive inheritance of their condition. Clinical characteristics vary slightly among the families. In four families from Turkey and Iran, the condition is due to homozygosity for mutations in the VLDLR gene encoding the very low density lipoprotein receptor (CAMRQ1 [MIM 224050]). Each of these four families harbors a different VLDLR mutation. In a fifth family, from Iraq, the condition is due to homozygosity for a missense mutation in the CA8 gene encoding carbonic anhydrase VIII (CAMRQ3 [MIM 613227]). In Family B, the first family described in the literature and also referred to as Uner Tan syndrome (Tan 2006), homozygosity mapping revealed a 7.1-Mb interval on chromosome 17p13, containing 192 genes and at least 20 pseudogenes, that segregates with the disease (CAMRQ2 [MIM 610185]) (Turkmen et al. 2006; Ozcelik et al. 2008). In order to identify the mutation responsible for CAMRQ2 in Family B, we targeted and fully sequenced the 7.1-Mb genomic interval and evaluated all variation in the region.
Results
Description of the affected family
Family B came to medical attention because of the unusual form of locomotion in five of the 19 siblings. A detailed clinical description, including video recordings and genetic mapping, was published elsewhere (Tan 2006; Turkmen et al. 2006; Ozcelik et al. 2008). Pedigree analysis suggested autosomal recessive inheritance. Linkage analysis and homozygosity mapping revealed a single locus on chromosome 17p between D17S1866 and D17S960. Illumina 300 Duo v2 BeadChip SNP genotype data of two of the affected individuals (05-984 and 05-987) revealed a single 6.8-Mb homozygous stretch between markers rs4617924–rs7338 (chr17: 114,669–6,917,703) and confirmed that chromosome 17p is the only region of interest (Supplemental Fig. 1).
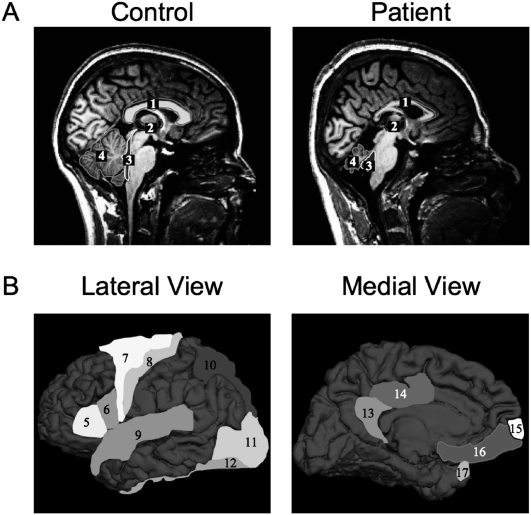
The phenotype was further characterized by magnetic resonance imaging (MRI) and morphometric analyses (Fig. 1). The most dramatic morphological differences were significant reductions in volume in the cerebellum and corpus callosum of the patient's brain (Fig. 1A). Both the cortex and the white matter of the cerebellum were significantly smaller in the patients. In contrast, the volume occupied by the caudate nucleus was significantly larger. Significant structural differences were also detected in the motor areas precentral gyrus and BA6 (increased mean curvature and gray matter volume) and motor speech areas pars opercularis and pars triangularis (increased cortical thickness and mean curvature) (Fig. 1B). A detailed account of the morphometric analyses is presented in Supplemental Figure 2 and Supplemental Table 1. Diffusion tensor imaging (DTI) and fiber tractography revealed moderate to high atrophy in superior, middle, and inferior cerebellar peduncles (Supplemental Fig. 3).
Figure 1.
MRI-based morphological analysis of brain from affected and unaffected individuals. (A) Midsagittal MRI scans of a healthy control individual (left) and affected relative from Family B (right). The highlighted regions show areas where volumetric differences are readily visible: corpus callosum (1), third ventricle (2), fourth ventricle (3), and cerebellum (4). (B) Cortical regions with significant differences in morphometric parameters are displayed on a reference cortex, from lateral and medial view: BA45 (5), BA44 (6), BA6 (7), precentral (8), superior temporal (9), superior parietal (10), lateral occipital (11), fusiform (12), isthmus cingulated (13), posterior cingulated (14), frontal pole (15), medial orbitofrontal (16), and temporal pole (17). Additional details are provided in Supplemental Figure 2 and Supplemental Table 1.
Targeted next-generation sequencing of the critical region
The critical region at chr17: 82,514–7,257,922 (hg19) was captured by NimbleGen 385K microarrays and sequenced with 454 Life Sciences (Roche) GS FLX in DNA of two of the affected individuals (05-985, 05-987) and two of the unaffected obligate carrier parents (05-981 father, 05-982 mother). An average of ∼400 Mb, yielding 46.3× haploid coverage, was sequenced from the captured DNA of each individual. An average of 79% of all reads from each sample mapped to the target region, representing 1275-fold to 2247-fold enrichment (Supplemental Table 2). On average, 99.4% of all targeted bases were covered by at least four reads (Supplemental Table 3).
In a parallel experiment, the same region from the DNA of another affected sibling (05-984) was captured with NimbleGen HD2 2.1M sequence capture microarrays and sequenced on an Illumina Genome Analyzer IIx. The captured region was enriched 123-fold, with 2.98 billion bases and 40.3 million reads obtained and 28% of reads mapped to the targeted region; 99.6% of targeted bases were covered by at least four reads. Combined sequence data for the three affected siblings yielded at least a fourfold coverage of 99.78% of all coding base pairs, 95.32% of intronic and UTR base pairs, and 91.36% of intergenic base pairs. The remaining 0.22% of coding regions with less than fourfold coverage was analyzed by Sanger sequencing (Supplemental Table 4).
With the 454 GS FLX platform, a total of 18,410 different variants were detected at high confidence (defined as in Hedges et al. 2009) in at least one sample (Supplemental Table 2). No additional functional variants were detected with the Illumina sequencing platform. Comparison of the sequence data from both platforms with Illumina 300 Duo v2 SNP genotype data indicated that the alleles were detected with sensitivity and specificity >99%. Heterozygous SNPs detected at the borders of the homozygous blocks of the affected individuals narrowed the region of homozygosity to 6.74 Mb (Supplemental Table 5). The Mendelian error rate, an indicator of call errors (Hedges et al. 2009), was calculated as 0.3%.
Of the 18,410 high-confidence variants, 17,281 were reported by dbSNP. For each nonsynonymous SNP compatible with the Mendelian transmission of the disease allele, the frequencies of homozygotes for each allele were accessed from public databases. With one exception, homozygosity at both alleles had been reported in control populations. The one exception, rs55916885, was at a nonconserved site and was predicted as tolerated by SIFT (Ng and Henikoff 2001) and Polyphen-2 (Sunyaev et al. 2001). Based on these observations, all previously reported nonsynonymous variants were excluded (Supplemental Table 6).
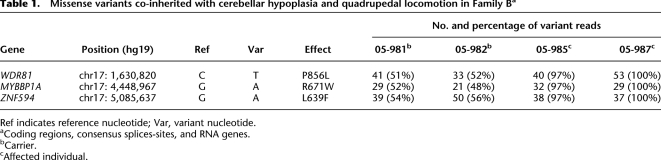
Of the 18,410 high-confidence variants, 1119 variants were both novel vis-á-vis dbSNP132 and present in both the affected siblings and their obligate carrier parents. These 1119 novel shared variants were classified by genomic context: coding sequence or flanking splice junctions (n = 20), 5′ UTR or 3′ UTR (n = 15), intronic (n = 689), or intergenic (n = 395). The 20 variants in the coding sequence or flanking splice junctions were genotyped in the family to evaluate cosegregation with the phenotype (Supplemental Table 7). Genotypes of three missense variants were consistent with the recessive inheritance of the disease allele in Family B: WDR81 p.P856L, MYBBP1A p.R671W, and ZNF594 p.L639F (Table 1). Of the 15 5′/3′ UTR variants, five cosegregated with the disease phenotype. Therefore, they were carried to a more detailed analysis, including evaluation of the protein interactions. None was found to interact with previously identified genes with cerebellar phenotypes, including CAMRQ-associated VLDLR and CA8 (Supplemental Table 8).
Table 1.
Missense variants co-inherited with cerebellar hypoplasia and quadrupedal locomotion in Family Ba
Identification of disease causing variant
MYBBP1A p.R671W could be excluded as the causal mutation for the disorder of Family B based on the genotypes of controls (Supplemental Table 9). In 214 unrelated healthy controls (428 chromosomes), 50 of whom were sampled from the same region of Turkey as Family B, 13 individuals were heterozygous for MYBBP1A p.R671W. This carrier frequency yields an allele frequency of 0.016 and an expected frequency of homozygotes of about one in 4000, far higher than the frequency of CAMRQ2, which occurs in only one extended family. In a second, independent series of 400 individuals of various European and Middle Eastern ancestries, MYBBP1A was fully sequenced in the context of whole-exome sequencing. Of these 400 individuals, two were homozygous for MYBBP1A p.R671W. Neither of these two homozygotes had any signs consistent with CAMRQ2. MYBBP1A p.R671W was therefore excluded as the allele responsible for the disorder of Family B.
ZNF594 p.L639F could be excluded as the causal mutation for the disorder based on conservation considerations. Residue 639 of ZNF594 is not well conserved: Two of 16 species sequenced have phenylalanine (F) at the orthologous site, strongly suggesting that phenylalanine at this site would also not be damaging in humans. A negative GERP score (−0.665) for the mutated nucleotide indicates that this site is probably evolving neutrally (Davydov et al. 2010). The variant is predicted as “benign” (PSIC score difference, 0.301) by PolyPhen (Sunyaev et al. 2001) and “damaging low confidence” (SIFT score, 0.04) by SIFT (Supplemental Table 10; Ng and Henikoff 2001). In addition, the human ZNF594 gene harbors polymorphic nonsense mutations at sites near the missense at L639F. ZNF594 p.E684X appeared in four of 118 Yoruban controls (rs114754534; allele frequency, 0.034), and ZNF594 p.Q681X appeared in one of 120 CEU controls (rs116878311; allele frequency, 0.0083) in the HapMap series.
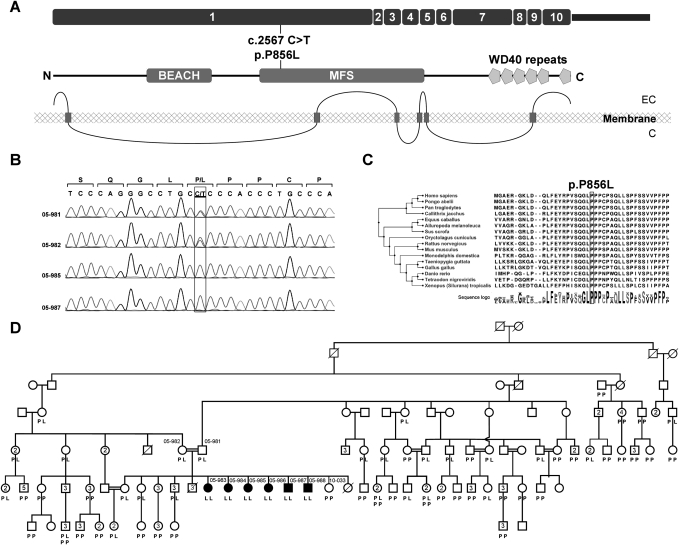
In contrast, WDR81 p.P856L (Fig. 2A,B; Supplemental Fig. 4) is both rare and alters a highly conserved site. This missense did not appear in any of the 549 individuals of the control series. WDR81 is a highly conserved protein throughout vertebrates, with no polymorphic stops in any sequenced species. In particular, proline at residue 856 is completely conserved in all known sequences of the WDR81 protein (Fig. 2C).
Figure 2.
Identification of the WDR81 mutation. (A) Genomic structure, predicted protein, and predicted transmembrane domains of WDR81 gene [(EC) extracellular, (C) cytosol]. (B) Confirmation of the missense mutation c.2567C>T (p.P856L) in WDR81 isoform 1 by Sanger sequencing. (C) Sequence homology of WDR81 protein in vertebrates. The box indicates the mutant amino acid. (D) Family B with affected individuals indicated by filled symbols and genotypes shown for WDR81 p.P856L.
The extended genealogy of Family B revealed consanguinity in several branches of the kindred (Fig. 2D), whose ancestors have migrated from a village on the Syrian side of the border with Hatay, Turkey, in the early 1950s. Approximately 240 individuals spanning seven generation could be ascertained. WDR81 p.P856L was genotyped in 177 members of the kindred spanning five generations. A single union of heterozygous carriers, 05-981 × 05-982, was observed whose children include the affected individuals of this study. None of the 172 unaffected individuals in the kindred is homozygous for WDR81 p.P856L. Genetic counseling is in progress for the 27 members of the family who are heterozygous carriers of the mutation. The status of WDR81 was evaluated in two different cohorts of the patients with neurodevelopmental/cerebellar phenotypes for whom the underlying genetic cause is still unknown. The first cohort consisted of 750 patients with structural cortical malformations or degenerative neurological disorders. By using the whole-genome genotyping data based on Illumina Human 370 Duo or 610K Quad BeadChips, we did not identify any patient with a cerebellar phenotype or ataxia phenotype to harbor a homozygous interval (≥2.5 cM) surrounding the WDR81 locus. Exome sequencing of the same group did not reveal any mutations, including compound heterozygous substitutions. In the second cohort of 58 probands, 12 had cerebellar hypoplasia with or without quadrupedal gait. No additional mutations in WDR81 were identified by Sanger sequencing of the entire coding regions.
Characterization of WDR81
WDR81 p.P856L at chr17: 1,630,820 (hg19) lies in exon 1 of WDR81 isoform 1 (ENST00000409644, NM_001163809.1, NP_001157281.1), the longest isoform of WDR81, containing 10 exons and encoding 1941 amino acids (Fig. 2A). Proline at this site was present in all species analyzed (Fig. 2C), including the most distantly related sequenced ortholog, the Tetraodon nigroviridis WDR81 protein, which is 47.8% identical and 57.2% similar and has a distance score of 0.76 compared with the human protein. WDR81 p.P856L was predicted to be “damaging” (SIFT score, 0) by SIFT (Ng and Henikoff 2001), “probably damaging” (PSIC score difference, 2.724) by PolyPhen (Sunyaev et al. 2001), and “under evolutionary constraint” (GERP score, 5.68) by GERP (Davydov et al. 2010).
The function of WDR81 is unknown, but clues can be derived from its structure. The conserved region of WDR81 that includes P856 is flanked on the N-terminal side by a BEACH (Beige and Chediak-Higashi) domain at amino acids 352–607. BEACH proteins have been implicated in membrane trafficking (Wang et al. 2000), synapse morphogenesis (Khodosh et al. 2006), and lysosomal axon transport (Lim and Kraut 2009). A BEACH domain is the major structural feature of neurobeachin, a scaffolding protein disrupted in a patient with autism (Volders et al. 2011). WDR81 p.P856L lies in a major facilitator superfamily (MFS) domain, a region characteristic of solute carrier transport proteins (Saier et al. 1999). The C terminus of WDR81 is composed of six WD-repeats that are likely constituents of a beta-propeller. Based on analysis by TMpred (www.ch.embnet.org/software/TMPRED_form.html), WDR81 is a transmembrane protein with six membrane-spanning domains, the most N-terminal at amino acids 45–66 and the other five at the C terminus of the protein, between amino acids 980 and 1815 (Fig. 2A). Supporting the likelihood that WDR81 is a transmembrane protein is the observation that WDR81 transcript expression is increased in membrane-associated RNA in contrast to cytoplasmic RNA (4.14 folds, P = 0.03, and 1.78 folds, P = 0.0002 in Gene Expression Omnibus [GEO] [http://www.ncbi.nlm.nih.gov/geo/] data set GSE4175) (Diehn et al. 2006).
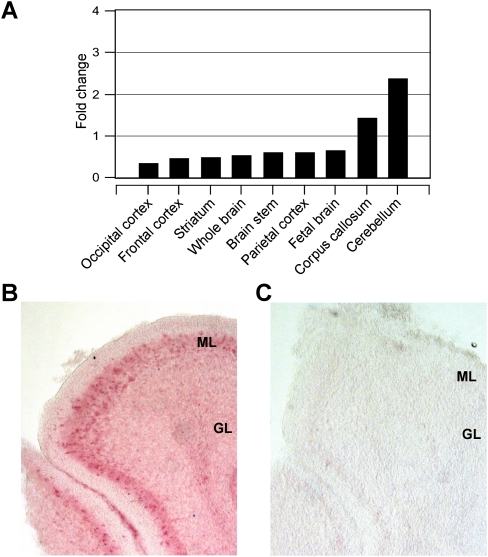
In order to assess a possible role for WDR81 in regulating motor behavior, we evaluated the expression profiles of human and mouse WDR81/Wdr81 isoform 1 in the brain. Human WDR81 isoform 1 transcript was expressed in all the tissues evaluated (Supplemental Fig. 5). In particular, all the brain tissues were positive for the transcript, with highest levels of expression in the cerebellum and corpus callosum (Fig. 3A). In the mouse brain at post-partum day P7, Wdr81 expression was observed in Purkinje cell layer in the cerebellum (Fig. 3B,C). The cerebellum is a crucial regulatory center for motor function.
Figure 3.
Expression pattern of WDR81 in brain. (A) Expression in human brain with highest levels in cerebellum and corpus callosum. (B) In situ hybridization of mouse embryonic brain revealing increased expression of Wdr81 in purkinje cells and molecular layer of cerebellum. (C) No hybridization was observed with the sense probe. (ML) Molecular layer, (GL) granular layer.
We examined the expression of WDR81 in the context of expression profiles of the early embryonic mouse brain (GSE8091) (Hartl et al. 2008). Differentially expressed genes within the day groups were filtered (one-way ANOVA test Bonferroni-corrected P < 0.001, n = 3611). From these profiles, we identified the subset of genes whose expression was highly correlated with that of WDR81 (R > 0.95, n = 670) and then used DAVID tools (Huang et al. 2009) to evaluate the predicted functions of this subset of genes. The subset of genes coexpressed with WDR81 was enriched for those involved in neuronal differentiation and neuronal projection, axonogenesis, and cell morphogenesis (Bonferroni-corrected P-values 2.3 × 10−11, 1.3 × 10−9, and 3.7 × 10−9, respectively). Among the genes coexpressed with WDR81 were those encoding prion protein, doublecortin (responsible for lissencephaly), and L1CAM (responsible for MASA syndrome) (Supplemental Table 11). WDR81 is not coexpressed with VLDLR and CA8, raising the possibility that WDR81 represents a different developmental regulatory pathway.
Discussion
The identification of genes responsible for human disease has been greatly facilitated with new technologies, particularly the targeted enrichment of the genome by solution capture, followed by genomic sequencing (Bilguvar et al. 2010). Despite these advances, demonstrating the causality for a mutation in the absence of two or more independent cases remains a challenge. This is particularly true when multiple variants, none of them with obvious effect on protein function, cosegregate with the phenotype in the family; the candidate gene encodes a previously uncharacterized protein with multiple isoforms, of which the critical mutation is on only one; and the candidate mutation is a missense. However, unique families and uncharacterized proteins exist, and precisely because of this reason, it becomes imperative to fully exploit genetics and genomics approaches to distinguish the causative mutation.
We describe here the discovery of a mutation associated with an extremely rare and genetically heterogeneous autosomal recessive phenotype in a unique consanguineous family (Tan 2006). The putative causative mutation could be distinguished from previously unknown rare polymorphisms in the same genomic region by analysis of conservation at all candidate variant sites, by the presence of polymorphic stops in the critical region of another candidate gene, and by genotyping ethnically matched unaffected individuals who would not be expected to carry homozygous mutations at the mutant site. We conclude that the WDR81 p.P856L mutation is the cause of cerebellar hypoplasia associated with quadrupedal locomotion in Family B.
WDR81 is an uncharacterized gene. It shows similarity with a host of genes, including NSMAF (neutral sphyngomyelinase activation associated factor), NBEA (neurobeachin), and LYST (lysosomal trafficking regulator). The LYST gene contains HEAT/ARM repeats, a BEACH domain, and seven WD40 repeats (Ward et al. 2000). Nearly all reported LYST mutations result in protein truncation and lead to Chediak-Higashi syndrome (CHS), which is characterized by accumulation of giant intracellular vesicles leading to defects in the immune and blood systems (Rudelius et al. 2006). Two patients with missense LYST mutations have been reported (Karim et al. 2002). Interestingly, these patients presented with neurological symptoms without immunological involvement. The LystIng3618/LystIng3618 mutant mouse harbors a missense mutation in the WD40 domain. Purkinje cell degeneration accompanied by age-dependent impairment of motor coordination without signs of lysosomal deficiency in immunological organs were characteristics of these animals (Rudelius et al. 2006).
Expression of WDR81 at high levels in the human cerebellum and corpus callosum and in the Purkinje cell layer of the mouse cerebellum is consistent with our observations of major structural abnormalities in these regions of the brain of affected individuals. Together, these observations suggest a possible role for WDR81 in motor behavior. Further work will be required to understand the normal biological function of WDR81 and the role of the mutation in causing cerebellar hypoplasia and quadrupedal locomotion. Genomic analysis of Family B demonstrates that WDR81 is highly likely to be critical to these developmental processes.
Methods
Human subjects
The institutional review boards of Bilkent, Hacettepe, Baskent, and Cukurova Universities approved the study (decisions: BEK02, 28.08.2008; TBK08/4, 22.04.2008; KA07/47, 02.04.2007; and 21/3, 08.11.2005, respectively). Written informed consent, prepared according to the guidelines of the Ministry of Health in Turkey, was obtained from all family members and control group subjects prior to the study. A total of 18 subjects participated in MRI scans. Six of them were from Family B, including four affected siblings (05-984, 05-986, 05-987, 05-988), one normal female sibling homozygous for the wild-type allele of the WDR81 p.P856L variant (10-033), and their carrier father (05-981). The remaining 14 participants were age- and sex-matched healthy controls. The two male patients (age, mean ± SD = 37.00 ± 4.24) were matched to seven male controls (age, mean ± SD = 35.14 ± 5.76), and the two female patients (age, mean ± SD=27.00 ± 4.24) were matched to seven female controls (age, mean ± SD = 28.57 ± 3.64). Family B members were scanned under sedation. For the healthy controls, no sedation was performed. Sedation was achieved by initial administration of midazolam (2 mg per subject), which was followed by propofol (120 mg) and fentanyl (50 mcg) administration intravenously. Hypnosis level was adjusted by 20 mg injections of propofol approximately every 10 min to eliminate somatic responses such as slight movements. Blood oxygen level and heart rate were monitored during the entire procedure. Eyelash reflexes were absent at all times. Neuromuscular blockade was not used.
Next-generation sequencing
NimbleGen 385K microarrays were produced to capture the critical region at chr17: 82,514–7,257,922 (hg19) using 7464 unique probes with a total probe length of 4,853,455 bp. Sequence Search and Alignment by Hashing Algorithm (SSAHA) (Ning et al. 2001) was used to determine probe uniqueness by NimbleGen (Roche NimbleGen). Sequence capture was conducted by the NimbleGen facility using 25 μg of input DNA. Captured DNA samples were subjected to standard sample preparation procedures for 454 GS FLX sequencing with Titanium series reagents. Four full 454 GS FLX runs were conducted for two affected individuals (05-985, 05-987) and their unaffected obligate carrier parents (05-981 father, 05-982 mother). Sequence data were initially mapped to human genome reference sequence and annotated using the GSMapper software package (Roche). Fold enrichment of the target region was calculated with the formula ∑REMTrm/STrm: ∑RMG/SG as described previously (REMTrm, number of reads mapped to target region; STrm, size of target region; RMG, number of reads mapped outside of the target region; SG, size of human genome) (Rehman et al. 2010). Variants were identified with ALLDiff and more stringent HCDiff approaches (Hedges et al. 2009). Annotation of variants was made by GSMapper software using the refGene table of the University of California, Santa Cruz (UCSC) Genome Browser (Fujita et al. 2010). Ensembl 62 genome annotation data for hg19 human genome assembly were extracted using the BIOMART data-mining tool for further analysis of intronic and intergenic variants in terms of hypothetical genes and splicing variants (Flicek et al. 2011). Novel variants were reported based on the SNPs included in the reference SNP database. For Illumina sequencing, a total of 6,184,539-bp-long unique probes were designed to target a 9-Mb genomic region spanning the disease locus (chr17:0–9,059,276; hg19) using a custom NimbleGen HD2 2.1M sequence capture microarray. Another affected individual was sequenced with the Illumina Genome Analyzer IIx. Illumina sequence data were mapped to the reference genome using MAQ tools (Li et al. 2008), and single nucleotide variants were determined with Samtools (Li et al. 2009). To determine indels, data were mapped with BWA (Li and Durbin 2010) and analyzed with Samtools. Sequence data were visually analyzed using the Integrative Genomics Viewer (IGV) (Robinson et al. 2011).
Array based genotyping
We conducted Illumina 300 Duo v2 BeadChip for two affected individuals (05-984, 05-987) according to the manufacturer's recommendations (Illumina). The image data were normalized, and the genotypes were called using data analysis software (Bead Studio, Illumina). Sex, inbreeding, and sibship were confirmed. The Mendelian compatibility of sequence variants was analyzed with PLINK (Purcell et al. 2007).
DNA sequencing
Confirmation of novel variants identified by next-generation sequencing was done with conventional capillary sequencing. The Primer3 software (Rozen and Skaletsky 2000) was used to design PCR primers for the amplification of candidate variants (Supplemental Table 12). Products were analyzed via gel electrophoresis and were sequenced using forward and reverse primers on an ABI 3130 XL capillary sequencing instrument (Applied Biosystems). Sanger sequence trace files were analyzed with the CLCBio Main Workbench software package (CLCBio Inc.).
Population screening
To distinguish the disease-causing variant from novel polymorphisms, a population screening approach was conducted for each candidate variant. Allele-specific PCR (AS-PCR) and restriction fragment length polymorphism (RFLP) analyses were performed (Supplemental Table 12) on 1098 chromosomes from a healthy control population. In addition, the first-, second-, and third-degree relatives of the affected family, amounting to 177 individuals, were sampled for genotype analysis. Sanger sequencing was performed to confirm all of the variants detected in the normal population using the above-mentioned methods. Racial distribution of the control group was 100% Caucasian, including 22% from southeastern Turkey.
Quantitative real-time RT-PCR analysis of WDR81 expression
First-strand cDNA was prepared from multi-tissue RNA panels (Clontech: 636567, 636643; Agilent: 540007, 540117, 540137, 540157, 540053, 540005, 540143, 540135) with RevertAid kit and random hexamer primers (Fermentas; K1622) after DNase I (Fermentas; EN0521) digestion. The PCR primers located in exon 1 and flanking the mutation site were designed using Primer3 software (Supplemental Table 13; Rozen and Skaletsky 2000). SYBR Green real-time PCR were realized according to standard protocols (BioRad; 170-8882) with 100% PCR efficiency. Each assay included minus RT and nontemplate controls. Ct values were normalized to GAPDH as an internal control. The data were analyzed using the Pfaffl method (Pfaffl 2001).
In situ hybridization
In order to examine the specific expression pattern of Wdr81 gene in the mouse brain, probes that contain the mutated region in human patients were prepared by PCR amplification of the region from mouse genomic DNA and subsequent cloning into plasmids. The riboprobes were synthesized by using Dig-labeled NTPs, and in situ hybridization experiments were performed as described (Tekinay et al. 2009). The Animal Ethics Committee of Bilkent University approved procedures for the tissue extraction and for in situ hybridization tests. Animals were group housed in a 12-h dark, 12-h light cycle. Embryo and P7 brain sections were prepared as described (Gong et al. 2003). Twenty-micrometer sagittal sections were taken with a cryostat (Leica). The antisense probe was prepared by PCR amplification from the mouse genomic DNA and subsequent cloning into pCR4-TOPO vector (Invitrogen). A modified version of pSK vector was used for cloning the sense probe of the same region. Digoxigenin (Dig)-labeled riboprobe was transcribed using Dig-NTP in the transcription reaction. Riboprobes were purified with Mini Quick Spin DNA columns (Roche) prior to hybridization. Sections were incubated at 60°C overnight in hybridization buffer containing 50% formamide, 5× SSC, 5× Denhardt's reagent, 50 μg/mL heparin, 500 μg/mL herring sperm DNA, and 250 μg/mL yeast tRNA. Hybridized sections were washed for 90 min with 50% formamide and 2× SSC at 60°C. Probes were detected with anti-Dig Fab fragments conjugated to alkaline phosphatase and NBT/BCIP substrate mixture (Tekinay et al. 2009).
Bioinformatics analyses
Homozygosity mapping analysis was performed using HomozygosityMapper software (Seelow et al. 2009). SIFT (Ng and Henikoff 2001) and PolyPhen (Sunyaev et al. 2001) tools were used to predict the functional impact of the variants. Genomic Evolutionary Rate Profiling (GERP) scores for each variant were obtained from the UCSC Genome Browser allHg19RS_BW track (Davydov et al. 2010). The PFAM protein domain search module of CLCMain Workbench V5.0 (CLCBio, Inc.) and ScanProsite (Gattiker et al. 2002) tools were used to predict domains and possible effects of the variant on protein product. Membrane spanning domains were predicted using TMpred software (www.ch.embnet.org/software/TMPRED_form.html). Homology searches were performed with CLCMain Workbench using appropriate modules (reference sequence accession codes for WDR81 orthologs are Ailuropoda melanoleuca, XP_002918082; Callithrix jacchus, XP_002747874; Danio rerio, XP_001921778; Equus caballus, XP_001502383; Gallus gallus, XP_415806; Monodelphis domestica, XP_001371487; Mus musculus, NP_620400; Oryctolagus cuniculus, XP_002718930; Pan troglodytes, XP_523527; Pongo abelii, XP_002826860; Rattus norvegicus, NP_001127832; Sus scrofa, XP_003131868; Taeniopygia guttata, XP_002194363; Tetraodon nigroviridis, CAG08933; Xenopus [Silurana] tropicalis, XP_002937192). Published microarray data sets of E9.5, E11.5, and E13.5 mouse brain tissue (GSE8091) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi) (Hartl et al. 2008) and processed with GeneSpring GX V11.1 software (Agilent Technologies). Data sets were grouped within day groups, and standard quality control and filtering analysis were performed (http://www.chem.agilent.com/cag/bsp/products/gsgx/manuals/GeneSpring-manual.pdf). Differentially expressed genes within the day groups were filtered using a one-way ANOVA test (Bonferroni-corrected P < 0.001). Genes that correlated with Wdr81 (R = 0.95 − 1.0) were obtained using the “Find Similar Entity Lists” module of the software. Functional annotation clustering was performed using the obtained gene list by DAVID tools (Huang et al. 2009). WDR81 differential expression in the GEO data sets was further investigated using the NextBio System, a web-based data-mining engine (Kupershmidt et al. 2010), and the GSE4175 (Diehn et al. 2006) data set was selected as a significant difference in membrane-associated RNA versus cytoplasmic RNA comparisons. Ensembl identifiers of the candidate genes and transcripts are as follows: WDR81 [ENSG00000167716; ENST00000409644], MYBBP1A [ENSG00000132382; ENST00000254718], and ZNF594 [ENSG00000180626; ENST00000399604].
MRI data acquisition and structural analysis procedures
MRI data were acquired using a three Tesla scanner (Magnetom Trio, Siemens AG) with a 12-channel phase-array head coil. A high-resolution T1-weighted three-dimensional (3D) anatomical-volume scan was acquired for each participant (single-shot turbo flash; voxel size = 1 × 1 × 1 mm3; repetition time [TR] = 2600 msec; echo time[TE] = 3.02 msec; flip angle = 8°; field of view [FOV] = 256 × 224 mm2; slice orientation = sagittal; phase encode direction = anterior-posterior; number of slices = 176; acceleration factor [GRAPPA] = 2). DTI data were acquired using a single-shot spin-echo EPI with a parallel imaging technique GRAPPA (acceleration factor 2). The sequence was performed with 30 gradient directions, and the diffusion weighting b-factor was set to 800 sec/mm2 (TR, 6400 msec; TE, 88 msec; in-plane resolution, 1 mm × 1 mm; slice thickness, 3.0 mm; 50 transverse slices; base resolution, 128 × 128). Structural analyses were performed with the Freesurfer image analysis package (http://surfer.nmr.mgh.harvard.edu/). The analyses involved intensity normalization, removal of nonbrain tissue, subcortical segmentation (Fischl et al. 2002), and identification of the white matter/gray matter boundary upon which cortical reconstruction and volumetric parcellation were performed. The cortex was then registered to a spherical atlas and parceled into units according to the gyral and sulcal structure based on the Desikan-Kilinay Atlas (Desikan et al. 2006) and the Destrieux Atlas (Destrieux et al. 2010). Next, using the same software, we performed morphometric analyses of cortical thickness, mean curvature, surface area, and volume for each unit of parcellation and computed the group differences. Significant differences between the groups are determined using two-tailed unpaired t-tests at an alpha level of 0.05. Fiber tracking was performed in MedINRIA (Toussaint et al. 2007). Fibers with FA < 0.3 were excluded from the analysis. Region of interests (ROIs) were drawn manually over cross-sections of superior, middle, and inferior cerebellar peduncles, using the MRI Atlas of Human White Matter as a reference (Oishi et al. 2010). ROIs were drawn at approximately corresponding locations for the patients and healthy controls. Fiber tracts were first limited to pass through these ROIs and were then subsequently refined using a recursive tracking technique (Toussaint et al. 2007). T1-weighted images were coregistered with DWI data using FSL (Smith et al. 2004; Woolrich et al. 2009). Final tracts were manually overlaid onto high-resolution T1-weighted images for illustration purposes.
Data access
Sequence data of the homozygous region has been deposited at the DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp/) under accession no. DRA000432. SNP genotype data have been deposited at the European Genome-Phenome Archive (EGA; http://www.ebi.ac.uk/ega/), which is hosted at the EBI, under accession no. EGAS00000000099.
Acknowledgments
We thank Dr. Mary-Claire King for innumerable discussions, suggestions, and critical reading of the manuscript. We also thank the members of Family B and their relatives for cooperation in this study. Dr. Alper Iseri and Dr. Bayram Kerkez kindly provided technical and logistic support. This work was supported by the Scientific and Technological Research Council of Turkey (TUBITAK-SBAG 108S036 and 108S355) and the Turkish Academy of Sciences (TUBA research support) to T.O., and the European Commission (PIRG-GA-2008-239467) and TUBA-GEBIP award to H.B.
Authors' contributions: S.G., A.B.T., K.D., H.B., and T.O. conceived and designed the experiments. S.G., H.U., K.D., and H.B. performed the experiments. S.G., A.B.T., K.D., H.B., K.B., H.U., A.O., E.A., T.K., M.G., and T.O. analyzed the data. O.E.O., A.N.B., H.T., M.T., and U.T. contributed patient materials. S.G. and T.O. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.126110.111.
References
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. 2010. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6: e1001025 doi: 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31: 968–980 [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E 2010. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 53: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Bhattacharya R, Botstein D, Brown PO 2006. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet 2: e11 doi: 10.1371/journal.pgen.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. 2002. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33: 341–355 [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2011. Ensembl 2011. Nucleic Acids Res 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Perlman S 2007. Clinical features and molecular genetics of autosomal recessive cerebellar ataxias. Lancet Neurol 6: 245–257 [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D 2010. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinformatics 1: 107–108 [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925 [DOI] [PubMed] [Google Scholar]
- Hartl D, Irmler M, Romer I, Mader MT, Mao L, Zabel C, de Angelis MH, Beckers J, Klose J 2008. Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics 8: 1257–1265 [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Burges D, Powell E, Almonte C, Huang J, Young S, Boese B, Schmidt M, Pericak-Vance MA, Martin E, et al. 2009. Exome sequencing of a multigenerational human pedigree. PLoS ONE 4: e8232 doi: 10.1371/journal.pone.0008232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Karim MA, Suzuki K, Fukai K, Oh J, Nagle DL, Moore KJ, Barbosa E, Falik-Borenstein T, Filipovich A, Ischida Y, et al. 2002. Apparent genotype–phenotype correlation in childhood, adolescent, and adult Chediak–Higashi syndrome. Am J Med Genet 108: 16–22 [PubMed] [Google Scholar]
- Khodosh R, Augsburger A, Schwarz TL, Garrity PA 2006. Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133: 4655–4665 [DOI] [PubMed] [Google Scholar]
- Kolb LE, Arlier Z, Yalcinkaya C, Ozturk AK, Moliterno JA, Erturk O, Bayrakli F, Korkmaz B, DiLuna ML, Yasuno K, et al. 2010. Novel VLDLR microdeletion identified in two Turkish siblings with pachygyria and pontocerebellar atrophy. Neurogenetics 11: 319–325 [DOI] [PubMed] [Google Scholar]
- Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, et al. 2010. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS ONE 5: e13066 doi: 10.1371/journal.pone.0013066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A, Kraut R 2009. The Drosophila BEACH family protein, blue cheese, links lysosomal axon transport with motor neuron degeneration. J Neurosci 29: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moheb LA, Tzschach A, Garshasbi M, Kahrizi K, Darvish H, Heshmati Y, Kordi A, Najmabadi H, Ropers HH, Kuss AW 2008. Identification of a nonsense mutation in the very low-density lipoprotein receptor gene (VLDLR) in an Iranian family with dysequilibrium syndrome. Eur J Hum Genet 16: 270–273 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S 2001. Predicting deleterious amino acid substitutions. Genome Res 11: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox A, Mullikin J 2001. SSAHA: A fast search method for large DNA databases. Genome Res 11: 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria AV, van Zijl PCM, Mori S 2010. MRI atlas of human white matter, 2nd ed Elsevier, Amsterdam [Google Scholar]
- Ozcelik T, Akarsu N, Uz E, Caglayan S, Gulsuner S, Onat OE, Tan M, Tan U 2008. Mutations in the very low-density lipoprotein receptor VLDLR cause cerebellar hypoplasia and quadrupedal locomotion in humans. Proc Natl Acad Sci 105: 4232–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Riazuddin S, et al. 2010. Targeted capture and next-generation sequencing identifies C9orf75, encoding Taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet 86: 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH 2007. New perspectives for the elucidation of the genetic disorders. Am J Hum Genet 81: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ 2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: Methods in molecular biology (ed. Krawetz S, Misener S), p. 365 Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Rudelius M, Osanger A, Kohlmann S, Augustin M, Piontek G, Heinzmann U, Jennen G, Russ A, Matiasek K, Stumm G, et al. 2006. A missense mutation in the WD40 domain of murine Lyst is linked to severe progressive Purkinje cell degeneration. Acta Neuropathol 112: 267–276 [DOI] [PubMed] [Google Scholar]
- Saier MH Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jähn PS, Lew K, Liu J, et al. 1999. The major facilitator superfamily. J Mol Microbiol Biotechnol 1: 257–279 [PubMed] [Google Scholar]
- Seelow D, Schuelke M, Hildebrandt F, Nürnberg P 2009. HomozygosityMapper: an interactive approach to homozygosity mapping. Nucleic Acids Res 37: W593–W599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, et al. 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23: S208–S219 [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W 3rd, Kondrashov AS, Bork P 2001. Prediction of deleterious human alleles. Hum Mol Genet 10: 591–597 [DOI] [PubMed] [Google Scholar]
- Tan U 2006. A new syndrome with quadrupedal gait, primitive speech, and severe mental retardation as a live model for human evolution. Int J Neurosci 116: 361–369 [DOI] [PubMed] [Google Scholar]
- Tekinay AB, Nong Y, Miwa JM, Lieberam I, Ibanez-Tallon I, Greengard P, Heintz N 2009. A role for LYNX2 in anxiety-related behavior. Proc Natl Acad Sci 106: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint N, Souplet JC, Fillard P 2007. MedINRIA: Medical image navigation and research tool by INRIA. In Proceedings of MICCAI'07 Workshop on Interaction in Medical Image Analysis and Visualization. Brisbane, Australia Lecture Notes in Computer Science, Vol. 4791. Springer, Berlin. [Google Scholar]
- Turkmen S, Demirhan O, Hoffmann K, Diers A, Zimmer C, Sperling K, Mundlos S 2006. Cerebellar hypoplasia and quadrupedal locomotion in humans as a recessive trait mapping to chromosome 17p. J Med Genet 43: 461–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkmen S, Guo G, Garshasbi M, Hoffmann K, Alshalah AJ, Mischung C, Kuss A, Humphrey N, Mundlos S, Robinson PN 2009. CA8 mutations cause a novel syndrome characterized by ataxia and mild mental retardation with predisposition to quadrupedal gait. PLoS Genet 5: e1000487 doi: 10.1371/journal.pgen.1000487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herberg FW, Laue MM, Wullner C, Hu B, Petrasch-Parwez E, Kilimann MW 2000. Neurobeachin: a protein kinase A-anchoring, beige/Chediak-higashi protein homolog implicated in neuronal membrane traffic. J Neurosci 20: 8551–8565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Griffiths GM, Stinchcombe JC, Kaplan J 2000. Analysis of the lysosomal storage disease Chediak–Higashi syndrome. Traffic 1: 816–822 [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, Smith SM 2009. Bayesian analysis of neuroimaging data in FSL. Neuroimage 45: S173–S186 [DOI] [PubMed] [Google Scholar]
- Volders K, Nuytens K, Creemers JW 2011. The autism candidate gene neurobeachin encodes a scaffolding protein implicated in membrane trafficking and signaling. Curr Mol Med 11: 204–217 [DOI] [PubMed] [Google Scholar]