Abstract
Copy-number variants (CNVs) form an abundant class of genetic variation with a presumed widespread impact on individual traits. While recent advances, such as the population-scale sequencing of human genomes, facilitated the fine-scale mapping of CNVs, the phenotypic impact of most of these CNVs remains unclear. By relating copy-number genotypes to transcriptome sequencing data, we have evaluated the impact of CNVs, mapped at fine scale, on gene expression. Based on data from 129 individuals with ancestry from two populations, we identified CNVs associated with the expression of 110 genes, with 13% of the associations involving complex, multiallelic CNVs. Categorization of CNVs according to variant type, size, and gene overlap enabled us to examine the impact of different CNV classes on expression variation. While many small (<4 kb) CNVs were associated with expression variation, overall we observed an enrichment of large duplications and deletions, including large intergenic CNVs, relative to the entire set of expression-associated CNVs. Furthermore, the copy number of genes intersecting with CNVs typically correlated positively with the genes' expression, and also was more strongly correlated with expression than nearby single nucleotide polymorphisms, suggesting a frequent causal role of CNVs in expression quantitative trait loci (eQTLs). We also elucidated unexpected cases of negative correlations between copy number and expression by assessing the CNVs' effects on the structure and regulation of genes. Finally, we examined dosage compensation of transcript levels. Our results suggest that association studies can gain in resolution and power by including fine-scale CNV information, such as those obtained from population-scale sequencing.
Copy-number variants (CNVs), or unbalanced structural variants, involving large deletions, insertions, and duplications, are among the least studied forms of genetic variation, although their net effect on the genome (in terms of affected base pairs) is higher than that of SNPs (Iafrate et al. 2004; Sebat et al. 2004; Tuzun et al. 2005; Redon et al. 2006; Korbel et al. 2007; Kidd et al. 2008; Conrad et al. 2010). CNVs have been associated with several disease phenotypes (Craddock et al. 2010), including systemic autoimmunity (Fanciulli et al. 2007), HIV susceptibility (Gonzalez et al. 2005), and psoriasis (Hollox et al. 2008). Recent reports have further reported associations of CNVs with gene expression variation, ascribing such associations to rare pathogenic CNVs (Lupski and Stankiewicz 2005) as well as to CNVs reaching appreciable allele frequencies in the population (McCarroll et al. 2006; Stranger et al. 2007). Specifically, a comprehensive survey by Stranger et al. (2007) reported a widespread association of large-scale CNVs with the expression of genes, an association frequently independent of SNPs.
Stranger and coworkers evaluated the effects of CNVs detected with two microarray platforms (bacterial artificial chromosome [BAC] arrays and 500k SNP arrays), which were used to ascertain CNVs with median sizes of 228 kb and 81 kb, respectively (Redon et al. 2006; Stranger et al. 2007). Technological advances, such as improvements in tiling microarray (Conrad et al. 2010) and sequencing technologies (1000 Genomes Project Consortium 2010), have recently led to “second-generation” CNV maps with markedly increased resolution and broadened CNV size range, with the latest study reporting CNVs of 50 bp to a 1 Mb in size (median size 730 bp) in more than 150 individuals (Mills et al. 2011). At the same time, the number of CNVs ascertained per genome has increased considerably, from less than a hundred (examined in Stranger et al. 2007) to several thousand per individual (Conrad et al. 2010; Mills et al. 2011). Advances in technology have further enabled the systematic distinction of different copy-number states in CNV regions, allowing the comprehensive genotyping of CNVs (McCarroll et al. 2008; Conrad et al. 2010; Park et al. 2010; Sudmant et al. 2010; Mills et al. 2011), which yields crucial information for associating CNVs with phenotypic data (Craddock et al. 2010). Furthermore, the mapping of CNV breakpoints has markedly improved in resolution, which enables relating CNVs to gene annotation at high resolution, such as to specific exons of a gene (Conrad et al. 2010; Kidd et al. 2010a; Pang et al. 2010; Mills et al. 2011). To our knowledge, no study has so far made use of these recent advances in technology and computational algorithms for comprehensively linking CNVs to gene expression data. Thus, our present understanding of the effect of CNVs on gene expression is incomplete.
Here we correlated CNVs recently discovered at fine resolution with genome-wide gene expression data. We thereby made use of recent advances in massive-scale transcriptome sequencing (RNA-seq) and reanalyzed data from two recent expression quantitative trait loci (eQTL) surveys that focused on SNPs. These two surveys examined lymphoblastoid cell lines (LCLs) from the HapMap project (International HapMap Consortium 2005): One study associated SNPs with gene expression data in LCLs derived from 60 unrelated Utah residents of Northern and Western European (CEU) ancestry (Montgomery et al. 2010); the other study associated SNPs with expression data in LCLs from 69 unrelated Yoruba individuals (YRI) from Nigeria (Pickrell et al. 2010). By analyzing CNV genotypes generated for the same HapMap individuals, our approach inferred associations between CNVs and the expression of more than a hundred genes, which included many novel CNV–gene expression associations. These results enabled us to evaluate relative influences of CNV type (e.g., deletion vs. duplication), CNV size, and overlap with coding regions (genic vs. intergenic) on such associations.
Results
Data set pre-processing and additional CNV genotyping
We obtained RNA-seq data in the form of Illumina GAII sequencing reads (with DNA bases called using standard Illumina software) from both aforementioned eQTL studies. We mapped the RNA-seq reads onto the human reference genome and further considered only such reads that displayed a unique (unambiguous) mapping position in the genome. Subsequently, we used a normalization procedure, described in the following, to obtain comparable expression measurements for both populations. In the CEU data set, which provides paired end sequences, we mapped on average 6.5 million read pairs onto the genome per individual. In the YRI data set, which provides single end sequences, we uniquely mapped on average 5.0 million reads onto the genome per individual. We related the mapped reads to a comprehensive set of protein-coding genes (see Methods) and normalized each data set to correct for influences on sample- and gene-specific read counts originating from: different overall numbers of mapped reads per individual; the individuals' sex (Stegle et al. 2010); and sequencing library-specific properties, such as differences in read lengths and GC-content biases (see Methods).
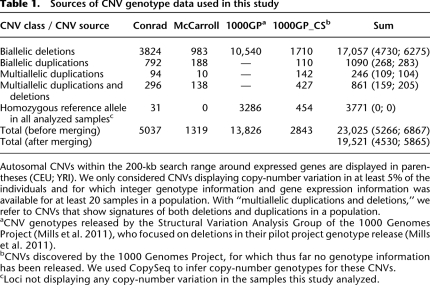
We further obtained comprehensive CNV genotype data by mining several published data sets assaying the same HapMap individuals. As these previous surveys focused, in part, on ascertaining different CNV types (e.g., deletions vs. duplications) (see Table 1), we combined these data sets to obtain a variant set that covers a wide CNV size spectrum. Specifically, the data sources comprised CNV (deletion and duplication) genotypes from two different microarray platforms, namely, genotypes for 1319 CNVs with a median size of ∼8 kb, inferred from a SNP/CNV hybrid array platform (McCarroll et al. 2008) and genotypes for 5037 CNVs (median ∼3 kb) from a high-resolution custom tiling array platform (Conrad et al. 2010). Furthermore, we used data based on population-scale sequencing: We included genotypes for 13,826 sequenced deletions (median size of ∼700 bp) that were discovered and genotyped by the 1000 Genomes Project's Structural Variation Analysis Group (Mills et al. 2011); furthermore, we analyzed the read depth of Illumina sequencing reads generated from the 1000 Genomes Project to extend the available list of CNVs with genotype information (see the Supplemental Material), by applying the CopySeq copy-number genotyping algorithm (Waszak et al. 2010). Specifically, we used CopySeq to infer CNV genotypes for 2843 CNVs (median ∼2 kb) from the 1000 Genomes Project that were released without CNV genotype information (Mills et al. 2011). We combined data from all four sources into a nonredundant CNV set with a merging approach that required both a genomic overlap of CNVs and genotype concordance for merging CNVs (see Methods) (Table 1).
Table 1.
Sources of CNV genotype data used in this study
Association of CNVs with gene expression phenotypes
Our approach to detect associations between expression and CNV, outlined in Figure 1, is analogous to previous eQTL studies that related RNA-seq data to SNP genotype information (Montgomery et al. 2010; Pickrell et al. 2010). Specifically, we related normalized RNA-seq read-count data to CNV genotype information in the form of copy-number genotypes, i.e., integer values from 0 to 10 reflecting the copy number of the genomic segment in question (see Methods). This enabled us to examine both biallelic CNVs (genomic segments with two possible allelic statuses, e.g., deletion and reference allele) and multiallelic CNVs (genomic segments with more than two possible allelic statuses, e.g., deletion, duplication, and reference allele; or several duplication alleles per locus leading to copy numbers of up to 10 at some loci, as a consequence of repeated locus duplication). Due to the high number of possible pairwise comparisons between CNVs and genes, we reduced the search space by focusing on CNV–gene pairs that would most likely result in robust associations. To this end, we removed rare CNVs as well as unexpressed or rarely expressed genes (Methods). Furthermore, we focused on proximal (i.e., putative cis) associations because, in the past, most strong eQTLs have been mapped close to their target gene (Stranger et al. 2007; Montgomery et al. 2010; Pickrell et al. 2010). Specifically, we limited our search to CNV–gene pairs separated by <200 kb (which we call the “search range”). Thus, we examined CNV–gene pairs on the basis of an operational definition for cis effects that in reality may also include short-distance trans effects. Furthermore, we excluded the sex-determining chromosomes from our analysis because of the imbalance of genes in males and females. With these filtering steps, our analysis considered 12,275 expressed genes and 4530 CNVs in the CEU samples, and 12,113 expressed genes and 5865 CNVs in the YRI samples (note that some genes had several CNVs in their search range, and vice versa). To test for association, we calculated Spearman correlation coefficients between copy-number genotypes and normalized expression values. We adjusted the resulting P-values for multiple testing to control the false-discovery rate (FDR) with a conservative threshold of FDR ≤ 10%.
Figure 1.
Relating CNVs to variation in gene expression. Flowchart of our approach for mapping CNV-associated eQTLs.
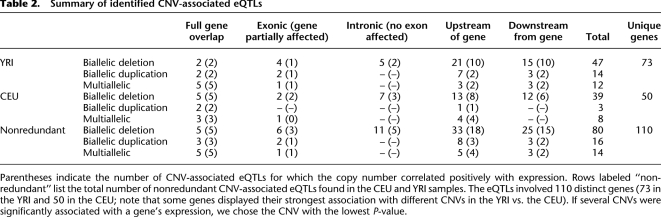
Applying this approach to the aforementioned HapMap samples yielded significant association between CNVs and expression for 50 and 73 genes in the CEU and YRI data sets, respectively (Table 2), or 110 distinct genes when combining the results from both data sets. In several cases, we identified more than one CNV associated with a given gene (Supplemental Table 3). Unless stated otherwise, the analyses below relate to the most strongly associated CNV–gene pairs, which we refer to as “CNV-associated eQTLs.” For ∼70% of all CNV-associated eQTLs, the distance between the respective CNV and gene was <100 kb, i.e., the CNV-associated eQTLs were enriched toward short-range associations (Supplemental Fig. 7). Despite the relative abundance of deletions in our CNV set (Table 1), CNV-associated eQTLs involving duplications (∼21% of all associated CNVs), including those involving multiallelic CNVs (∼13% of all associated CNVs), were enriched compared with deletions among all CNVs associated with expression (Supplemental Table 7). This may be attributable to an abundance of large, gene-duplicating variants in our data, as we discuss further below.
Table 2.
Summary of identified CNV-associated eQTLs
Overlap of CNV-associated eQTLs between populations and comparison with previous surveys
To assess the robustness of our CNV–expression associations, we first examined to what extent CNV-associated eQTLs were independently observed in both populations, because such observations would increase statistical confidence. We observed a strong (>32-fold) enrichment of CNV–gene pairs that were both identified as significantly associated in the YRI and the CEU samples (Supplemental Fig. 5). Furthermore, when analyzing our results at the level of identified genes, rather than at the level of identified CNV–gene pairs, we found that 26% of the genes corresponding to CNV-associated eQTLs in the CEU were also observed in the YRI. By comparison, 18% of the eQTLs identified in the YRI were also observed in the CEU. We may assume that the portion of CNV-associated eQTLs shared between both populations is actually higher, because small sample sizes, which limit the detection power (Gilad et al. 2008), may lead to false negatives in eQTL-mapping studies. Hence, we focused on genes that were expressed in both populations and evaluated how often CNV-associated eQTLs inferred in one population were evident in the other population, requiring a correlation with the same sign (e.g., positive correlation in both populations, or negative correlation in both populations) together with an unadjusted Spearman correlation P-value of P < 0.05. This analysis revealed markedly higher overlaps (namely, 42% of the genes in the CEU were also found in the YRI, and 46% for the reciprocal comparison). These results add further confidence to the association signals inferred by our approach.
We also compared the results from our survey to previous surveys. We first examined the three genes (i.e., UGT2B17, GSTM1, and GSTT1) reported as associated with expression variation by McCarroll et al. (2006), which were also expressed in at least one population in our study: All three were detected as CNV-associated eQTLs in our data. Furthermore, we compared our results to the results of Stranger et al. (2007), who identified CNV–expression associations in individuals from four different populations, including individuals from the CEU and YRI populations. In our comparison, we considered CNV-associated eQTLs that Stranger et al. reported in the CEU and YRI. Specifically, Stranger et al. (2007) reported these eQTLs in terms of “CNV clones,” which they associated with genes that corresponded to 42 unique gene identifiers in our gene set (Supplemental Material). Nine of these genes were also identified as eQTLs in our study. We further assessed the overlap by limiting the comparison to genes that were considered expressed in our study and that were within a 200-kb search range with respect to the CNV sets of both our survey as well as that of Stranger et al. (2007). Following these filtering steps, out of the 13 remaining genes from Stranger et al. (2007), six (46%) were identified by our approach. Thus, our approach recovers many previously identified CNV-associated eQTLs and also detects a number of new associations.
Analysis of CNV-associated eQTLs: Categorization according to gene overlap and functional categories
We continued by analyzing the inferred associations in detail. Making use of the high resolution of our CNV set, we first categorized CNV-associated eQTLs according to the type of overlap between the CNV and its associated expressed gene. We distinguished between CNVs overlapping their associated genes entirely and those leading to partial gene overlap with affected exonic sequences (i.e., gene disruption), as well as those affecting nonexonic regions (i.e., intronic and intergenic categories). In ∼20% of our CNV-associated eQTLs, the CNVs intersected exonic sequences of the genes or contained genes entirely (Table 2). In the CEU and YRI data set, CNVs associated with the expression of five and two genes, respectively, involved full gene deletions (for a specific example, see Fig. 2). This included a gene of potential biomedical relevance, i.e., the CDK11A (CDC2L2) gene (Supplemental Fig. 1), which has been associated with type 2 diabetes in Asians on the basis of SNPs (Li et al. 2007). Our results suggest that follow-up studies should consider CDK11A gene deletion as a possible causative variant.
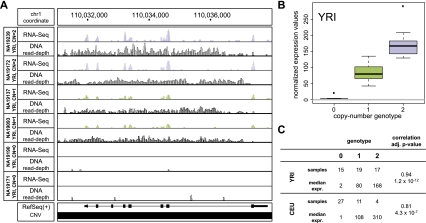
Figure 2.
Association of a deletion with gene expression. (A) A biallelic deletion (chr1:110,024,361–110,046,935) fully deleting the GSTM1 gene was found to be significantly associated with GSTM1 transcript level in the YRI and CEU samples. RNA-seq tracks display gene expression values following normalization. DNA read-depth tracks were generated based on population-scale sequencing reads (1000 Genomes Project Consortium 2010). (B) Correlation between copy-number genotype and normalized gene expression values for GSTM1, computed based on the YRI samples. (C) Summary of observed sample abundance and median normalized expression values for each copy-number state in the YRI and CEU samples.
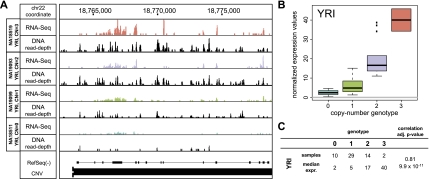
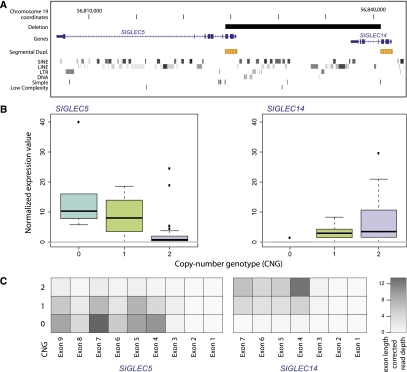
There were also several gene duplications among our CNV-associated eQTLs (Table 2), for example, the PI4KAP1 gene, a gene within a multiallelic CNV region displaying both duplication and deletion alleles (Fig. 3). Furthermore, at several gene loci, the associated CNVs intersected partially with genes, affecting at least one of their exons (Table 2). A peculiar example is the inferred association between the expression of SIGLEC5 and a deletion intersecting several of its exons. This deletion fuses SIGLEC5 with the paralogous SIGLEC14 gene, resulting in an in-frame gene hybrid, with gene expression patterns supporting the formation of an expressed SIGLEC14/5 fusion gene that acquired upstream regulatory elements from SIGLEC14 (Fig. 4; Supplemental Fig. 2).
Figure 3.
Association of a multiallelic CNV involving deletion and duplication alleles with gene expression. (A) A multiallelic CNV (chr22:18,763,501–18,789,830) entirely overlapping the PI4KAP1 gene locus was found to be significantly associated with PI4KAP1 expression. (B) Correlations between copy-number genotype and normalized gene expression values for PI4KAP1 (YRI samples). (C) Summary of observed sample abundance and median normalized expression value for each copy-number state.
Figure 4.
Detailed expression analysis for a CNV-associated eQTL involving a polymorphic fusion gene. The expression of the adjacent genes SIGLEC5 and SIGLEC14 is affected by a deletion that partially disrupts both genes, resulting in the “SIGLEC14/5” gene hybrid. (A) SIGLEC14/5, which displays >99.8% coding sequence identity with SIGLEC5, encompasses exons 1–3 as well as upstream regulatory sequences from SIGLEC14, and further contains exons 4–9 from SIGLEC5 (Supplemental Fig. 2). We identified a CNV-associated eQTL involving the biallelic deletion and SIGLEC5 in both populations and analyzed the expression patterns of the locus making use of published nucleotide resolution breakpoint information for the deletion (Yamanaka et al. 2009). (B) The observed negative correlation between copy number and expression can plausibly be explained by the comparably higher expression of SIGLEC14 in the absence of the deletion, and further by the juxtaposition of SIGLEC14/5 with the SIGLEC14 upstream (promoter) region. Because SIGLEC14/5 displays high sequence identity with SIGLEC5, transcripts originating from the fusion gene were mostly mapped to the SIGLEC5 gene locus. Although the fusion gene is not represented in the reference genome, RNA-seq-based expression measurements at the locus were allele-specific in the presence of the homozygous reference allele (copy number [CN] = 2), and also in the presence of the homozygous deletion (CN = 0). Namely, displayed read counts for CN = 0 stem from SIGLEC14/5; for CN = 2, depicted read counts were unambiguously assigned to either SIGLEC14 or SIGLEC5 (the latter is expressed at very low level). By comparison, for CN = 1, we measured a mixture of reads originating from all three genes. (C) Read counts are shown in the form of a heat map for YRI samples, based on the raw number of RNA-seq reads that uniquely map to the reference genome at SIGLEC5 and SIGLEC14 loci. Few reads could be uniquely mapped into exons 1–3, since these fall into a segmental duplication and thus lack unique sequence. We found the deletion genotype and the expression of SIGLEC14 to be positively correlated (Spearman rank correlation P-value < 0.003 for YRI and < 0.05 for CEU), as expected as the deletion disrupts most transcript sequence unique to SIGLEC14.
CNVs intersecting exonic sequences are particularly relevant, since for such CNVs a direct effect on gene expression is expected. Specifically, one would expect that the expression of genes overlapping CNVs correlates positively with copy number (with gene deletions and gene disruptions being associated with a relative expression decrease, and gene duplications being associated with an expression increase) (for examples, see Figs. 2, 3). Our data confirm this expectation: Namely, expression-associated CNVs disrupting, deleting, or duplicating genes typically displayed the expected positive correlation (Supplemental Fig. 6), with a few exceptions, which we examined in detail. First, the aforementioned SIGLEC5 gene displayed a negative correlation between its expression and the copy-number genotype of the gene-intersecting deletion. The fusion of SIGLEC5 with the SIGLEC14 gene and the resulting juxtaposition of the fusion gene with the SIGLEC14 promoter region can plausibly serve to explain the observed negative correlation (for details, see Fig. 4). We also examined in further detail the association between a CNV partially disrupting the ULK1 gene, for which we identified a negative correlation with the gene's expression, and obtained evidence, based on read-depth analysis, that the CNV boundaries were mis-annotated and that the ULK1 coding regions hence were not contained within the CNV (see the Supplemental Material).
By comparison, for CNVs not affecting exonic sequences, we had no a priori expectation on the sign of correlations, and, in fact, we observed a mixture of positive and negative correlations between copy-number genotype and expression (Supplemental Figs. 3, 4; Table 2). In these cases, the CNVs, or other linked variants, may contribute to the observed variation in expression through regulatory mechanisms.
To assess the implications of CNV–expression associations on gene function, we examined the set of CNV-associated eQTLs with respect to Gene Ontology (GO) and KEGG functional category classifications. These analyses revealed a significant enrichment for functional gene categories related to immunity (Supplemental Table 1). While relative enrichments of immune-related functions among CNVs have been reported before (e.g., Conrad et al. 2010 and references therein; Mills et al. 2011), our analysis of gene functional categories, which used a gene universe that excluded genes not expressed in LCLs, shows that enrichments in immune-related functions carry through to the level of CNV–expression associations. Such enrichment may be of particular relevance given that several previous studies have related CNVs affecting immunity-related genes with disease phenotypes, including genes that our approach identified as CNV-associated eQTLs: namely, the innate immunity gene APOBEC3B, for which CNVs have also been associated with HIV susceptibility (An et al. 2009); and CCL4L1, a chemokine gene associated with transplant rejection.
Novelty of our reported eQTLs and overlap with SNP-focused eQTL studies
A relevant question relates to the extent at which CNVs and SNPs are jointly or independently associated with a heritable trait, such as with gene expression, given that SNPs are the form of genetic variation primarily ascertained in genome-wide association studies. Since CNVs often represent ancestral mutations that are in linkage disequilibrium (LD) with SNPs (McCarroll et al. 2006), SNPs that tag nearby CNVs could plausibly be used as markers for identifying CNV-associated eQTLs (hence, an abundance of suitable “tag SNPs” might make a separate ascertainment of CNVs unnecessary). Since the expression data we used were originally applied to map eQTLs using SNPs, we compared our results to these previously mapped SNP-based eQTLs to obtain a point estimate for the fraction of our CNV-associated eQTLs that could be detected on the basis of SNPs. Altogether, 53 (48%) out of the 110 genes identified by our approach were recently observed as eQTLs on the basis of tag SNPs, using the same expression data (Montgomery et al. 2010; Pickrell et al. 2010). Namely, of the 50 genes whose expression we found to be associated with CNVs in the CEU population, 24 (48%) were also shown to be associated with SNPs by Montgomery et al. (2010), while 26 were only associated in our study. Furthermore, of the 73 genes we found in the YRI population, 35 (48%) were also reported by Pickrell et al. (2010), and 38 were only associated in our study. We also compared our results with the comprehensive list of loci described in the eQTL browser (Pickrell et al. 2010), a database summarizing results from seven SNP-focused eQTL studies that examined distinct cell and tissue types (Myers et al. 2007; Schadt et al. 2008; Veyrieras et al. 2008; Dimas et al. 2009; Montgomery et al. 2010; Pickrell et al. 2010; Zeller et al. 2010), and further from a study analyzing both SNP eQTLs and CNV-associated eQTLs in LCLs (Stranger et al. 2007). These comparisons showed that 32 out of the 110 genes (30%) that our approach identified were not previously reported in conjunction with eQTLs in any study included in the eQTL browser. Thus, our CNV-focused study has inferred several novel eQTLs.
We further evaluated what fraction of the CNV-associated eQTLs were correlated with a “tag SNP” that was equally, or better, associated with expression than the CNV in question, making use of the high-quality SNP genotype data that were recently released by the 1000 Genomes Project (see the Supplemental Material). Specifically, we calculated correlations between 1000 Genomes Project SNPs and normalized expression values, using the same criteria as described above for CNVs (i.e., focusing on the 200-kb search range defined by our CNV set). This analysis revealed that 57% of expressed genes in our list of CNV-associated eQTLs displayed a higher correlation with their most strongly associated CNV than with any SNP in the search range (Supplemental Table 2). Furthermore, the tendency of genes to display their highest correlation with CNVs, rather than with SNPs, was markedly more pronounced for CNVs overlapping exons: That is, all (10/10) of the genes that were deleted or duplicated displayed a higher correlation with these CNVs than with any SNP (Supplemental Table 8). This is compatible with the notion that CNVs affecting exonic sequence typically contribute themselves to the genes' expression. Furthermore, despite the CNVs typically showing higher correlations with gene expression than SNPs, 50% of the CNVs identified in the CEU and 44% of the CNVs identified in the YRI had strongly correlated SNPs (r2 > 0.8) that may serve as a tag for identifying these eQTLs in a SNP-focused survey.
Large CNVs preferentially contribute to CNV-associated eQTLs
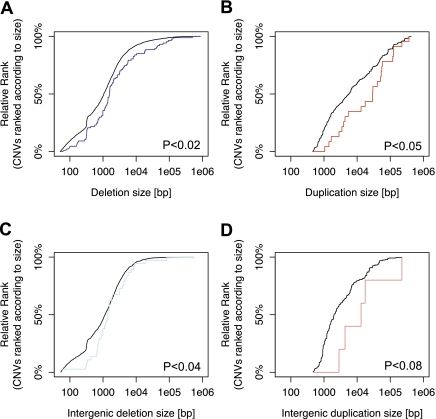
The wide CNV size spectrum available to this study, with genotyped CNVs from 50 bp up to megabase-pair level in size, enabled us to evaluate the influence of CNV size on the association between CNVs and expression. We evaluated to what extent small CNVs contributed to our list of CNV-associated eQTLs compared with large CNVs, by examining the spectrum of CNV sizes separately for deletions and duplications (Fig. 5). Overall, the median size of CNVs significantly associated with expression was in the lower kilobase range (<4 kb). Many expression-associated CNVs were below 1 kb in size (32% CEU, 23% YRI), and thus in a size range that recent technological advances (e.g., high-throughput DNA sequencing and tiling microarray technology) have made amenable to systematic analysis (Fig. 5AB). Despite the abundance of these relatively small CNVs, however, we observed a relative enrichment of large CNVs, including large deletions (Fig. 5A) and large duplications (Fig. 5B), among the CNV-associated eQTLs. This enrichment was significant, even when controlling for the fact that large CNVs are, based on our criteria (see Methods), more often present in the herein defined search range of genes than small CNVs (Fig. 5AB; Supplemental Table 6). The enrichment of large CNVs among CNV-associated eQTLs was evident also when limiting our analysis to CNV data from a single data source (e.g., tiling array based CNVs, or population-scale sequencing-based CNVs).
Figure 5.
Enrichment of large deletions and duplications among CNV-associated eQTLs. (A) Enrichment of large deletions among expression-associated variants. (Blue line) Cumulative distribution functions of the size of expression-associated deletions. (Gray line) The size distribution of the entire list of deletions in the search range of our survey. The distribution is adjusted for the fact that large variants have a higher chance to be within the search range of a gene than small variants, by considering each variant as many times as there are expressed genes in its search range. The P-values shown are based on Kolmogorov-Smirnov (KS) tests. (B) Significant enrichment of large duplications (red line) among expression-associated variants. (Gray line) The size distribution of duplications considered in our survey. (C) Significant enrichment of large intergenic deletions (light blue line) among expression-associated variants. (Gray line) The size distribution of intergenic deletions considered in our survey. (D) Marginally significant enrichment of large intergenic duplications (light red line) among expression-associated variants. (Gray line) The size distribution of intergenic duplications considered in our survey.
One plausible explanation for the enrichment of large CNVs among CNV-associated eQTLs is the abundance of gene-intersecting CNVs among our eQTLs, since gene-intersecting CNVs are comparably large and since CNVs overlapping genes are expected to typically affect expression. Interestingly, however, we observed an enrichment among our CNV-associated eQTLs also for large intergenic CNVs (Fig. 5CD; Supplemental Table 6), specifically for deletions (the enrichment we observed of intergenic duplications was marginally significant). Thus, the overlap with genes may only in part account for the preferential association of large CNVs with expression. We note that the enrichment of large CNVs, including large intergenic CNVs, suggests that these CNVs more frequently contribute to expression variation. (They might do so by changing a gene's copy number, by affecting DNA regulatory regions through positioning effects [Kurth et al. 2009; Ricard et al. 2010], or by altering the copy number of a regulatory site [Kasowski et al. 2010].) Furthermore, the general abundance of large (often gene-overlapping) CNVs among duplications may account for the observed relative abundance of duplications among CNV-associated eQTLs (Fig. 5B; Supplemental Table 7).
Evaluation of possible dosage-compensation effects associated with gene deletions
Especially for CNVs fully overlapping a gene, we expect a direct effect on expression level—therefore, expression should be proportional to copy number, unless gene dosage variation is compensated by buffering or feedback-regulation mechanisms (Deng and Disteche 2010). Such compensation for gene dosage was, for example, used to explain the observation that less than a third of all genes on chromosome 21 are overexpressed in the case of Down syndrome caused by trisomy 21 (Ait Yahya-Graison et al. 2007). Recent reports further noted that dosage compensation may explain why some CNV regions displayed limited correlation between copy number and expression in rodent species (for review, see Henrichsen et al. 2009). An evaluation of the magnitude with which CNVs typically influence expression levels in humans is highly relevant for deducing their effects on phenotypes.
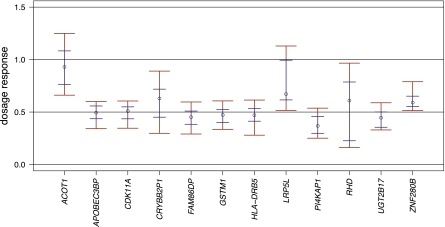
To examine this effect in detail, we assessed to what extent human genes affected by the CNVs in our list showed signs of dosage compensation. Specifically, we analyzed 18 deletions of expressed genes for which detailed inspection of the locus confirmed the evidence for full gene deletion (see the Supplemental Material). We assessed in these loci whether transcript levels associated with a heterozygous deletion (copy number [CN] = 1) were markedly higher than half of the level observed for the homozygous reference (CN = 2), which would be indicative of dosage compensation. Specifically, we computed point estimates for relative expression levels by dividing the median of the normalized expression values obtained for individuals with CN = 1 by the median of the expression from individuals with CN = 2, and then used bootstrapping to define confidence intervals (see the Supplemental Material). Twelve genes displayed tight confidence intervals (i.e., such that they enabled us to discriminate between a relative expression level of ∼0.5 and ∼1) (see the Supplemental Material). In nine loci, such as the CDK11A, GSMT1, and UGT2B17 loci, we found the correlation between copy number and expression to be compatible with negligible dosage compensation, because these loci displayed a relative expression level of ∼0.5 relative to the homozygous reference. Three loci (ACOT1, LRP5L, ZNF280B), however, displayed confidence intervals indicative for dosage compensation (Fig. 6; Supplemental Table 9), and buffering or feedback regulation mechanisms may contribute to transcript levels of these genes. While we envision that future studies will also examine the impact of dosage compensation on duplications, such analysis is currently complicated by uncertainties about the functionality of DNA regulatory elements (e.g., promoters and enhancers) in duplication alleles.
Figure 6.
Evaluation of dosage compensation in gene deletion loci. The figure displays relative normalized expression values of samples with copy number (CN) = 1 relative to samples with CN = 2. The circles mark the ratio between the median expressions of samples with CN = 1 and samples with CN = 2. The error bars indicate bootstrap confidence intervals for 68% (short blue horizontal lines) and 95% (long red horizontal lines).
Discussion
We have examined the impact of CNVs covering a wide size spectrum on gene expression, motivated by the concept that variation in expression may serve as a model for phenotypic variation (Myers et al. 2007; Stranger et al. 2007; Schadt et al. 2008; Veyrieras et al. 2008). Our approach revealed more than a hundred associations between CNVs and gene expression, confirming several previously identified associations and also revealing several novel ones. Therefore, our analysis benefitted from recent advances in analysis approaches and DNA sequencing technology. We assessed a CNV size spectrum from 50 bp up to a megabase pair, which facilitated evaluation of the impact of CNV size. Furthermore, comprehensive copy-number genotype information enabled us to ascertain associations between distinct CNV types; as an example, we reported that approximately a fifth of our CNV–expression associations involved duplications, and ∼13% involved multiallelic CNVs. The CNV data set resolution further enabled us to elucidate the impact of CNVs on expression in detail. For example, while our results suggest that CNVs with impact on expression frequently intersect with gene sequences, most associations that we reported involved intergenic CNVs, and these CNVs were often relatively small (i.e., <4 kb in size). Nonetheless, our analysis shows that large duplications and deletions, including large intergenic CNVs, are generally enriched among CNV-associated eQTLs. This suggests that CNV–expression associations involving large CNVs are particularly frequently attributable to the CNVs themselves, rather than to a linked variant (conversely, this is likely to be less often the case for small CNVs).
Furthermore, when categorizing CNVs according to gene overlap, we observed that CNVs intersecting genes typically affect genes in the expected direction, i.e., involving a positive correlation between gene copy number and expression level, as was also observed previously in an assessment of the impact of large-scale (mostly >100 kb) CNVs on expression (Stranger et al. 2007). On the other hand, we found that associations of CNVs in intergenic regions displayed a mixture of positive and negative correlations. Further analyses allowed us also to elucidate rare, unexpected negative correlations between gene copy number and expression. Several instances of unexpected negative correlations have been observed before (e.g., Stranger et al. 2007; Henrichsen et al. 2009), and our study shows how detailed analyses of fine-resolution CNV data can help to clarify the basis of such intuitively unanticipated correlations.
We also evaluated the effect of gene dosage reduction (i.e., heterozygous gene deletions) on transcript levels. While the expression of some genes may be affected by dosage changes, our results suggest that dosage compensation in human cells may be less pronounced than in Drosophila melanogaster, where autosomal genes are typically partially compensated for dosage, with relative expression levels (heterozygous deletion vs. homozygous reference allele) of ∼0.75 (Zhang et al. 2010). Other human cell types (i.e., non-LCL cells) may show a different propensity for dosage compensation, since compensatory loops may differ between tissues and developmental time points (Chaignat et al. 2011).
Half (48%) of the genes we identified in the context of CNV-associated eQTLs were recently associated on the basis of SNPs with the same expression data (Montgomery et al. 2010; Pickrell et al. 2010). This portion is higher than the fraction of genes associated with CNVs by Stranger and coworkers that also displayed a significant SNP association (18%; see Stranger et al. 2007). The comparably higher overlap observed in our study is, however, not surprising, given the increased density at which CNVs and SNPs can now be ascertained in the genome (Supplemental Table 4). Regardless of the overall level of correlation between CNV genotypes and genotypes of nearby SNPs, the relatively strong association we observed among CNVs' overlapping genes and these genes' expression is explainable either by the CNVs frequently representing causative variants or by CNV genotypes displaying a slightly reduced genotyping error rate compared with SNP genotypes (which also would cause CNVs to be more strongly associated). Strikingly, we found an abundance of gene-duplicating and gene-deleting CNVs among the variants that were particularly strongly associated with expression. Such an abundance of plausible causative variants underlines the value of an independent assessment of CNVs in association studies, and we conclude that inclusion of CNVs increases the power for identifying eQTL loci and further facilitates their “fine mapping” to the actual functional variant.
Our approach focused on proximal effects, defined by a search range of 200 kb, and did not consider associations between distal CNVs and expression variation (i.e., most trans effects). In this regard, for ∼70% of all CNV-associated eQTLs, the distance between the respective CNV and gene was <100 kb, such that CNV effects on expression were enriched toward short-range signals. Not surprisingly, given the abundance of deletions in our CNV sources, most associations we found involved deletions, despite an enrichment of duplications among CNV-associated eQTLs. We envision that future surveys will strengthen their focus on simultaneously ascertaining structure, content and location of duplications, on the basis of recently developed approaches (Kidd et al. 2010b; Sudmant et al. 2010), to facilitate inferring associations for an increased number of duplications. This will also enable crucial analyses relating to the integrity of regulatory regions in duplications and will further facilitate the discrimination of intact gene duplicates from pseudogenized duplicates (Korbel et al. 2008). Regardless of these expected coming advances, our results suggest that studies that aim to assess relationships between genetic variation and heritable traits will benefit from considering CNVs discovered and genotyped with the latest technologies.
Methods
CNV genotype sources
SNP/CNV hybrid microarray based genotypes for 1319 CNVs (McCarroll et al. 2008) and custom microarray-based genotypes for 5037 CNVs (Conrad et al. 2010) were obtained from the Database of Genomic Variants (Iafrate et al. 2004). Population-scale sequencing based genotypes for 13,826 deletions (Mills et al. 2011) were obtained from the 1000 Genomes Project's website (http://1000genomes.org). We further inferred genotypes for an additional 2843 CNVs using the CopySeq genotyping algorithm (see the Supplemental Material; Supplemental Table 5), by using CopySeq on population-scale sequencing-based CNVs from Mills et al. (2011) that were released without genotype information. We assessed all CNV genotypes in the form of copy-number genotypes, i.e., integer values reflecting the absolute copy number (CN) of a genomic segment in question, with, for example, homozygous and heterozygous deletions reflected by CN = 0 and CN = 1, respectively, and duplications reflected by CN = 3 or higher.
CNV data set merging
CNVs from different sources that overlapped in their genomic coordinates were merged if they exhibited a perfect (100%) genotype concordance across all available samples (i.e., we did not merge overlapping CNVs that displayed discordant genotypes to avoid discarding significant associations between CNVs and expressed genes). We merged CNVs by keeping the breakpoint annotations from the CNV source generated at higher resolution; to this end, we used the median CNV size of each source as a guideline for ranking CNVs by resolution. According to this criterion, the CNVs based on population-scale sequencing were assumed to have a higher resolution than the array-based CNVs; furthermore, the custom microarray-based CNVs were assumed to have a higher resolution than the SNP/CNV hybrid array-based CNVs.
RNA-seq data retrieval and read mapping
RNA-seq reads were obtained from recent publications (Montgomery et al. 2010; Pickrell et al. 2010). We aligned all reads onto the human reference genome (hg18) using GSNAP (2011-03-28.v3) mapping software (Wu and Nacu 2010) with default parameters and retrieved the number of mapped reads for our set of 19,950 protein-coding genes (based on Ensembl Build54). We discarded all reads that mapped to more than one position with a score of ≥20 as nonspecific (“ambiguous”).
RNA-seq quality filtering and normalization
We normalized the RNA-seq data by first correcting all data sets for GC content effects as in Pickrell et al. (2010). We further scaled the number of reads in each individual according to a normalization scheme provided by DESeq (Anders and Huber 2010). Next, we corrected the YRI data for sequencing center/protocol based-effects (e.g., distinct read lengths used), as previously described (Pickrell et al. 2010). Fourth, all YRI and CEU read data were corrected for sex effects using a method previously developed to correct for sequencing center-based effects (Pickrell et al. 2010). The normalized read counts were then used as “normalized expression values.”
Filtering expressed genes and common CNVs
We limited our analysis to expressed genes, which we defined as those displaying a normalized expression value of at least 20 in at least 5% of the samples (i.e., in at least four YRI samples or in at least three CEU samples). (The normalized expression value was scaled in such a way that the expression value 20 corresponded to 20 reads mapping onto a gene of interest in a sample sequenced at average sequencing depth.) We further removed CNVs displaying copy-number variation in very few samples, by excluding CNVs with an inferred genotype that differed from the most common genotype in <5% of the individuals for which gene expression data were available. Last, we considered CNVs only if integer copy-number genotype information and gene expression information were available for at least 20 samples in a population.
Inference of associations between CNVs and expressed gene loci
We calculated pairwise association values between transcript levels and CNVs by computing Spearman rank correlation coefficients between vectors of normalized gene expression values measured across all samples from a population (CEU or YRI) with vectors of copy-number genotypes measured in the same samples. We limited our analysis to a search range of 200 kb by evaluating CNV–gene pairs only if the distance between the closest coordinates corresponding to the annotated genomic CNV and transcript coordinates was ≤200 kb. To assess the significance of these correlations, we calculated P-values, which we adjusted for multiple hypothesis testing according to (Benjamini and Hochberg 1995) by controlling the false discovery rate (FDR) at 10%. Whenever we report an association between a gene and a CNV in the text, we are referring to the CNV with the lowest P-value with the gene in question (if not stated differently).
Acknowledgments
We thank Megumi Onishi-Seebacher and Robert Weatheritt for comments on the manuscript, and all members of the Korbel group for valuable discussions. We further thank the high-performance computing facilities of the EMBL IT Service Unit for technically supporting the study. The study was supported by an Emmy Noether Fellowship from the German Research Foundation (DFG) to J.O.K.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122614.111.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Creau N, Blehaut H, et al. 2007. Classification of human chromosome 21 gene-expression variations in Down syndrome: Impact on disease phenotypes. Am J Hum Genet 81: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Johnson R, Phair J, Kirk GD, Yu XF, Donfield S, Buchbinder S, Goedert JJ, Winkler CA 2009. APOBEC3B deletion and risk of HIV-1 acquisition. J Infect Dis 200: 1054–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Chaignat E, Yahya-Graison EA, Henrichsen CN, Chrast J, Schutz F, Pradervand S, Reymond A 2011. Copy number variation modifies expression time courses. Genome Res 21: 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Disteche CM 2010. Genomic responses to abnormal gene dosage: The X chromosome improved on a common strategy. PLoS Biol 8: e1000318 doi: 10.1371/journal.pbio.1000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Arcelus MG, Sekowska M, et al. 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325: 1246–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI, et al. 2007. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet 39: 721–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet 24: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. 2005. The influence of CCL3L1 gene–containing segmental duplications on HIV-1/AIDS susceptibility. Science 307: 1434–1440 [DOI] [PubMed] [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A 2009. Copy number variants, diseases and gene expression. Hum Mol Genet 18: R1–R8 [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, et al. 2008. Psoriasis is associated with increased β-defensin genomic copy number. Nat Genet 40: 23–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2005. A haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. 2010. Variation in transcription factor binding among humans. Science 328: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE 2010a. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, Alkan C, Malig M, Ventura M, Giannuzzi G, et al. 2010b. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods 7: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Kim PM, Chen X, Urban AE, Weissman S, Snyder M, Gerstein MB 2008. The current excitement about copy-number variation: how it relates to gene duplications and protein families. Curr Opin Struct Biol 18: 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Klopocki E, Stricker S, van Oosterwijk J, Vanek S, Altmann J, Santos HG, van Harssel JJ, de Ravel T, Wilkie AO, et al. 2009. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat Genet 41: 862–863 [DOI] [PubMed] [Google Scholar]
- Li Y, Wu G, Zuo J, Gao J, Chang Y, Fang FD 2007. Genetic variations of the CDC2L2 gene are associated with type 2 diabetes in a Han Chinese cohort. Diabetes Metab Res Rev 23: 455–461 [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P 2005. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1: e49 doi: 10.1371/journal.pgen.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. 2006. Common deletion polymorphisms in the human genome. Nat Genet 38: 86–92 [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. 2008. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40: 1166–1174 [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. 2007. A survey of genetic human cortical gene expression. Nat Genet 39: 1494–1499 [DOI] [PubMed] [Google Scholar]
- Pang AW, MacDonald JR, Pinto D, Wei J, Rafiq MA, Conrad DF, Park H, Hurles ME, Lee C, Venter JC, et al. 2010. Towards a comprehensive structural variation map of an individual human genome. Genome Biol 11: R52 doi: 10.1186/gb-2010-11-5-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, Kim S, Lee S, Suh D, Hong D, Kang HP, et al. 2010. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 42: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, Molina J, Chrast J, Gu W, Gheldof N, Pradervand S, Schütz F, Young JI, Lupski JR, Reymond A, et al. 2010. Phenotypic consequences of copy number variation: Insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol 8: e1000543 doi: 10.1371/journal.pbio.1000543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6: e107 doi: 10.1371/journal.pbio.0060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Stegle O, Parts L, Durbin R, Winn J 2010. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 6: e1000770 doi: 10.1371/journal.pcbi.1000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4: e1000214 doi: 10.1371/journal.pgen.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stutz AM, Schlattl A, Lancet D, Korbel JO 2010. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol 6: e1000988 doi: 10.1371/journal.pcbi.1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka M, Kato Y, Angata T, Narimatsu H 2009. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19: 841–846 [DOI] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. 2010. Genetics and beyond—The transcriptome of human monocytes and disease susceptibility. PLoS ONE 5: e10693 doi: 10.1371/journal.pone.0010693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol 8: e1000320 doi: 10.1371/journal.pbio.1000320 [DOI] [PMC free article] [PubMed] [Google Scholar]