Abstract
Epigenetic mechanisms are important regulators of cell type–specific genes, including miRNAs. In order to identify cell type–specific miRNAs regulated by epigenetic mechanisms, we undertook a global analysis of miRNA expression and epigenetic states in three isogenic pairs of human mammary epithelial cells (HMEC) and human mammary fibroblasts (HMF), which represent two differentiated cell types typically present within a given organ, each with a distinct phenotype and a distinct epigenotype. While miRNA expression and epigenetic states showed strong interindividual concordance within a given cell type, almost 10% of the expressed miRNA showed a cell type–specific pattern of expression that was linked to the epigenetic state of their promoter. The tissue-specific miRNA genes were epigenetically repressed in nonexpressing cells by DNA methylation (38%) and H3K27me3 (58%), with only a small set of miRNAs (21%) showing a dual epigenetic repression where both DNA methylation and H3K27me3 were present at their promoters, such as MIR10A and MIR10B. Individual miRNA clusters of closely related miRNA gene families can each display cell type–specific repression by the same or complementary epigenetic mechanisms, such as the MIR200 family, and MIR205, where fibroblasts repress MIR200C/141 by DNA methylation, MIR200A/200B/429 by H3K27me3, and MIR205 by both DNA methylation and H3K27me3. Since deregulation of many of the epigenetically regulated miRNAs that we identified have been linked to disease processes such as cancer, it is predicted that compromise of the epigenetic control mechanisms is important for this process. Overall, these results highlight the importance of epigenetic regulation in the control of normal cell type–specific miRNA expression.
MicroRNAs (miRNA) are short single-stranded RNA molecules that regulate gene expression at the post-transcriptional level by inhibiting translation of target mRNAs or by stimulating their degradation. Mature miRNAs are processed from hairpin precursors that are either encoded by dedicated miRNA genes that are often found in clusters, or they reside in the introns of protein-coding genes and are processed following transcription of the host gene. According to current estimates, there are >1000 miRNAs expressed from over 500 transcriptional units (miRNA genes) encoded in the human genome. These miRNAs control, in part, the expression of about two-thirds of human genes (Friedman et al. 2009). miRNAs are involved in determination of cell identity and their expression is often deregulated in cancer (Peter 2009); however, relatively little is known about how their expression is regulated. Evidence is emerging that similar to protein-coding genes, epigenetic mechanisms play an important role in this process (Iorio et al. 2010).
Epigenetic mechanisms involve DNA methylation and post-translational modifications of chromatin proteins, including histones. 5-methylcytosine residues are a feature of transcriptionally silent heterochromatin, and this epigenetic mark is indispensable for mammalian development; it participates in X chromosome inactivation, gene imprinting, and control of cell type–specific gene expression patterns (Bird 2002; Lister et al. 2009; Laurent et al. 2010). Methyl groups are deposited on CpG cytosine residues by de novo DNA methyltransferases, DNMT3A and DNMT3B, and then maintained by DNA methyltransferase DNMT1 (Jaenisch and Bird 2003; Miranda and Jones 2007). In addition to DNA methylation, there exist a number of post-translational modifications on histones that act as positive or negative epigenetic regulators of gene expression. Two major repressive histone marks commonly present in euchromatin regions are trimethylation of histone H3 at lysine 27 (H3K27me3) and dimethylation of histone H3 at lysine 9 (H3K9me2). The H3K27me3 modification is deployed by the histone methyltransferase EZH2, a part of the polycomb repressive complex 2 (PRC2) (Simon and Kingston 2009). The H3K9me2 repressive histone modification is deposited by G9a, a member of the H3K9-specific histone methyltransferases (Tachibana et al. 2002). Two major permissive marks are trimethylation of histone H3 at lysine 4 (H3K4me3) and acetylation of histone H3 at multiple lysine residues (H3Ac) (Liang et al. 2004). Recent evidence suggests that epigenetic control is also involved in the regulation of miRNA gene expression in both normal and cancer cells (Bueno et al. 2008; Kozaki et al. 2008; Lujambio et al. 2008; Vrba et al. 2010).
To more fully understand the role of epigenetic mechanisms in the regulation of normal cell type–specific miRNA expression, we studied three different isogenic pairs of human mammary epithelial cells (HMEC) and human mammary fibroblasts (HMF) derived from reduction mammoplasty tissue (Garbe et al. 2009). These normal, previously characterized (Garbe et al. 2009; Novak et al. 2009) finite lifespan cells represent two differentiated cell types typically present within a given organ, each with a distinct phenotype and a distinct epigenotype. The analysis of multiple genotypes allowed us to assess interindividual variability in miRNA expression and epigenetic marks and more precisely identify miRNA genes targeted by epigenetic regulation. To identify cell type–specific miRNAs regulated by epigenetic mechanisms in normal cells, we integrated miRNA expression data obtained by high-throughput sequencing of small RNA libraries with the epigenetic profiles of their miRNA promoter regions obtained using a custom designed miRNA tiling microarray. Results from this analysis showed that miRNA gene expression and epigenetic state display high interindividual concordance within a given cell type; however, intercell-type concordance was lower, with 13% of expressed miRNAs showing >10-fold difference in expression between the two normal mammary cell types. We found that a majority of these cell type–specific miRNAs are regulated by epigenetic mechanisms in normal cells; 38% were subject to DNA methylation-mediated repression of their promoter in the normal nonexpressing cells, while 58% were subject to H3K27me3-mediated repression of their promoter in normal nonexpressing cells. Overall, there is limited overlap of these two repressive marks at individual miRNA promoters, although a few notable exceptions, MIR10A, MIR10B, and MIR205 appear to be under dual epigenetic repression by both DNA methylation and H3K27me3 in normal nonexpressing cells. In other cases, DNA methylation and H3K27me3 independently target individual miRNA clusters in order to repress complete miRNA families. These results indicate that a significant fraction of cell type–specific miRNAs are regulated at the epigenetic level and that these miRNAs are likely to be important targets in human diseases caused by epigenetic dysfunction.
Results
We determined miRNA expression in normal human mammary epithelial and fibroblast cells by high-throughput sequencing of small RNA libraries prepared from three isogenic pairs of HMEC, and HMF from specimens 48, 184, and 240. About 3.7 million reads in each library were aligned to annotated miRNA regions. Detailed information about sequencing data quality is provided in Supplemental Figures 1 and 2 and Supplemental Table 1. This miRNA transcriptome sequencing data showed that 392 of 703 (mirbase 13) mature miRNAs were expressed in HMEC and/or HMF at levels that ranged from zero to one million counts per library, demonstrating a large dynamic range of miRNA expression. The expression level of these 392 miRNAs for each of the six samples is provided in Supplemental Data 1. Results also revealed a striking interindividual concordance in miRNA expression within a cell type, with a correlation of 0.94–0.99 for HMEC and 0.97–0.98 for HMF (Supplemental Fig. 3).
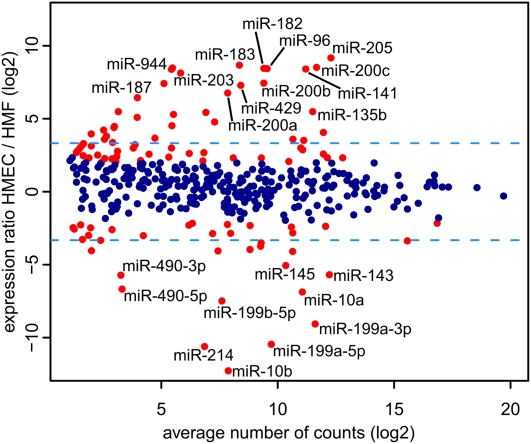
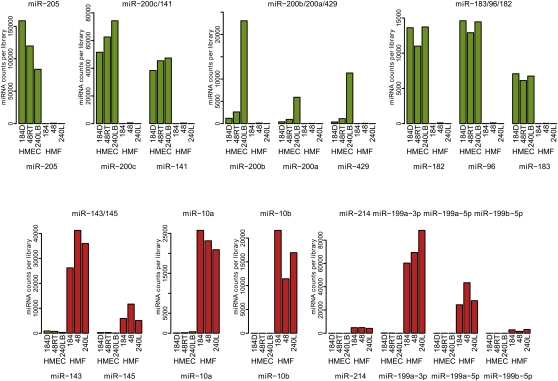
Intercell-type concordance was considerably lower than interindividual with correlations between HMEC and HMF ranging from 0.75 to 0.84, indicating a substantial population of miRNAs expressed in a cell type selective fashion. Twenty seven percent (104/392) of the expressed miRNAs showed at least fourfold (P < 0.05) difference in expression, with 68 showing HMEC selective expression, and 36 showing HMF selectivity (Fig. 1). Thirteen percent (50/392) of the expressed mature miRNAs showed at least a 10-fold difference in expression between HMEC and HMF. This fraction was considered cell type–specific, with 32 miRNAs being HMEC specific and 18 being HMF specific. Figure 2 shows quantitative expression results for several representative cell type–specific miRNAs that illustrate the strong interindividual concordance in the cell type–specific expression as well as the magnitudes of difference that can be seen in miRNA expression between the different cell types (exceeding three orders). Examples of mammary epithelial specific miRNAs include miR-205, the two clusters forming the miR-200 family, and the miR-183/96/182 cluster (Fig. 2, top). The miR-200b/200a/429 cluster of miR-200 family is the only tissue-specific cluster that displays substantial variability between individual HMEC genotypes. Mammary fibroblast–specific miRNAs are represented by the miR-10 family, the miR-199 family (and miR-214), and the miR-143/145 cluster (Fig. 2, bottom). To verify miRNA sequencing data we have analyzed the expression of 10 selected tissue-specific miRNAs by real-time PCR (Supplemental Fig. 4). The real-time PCR data are in high concordance with miRNA sequencing data, including interindividual variability of miR-200b. Overall, these miRNA expression results indicate that normal human mammary cells show strong cell type–specific patterns of a substantial fraction of miRNAs.
Figure 1.
Differential miRNA expression between HMEC and HMF obtained from three independent pairs of samples. The y-axis displays the HMEC to HMF expression ratio, the x-axis displays the average expression of miRNAs; both axes are in logarithmic scale. (Red) Differentially expressed miRNAs (at least fourfold difference in expression, P-value ≤0.05). (Blue dashed horizontal lines) Tenfold differences in expression. Several cell type–specific miRNAs are marked.
Figure 2.
Expression data for selected cell type–specific miRNA genes obtained by sequencing of small RNA libraries from isogenic pairs of human mammary epithelial and fibroblast cells. The normalized counts for each sample are displayed. (Green) Individual HMEC samples; (red) their paired HMF samples. The expression levels of the 392 detected mature miRNAs across all six samples are presented in Supplemental Data 1.
Since epigenetic mechanisms play a role in the control of cell type–specific protein-coding genes, we performed a broad epigenomic analysis to identify epigenetically regulated miRNA genes. A custom miRNA gene tiling microarray (described in detail in Methods) was used to probe the epigenetic state of miRNA promoters in each of the three pairs of isogenic HMEC–HMF. 5-methylcytosine or chromatin-immunoprecipitated genomic fractions and their respective inputs were hybridized to the microarrays. Then, miRNA targets of DNA methylation, the repressive histone marks H3K27me3 and H3K9me2, and the permissive histone marks of H3Ac and H3K4me3 were identified. The H3K4me3 mark is present on active promoters (Heintzman et al. 2007), reaching a maximum peak at ∼500 bp downstream from the transcription start site (Guenther et al. 2007) and has been extensively used to identify these elements (Marson et al. 2008; Ozsolak et al. 2008). Therefore, we used the results from our H3K4me3 analysis to identify and refine the promoter regions of miRNA genes expressed in normal HMEC and HMF (100 or more reads across the libraries). The positions of the identified promoter regions for 169 miRNA genes and gene clusters including 232 miRNA-coding regions are provided in Supplemental Data 2, and it is these 169 regions that were evaluated for epigenetic state. These 169 regions include promoters of 24 miRNA gene clusters encoding 37 out of 50 tissue-specific miRNAs. The remaining 13 tissue-specific miRNAs were either expressed in lower than 100 counts across libraries or there was no significant H3K4me3 enrichment peak in the region covered by microarray, and promoter regions were therefore not predicted for these miRNAs.
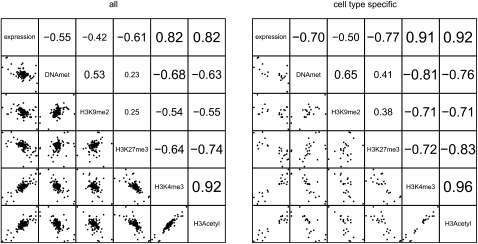
Figure 3 shows correlations that exist between miRNA expression and the miRNA promoter's epigenetic state. Specifically, either all expressed miRNA genes, or those 10-fold differentially expressed were compared to discover a correlation to the DNA methylation or histone modification state of their respective promoters. The comparisons show that for cell type–specific miRNAs there is an increase in correlation between miRNA expression and the various epigenetic marks at their promoter regions. As expected, the correlation was much lower when we analyzed only promoters of miRNAs not differentially expressed (Supplemental Fig. 5). With respect to permissive histone marks, H3Ac and H3K4me3 display strong positive correlations with miRNA expression (Fig. 3), as well as with each other, consistent with a transcriptionally active state. These permissive histone marks also display strong negative correlations with DNA methylation and the repressive histone modifications, H3K27me3 and H3K9me2.
Figure 3.
Differences in HMEC/HMF miRNA expression correlated with differences in epigenetic marks at their promoters. We used data from all probes within a 2-kb region centered on the predicted TSS region, and calculated the correlation of difference in enrichment of individual epigenetic marks between the cell types. (Left) Correlations for all 169 miRNA genes and gene clusters, where the promoter was predicted; (right) 24 cell type–specific miRNA genes only. The numbers show correlation coefficients for individual pairs of epigenetic marks.
Repressive epigenetic marks in miRNA promoter regions show increasingly strong negative correlations as the levels of cell type differential expression increases. In miRNAs that show a >10-fold difference in expression, DNA methylation and H3K27me3 show inverse correlations with an miRNA expression of −0.70 and −0.77, respectively, suggesting that a number of these miRNAs are epigenetically regulated (Fig. 3). H3K9me2 shows a weaker negative correlation (−0.50) that is more closely associated with DNA methylation than with H3K27me3, supporting the functional linkage between H3K9me2 and DNA methylation (Epsztejn-Litman et al. 2008). While DNA methylation and H3K27me3 both have strong inverse correlations with permissive histone marks and miRNA expression state, these two marks do not correlate well with each other (0.41), supporting their largely independent mechanisms and distinct targets of epigenetic repression. Taken together, these associations suggest that a subset of cell type–specific miRNA genes are repressed by DNA methylation/H3K9me2 and others by H3K27me3, while only a small fraction of these miRNA genes are likely to be repressed by both epigenetic marks. In addition to promoter regions, we also analyzed the correlation between repressive epigenetic marks and miRNA expression at miRNA hairpin encoding regions downstream from their promoters (Supplemental Fig. 6). In these regions, H3K27me3 correlates negatively with expression at the level similar to that observed in promoter regions. DNA methylation, however, does not correlate with expression in hairpin coding regions. These results indicate that DNA methylation plays a promoter-centric repressive role, in contrast to the polycomb-specific H3K27me3 that spreads over the whole regions of repressed genes.
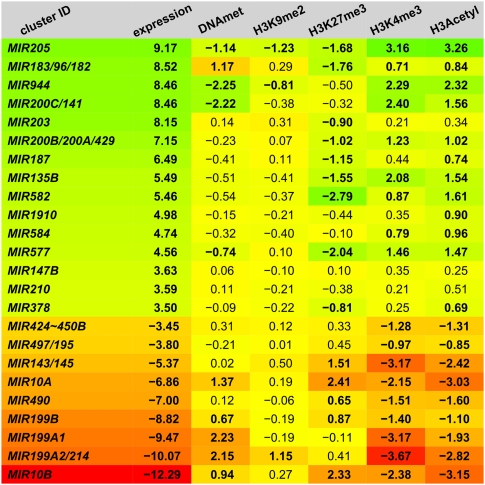
We further focused on the promoters of 24 miRNA genes and gene clusters expressing 37 cell type–specific miRNAs (Fig. 4). Eighteen of these promoters (75%) show cell type–specific patterns of DNA or H3K27 methylation that are linked to the transcriptional repression of their associated miRNAs in the nonexpressing cell type, while six promoters (25%) show no substantial cell type–specific differences in repressive epigenetic marks. Nine of 24 promoters (38%) are repressed by DNA methylation, 14 of 24 promoters (58%) are differentially occupied by polycomb-specific mark H3K27me3, and interestingly, five of the 18 epigenetically targeted miRNA promoters possess differences in both, DNA methylation and H3K27me3, suggesting a dual epigenetic repression of these miRNAs. Examples of this small fraction of miRNA genes under dual epigenetic repression by DNA methylation and H3K27me3 include the HMEC-specific MIR205 and the HMF-specific MIR10A and MIR10B.
Figure 4.
Cell type–specific miRNA genes (clusters), their difference in expression, and differences in occupation of their promoter regions by five epigenetic marks. The data are presented as log2 (fold difference HMEC/HMF). The color scale indicates whether the data behave as expected for HMEC-specific miRNAs (green) or HMF-specific miRNAs (red). The MIR424∼450B is the label for the MIR424/503/542/450A2/450A1/450B cluster.
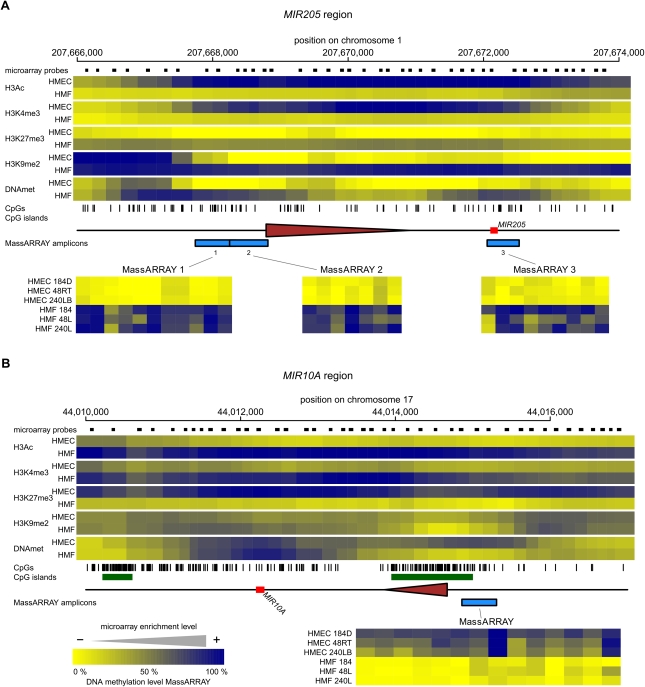
miR-205 is an HMEC-specific miRNA that is functionally related to the miR-200 family. The MIR205 gene has an increased CpG density at its regulatory region; however, it does not have a CpG island according to the original criteria (Gardiner-Garden and Frommer 1987). H3K4me3 and H3Ac are present in miR-205 expressing HMEC cells, but are absent in nonexpressing HMF (Fig. 5A). In contrast, the repressive epigenetic marks of DNA methylation, H3K9me2 and H3K27me3, are all present in nonexpressing HMF, but are absent in expressing HMEC. To increase the resolution and confirm the DNA methylation data from the microarray analysis, we analyzed certain regions in more detail using MassARRAY technology (Fig. 5A, bottom). The results from MassARRAY are in agreement with the microarray data, showing very low levels of methylation in all HMEC samples and high levels of DNA methylation in HMF. Therefore, in mammary fibroblasts, the MIR205 gene appears to be under a dual epigenetic repression by both DNA methylation (linked to H3K9me2) and H3K27me3.
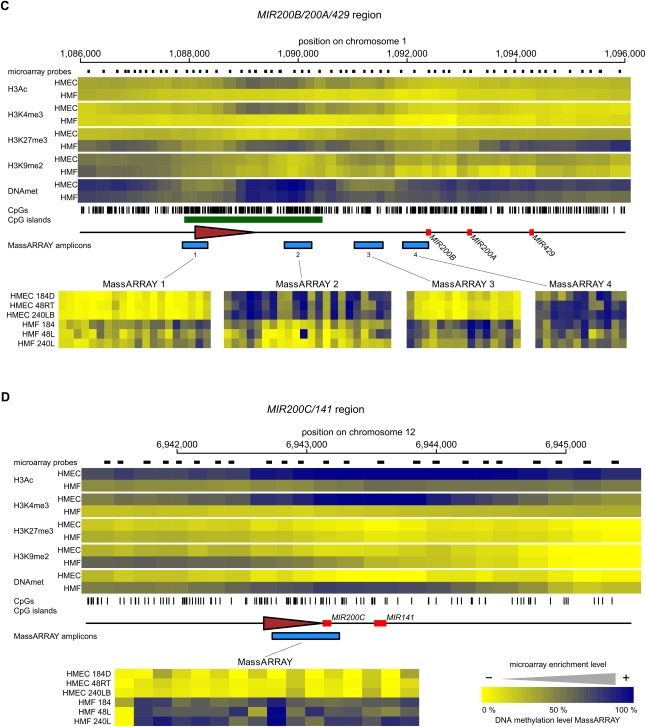
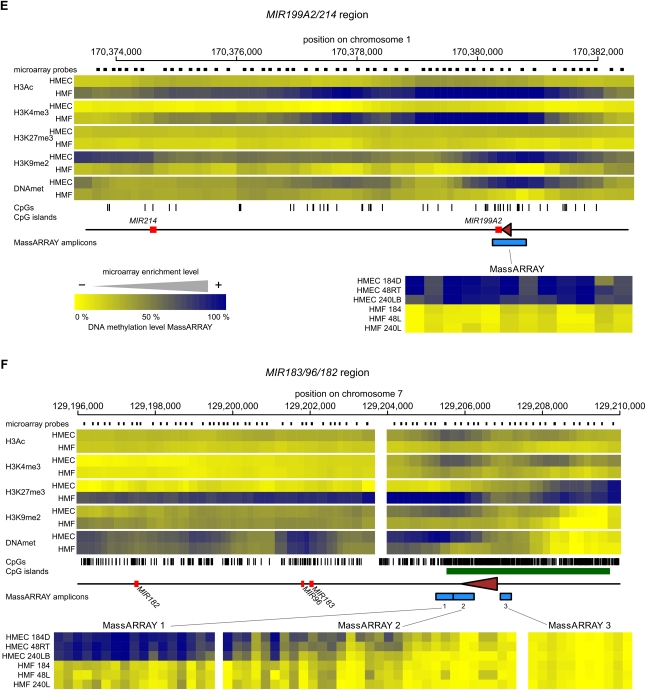
Figure 5.
The epigenetic state of cell type–specific miRNA genes in HMEC and HMF. Panels A–F show data for MIR205, MIR10A, MIR200B/200A/429, MIR200C/141, MIR199A2/214, and MIR183/96/182 miRNA genes, respectively. (Top part of each panel) The enrichment of individual epigenetic marks through the regions in a heatmap form, with yellow indicating no enrichment and blue color indicating high enrichment. The data for HMEC and HMF samples are averages from three genotypes of HMEC and HMF, respectively. The chromosomal positions are according to the hg18 human genome assembly. The miRNA hairpin coding regions are displayed as red rectangles and predicted transcription start site regions as brown triangles. Small black rectangles at the top lane indicate positions of individual microarray probes. The tics at the bottom indicate positions of individual CpG dinucleotides. CpG islands according to UCSC are displayed as green rectangles when present. The blue rectangles at the bottom lane indicate positions of regions analyzed for DNA methylation by MassARRAY technology. Results from MassARRAY for individual regions indicated are shown at the bottom part of each panel. The level of methylation of individual CpG units within the MassARRAY amplicon is displayed as a heatmap with the lowest methylation (0%) in yellow and the highest methylation (100%) in blue. The individual samples are labeled on the left.
A similar dual epigenetic control is seen in the HMF-specific MIR10A and MIR10B miRNA genes located within the HOXB and HOXD gene clusters, respectively. In MIR10A and MIR10B-expressing fibroblasts, H3K4me3 and H3Ac are present at MIR10A and MIR10B promoters, but are absent in nonexpressing epithelial cells. In contrast, the repressive H3K27me3 is present throughout the whole region of both the MIR10A and MIR10B genes in nonexpressing HMEC, but is absent in expressing HMF (Fig. 5B; Supplemental Fig. 7A), consistent with the knowledge that HOX gene clusters are polycomb targets (Simon and Kingston 2009). In addition to the repressive H3K27me3 mark in HMEC, the MIR10A and MIR10B gene promoters also show HMEC-specific DNA methylation (Fig. 5B; Supplemental Fig. 7A) along with low levels of H3K9me2. These results are similar to those seen for MIR205, suggesting that normal cells repress some miRNA genes using multiple epigenetic mechanisms.
Most epigenetically targeted cell type–specific miRNAs are repressed in association with either H3K27Me3 or DNA methylation, and these distinct repressive epigenetic states can even be seen in different members of the same miRNA families. One example is the MIR200 family, an epithelial-specific miRNA family repressed in mesenchymal cells. In this case, both members of the MIR200 family have their promoter region occupied by H3K4me3 and H3Ac in expressing epithelial cells. In contrast, nonexpressing HMF are devoid of these permissive marks and instead, each member of the MIR200 family is targeted by different repressive epigenetic marks in these cells. The promoter for the MIR200B/200A/429 cluster is occupied by H3K27me3, (Fig. 5C) indicating that polycomb is responsible for the fibroblast cell type–specific repression of this miRNA gene. DNA methylation of the MIR200B/200A/429 gene has a complex pattern. MassARRAY analysis revealed that the transcription start region located in the distal part of the CpG island is unmethylated in HMEC and has a low level of methylation in HMF (Fig. 5C). The other analyzed regions downstream from the TSS, however, show that DNA methylation alternates between nonexpressing and expressing cells in this particular H3K27me3 target gene (Fig. 5C). The proximal part of the CpG island shows more DNA methylation in expressing HMEC samples. Then follows the region methylated more in nonexpressing fibroblasts, and finally, the area just upstream of the MIR200B hairpin coding region shows an intermediate level of methylation in both cell types. Therefore, DNA methylation does not seem to play any conclusive role in normal tissue-specific repression of the MIR200B/200A/429 cluster. In contrast to the MIR200B/200A/429 cluster, we have previously shown DNA methylation and H3K9Me2 to be important in the repression of the MIR200C/141 members of the family (Vrba et al. 2010), and these earlier observations are further confirmed by the microarray analysis in the present studies (Fig. 5D). The DNA of the whole MIR200C/141 region is unmethylated in expressing HMEC and the microarray data show that DNA methylation in HMF reaches a maximum in the region just downstream from the TSS. The lack of H3K27me3 enrichment in the MIR200C/141 region in both mammary cell types indicates that this important cell type–specific miRNA cluster is not a polycomb target. Since both clusters of the MIR200 family share a high level of expression in epithelial cells, but no expression in fibroblasts, the distinct repressive epigenetic marks observed for the members of the MIR200 family suggest that distinct epigenetic mechanisms are involved in their respective repression.
Similar multifaceted forms of epigenetic repression of miRNA families are seen in the fibroblast-specific MIR199 family, whose members consist of the MIR199A2/214 cluster, MIR199A1 and MIR199B. These three miRNA genes of the MIR199 family are located at homologous positions on the antisense strands of introns of the three members of the DNM gene family. The MIR199A2/214 cluster is located in DNM3, MIR199A1 in DNM2, and MIR199B in DNM1, indicating coevolution of the MIR199 family together with the DNM gene family. Similar to HMEC-specific miRNA genes MIR200C/141 and MIR205, there is an increased CpG density in the promoters of the MIR199 genes; however, it does not meet the original CpG island definition criteria. The miRNA promoters are occupied by H3K4me3 and H3Ac in expressing fibroblasts, but these marks are absent in epithelial cells. Instead, in nonexpressing epithelial cells the MIR199A2/214 cluster is DNA methylated and H3K9me2 is present, but significant levels of H3K27me3 were not detected, indicating that this region is not a polycomb target (Fig. 5E). In contrast, only DNA methylation-linked repression was detected in the MIR199A1 regulatory region in epithelial cells, (Supplemental Fig. 7B), while MIR199B utilizes a dual epigenetic repression, that of DNA methylation in combination with H3K27me3 (Supplemental Fig. 7C). Overall, these results suggest that expression of the three members of the fibroblast-specific MIR199 family is repressed predominantly by DNA methylation, and to a lesser extent, by H3K27me3.
A surprising cell type–specific miRNA gene with respect to repressive epigenetic marks is the MIR183/96/182 gene. The entire MIR183/96/182 region is heavily occupied by H3K27me3 in nonexpressing HMF, but H3K27me3 is absent in expressing HMEC (Fig. 5F), suggesting that the polycomb repression is the critical repressive epigenetic mechanism regulating the MIR183/96/182 cluster gene. DNA methylation shows an unexpected phenomenon in this region. Although the TSS region is DNA methylation-free for both cell types, downstream from the TSS at the proximal end of the CpG island there exists a differentially methylated region that is DNA methylation-free in all three nonexpressing HMF samples and heavily methylated in all three expressing HMEC samples (Fig. 5F). MassARRAY analysis confirmed the microarray data for all three genotypes. Thus, the MIR183/96/182 cluster is a cell type–specific miRNA gene repressed by polycomb, where DNA methylation within the promoter region inversely correlates with H3K27me3 and positively correlates with expression (Fig. 4). Since the DNA hypermethylation in HMEC is located downstream from the TSS, in the area that is most occupied by polycomb-specific mark H3K27me3 in nonexpressing HMF (Fig. 5F), it may represent a case of antagonism between DNA methylation and polycomb repression, as was recently described in a mouse model (Wu et al. 2010).
Taken together, our data show strong tissue-specific expression of a substantial fraction of miRNAs between mammary epithelial cells and mammary fibroblasts, exceeding three orders of magnitude for some miRNAs. Most of the promoters of these tissue-specific miRNA genes are occupied by H3K27me3 or DNA methylation, or both of these marks in nonexpressing cells, indicating that the epigenetic state of miRNA promoters plays an important role in the cell type–specific control of miRNA expression.
Discussion
In this study, we sought to identify cell type–specific miRNAs regulated by epigenetic mechanisms. To this end, we analyzed the expression levels and the epigenetic state of miRNA genes in three isogenic pairs of normal, finite lifespan HMEC and HMF—two predominant differentiated cell types of ectodermal and mesodermal origin, respectively, found in mammary tissue. miRNA expression and epigenetic state showed strong interindividual concordance within a given cell type, but significant expression and epigenetic differences were found between the different cell types. We found 13% of expressed miRNAs to be expressed in a cell type–specific fashion (>10-fold difference between the two cell types), including several known to be important for maintaining cell-specific phenotypes. The differential expression of a majority of these cell type–specific miRNAs was linked to cell type–specific differences in the epigenetic state of their promoters, indicating that epigenetic mechanisms play an important role in the regulation of cell type–specific miRNA genes.
To identify miRNA promoters utilized by HMEC and HMF (Supplemental Data 2), we used H3K4me3 ChIP–chip data that we obtained from these cells, since this mark is exclusively present at promoters (Heintzman et al. 2007). We used the complementary miRNA expression data obtained from these same cells to limit promoter identification to only those miRNA expressed in at least one of the mammary cell types, allowing for precise identification of miRNA promoters relevant to human mammary cells. Our results revealed that the promoters of cell type–specific miRNAs mostly lacked H3K4me3 enrichment in the inactive promoters of the nonexpressing cell type (Fig. 5A,C–E), suggesting that these represent the epigenetically labile miRNAs of normal, nondiseased cells. Similar results were reported previously for tissue-specific protein-coding genes (Guenther et al. 2007). Finally, the H3K4me3 ChIP–chip results further suggest that H3K4me3-directed prediction of promoters for miRNA genes, and likely all genes in general, will be the most accurate if derived from cells known to express the genes of interest.
Of the 24 cell type–specific miRNA genes, three-quarters (75%) were found to be repressed by H3K27me3 or DNA methylation in the nonexpressing cell type, with 58% of the cell type–specific promoters differentially occupied by H3K27me3, 38% differentially occupied by DNA methylation, and 21% differentially occupied by both H3K27me3 and DNA methylation. In miRNAs repressed by H3K27me3, we find this mark extends beyond the promoter and encompasses larger regions that include the miRNA gene body, consistent with recent findings demonstrating a spreading of the H3K27me3 mark from the promoter during differentiation of human embryonic stem cells (Hawkins et al. 2010). Interestingly, when we extend our analysis beyond only miRNA promoters differentially active in mammary cells by integrating our H3K27me3 ChIP–chip data with all of the predicted miRNA promoter data and known miRNA hairpin coding regions, we found that ∼27% of all miRNA promoters, as well as miRNA hairpin coding regions, are enriched for H3K27me3 (Supplemental Fig. 8). In contrast, it has been estimated that only 9% of promoters of protein-coding genes are occupied by H3K27me3 (Bracken et al. 2006). Thus, H3K27me3, a polycomb mark responsible for regulating genes involved in cell identity and differentiation, shows an approximately threefold bias toward miRNA compared with protein-coding genes, providing further support for an important role for miRNAs as regulators of cell fate and phenotype and H3K27me3 as an important regulator of cell type–specific miRNA expression.
Cell type–specific DNA methylation of miRNA promoters was correlated with the presence of the H3K9me2 mark and the repression of the associated miRNAs, further supporting the functional linkage between these two repressive epigenetic marks (Epsztejn-Litman et al. 2008). The DNA methylation mark was found to be centered on the promoter regions of the miRNA and did not extend far upstream or downstream, in contrast to what was observed with the H3K27me3 repressive mark. In addition, a common feature of DNA methylation-repressed miRNA genes was the absence of a CpG island in the promoter region; in the case of cell type–specific miRNA genes with CpG island promoters, the H3K27me3 mark is most often responsible for transcriptional repression (Fig. 5A–F).
The weakest correlation between epigenetic marks was between DNA methylation/H3K9me2 and H3K27me3. The limited overlap between DNA methylation/H3K9me2 and H3K27me3 in miRNA promoters likely reflects the largely independent nature of these two epigenetic repressive pathways in normal cells. Despite this limited overlap there are miRNAs where both marks appear to be involved in their cell type–specific repression. The coincidence of DNA methylation and H3K27me3 on the same miRNA promoters may represent mutual fail-safe mechanisms, so that disruption of one epigenetic mechanism is not sufficient to initiate aberrant activation of the miRNA. If this is the case, then, miRNA targets of this dual epigenetic repression may be critical to maintain cell integrity/identity, and their compromise could be involved in the genesis of human disease states, including cancer. Two examples of miRNAs that support the possible importance of dual epigenetic repression are miR-205, miR-10a, and miR-10b, since their dysregulation has already been linked to a variety of different cancers (Gregory et al. 2008b; Ma and Weinberg 2008; Greene et al. 2010; Lund 2010). For example, the loss of epithelial-specific miR-205 in cancers of epithelial origin has been linked to the acquisition of aggressive tumor phenotypes (Baffa et al. 2009; Iorio et al. 2009; Tellez et al. 2011), while gain of expression of fibroblast-specific miR-10 family members by cancers of epithelial origin have similarly been linked to aggressive tumor phenotypes (Ma et al. 2007; Baffa et al. 2009; Tian et al. 2010).
miR-205 participates in the maintenance of the epithelial phenotype, is expressed in epithelial cells but not in fibroblasts, and is related to the miR-200 family (Gregory et al. 2008a; Park et al. 2008). Expression of miR-205 and the miR-200 family members appears incompatible with a mesenchymal phenotype, and since miR-205 is expressed at much higher levels than the miR-200 family members in HMEC, it is possible that dual epigenetic repression by DNA methylation and H3K27me3 in fibroblasts is necessary to prevent any inappropriate miR-205 expression. miR-205 along with the miR-200 family target the mesenchymal-specific transcriptional repressors ZEB1 and ZEB2 (Gregory et al. 2008a). Possibly, the total level of the miR-200 family and miR-205 secures an epithelial phenotype, and complete epigenetic silencing of MIR205 is necessary to allow ZEB1 and ZEB2 to be expressed at levels sufficient to direct the mesenchymal phenotype.
Similar to MIR205, we have found that the closely related MIR200 family members are also epigenetically regulated in a cell type–specific fashion. We and others have previously shown that the MIR200 family members MIR200C/141 have a permissive epigenetic state in expressing epithelial cells, while nonexpressing fibroblasts display a repressed epigenetic state driven by DNA methylation and H3K9me2, but not H3K27me3 (Vrba et al. 2010). In contrast, the fibroblast-specific repression of the other MIR200 family members MIR200B/200A/429 is linked to the H3K27me3 mark, and DNA methylation and H3K9me2 do not appear to play a decisive role in its repression (Fig. 5C). Therefore, the two genes of the MIR200 family that likely arose from a single common ancestor have acquired different mechanisms of epigenetic regulation during evolution. Overall, these results suggest that complete epigenetic transcriptional repression of individual miRNAs may require complementary epigenetic mechanisms to prevent spurious transcriptional activity. Similarly, our results also suggest that complementary epigenetic repressive mechanisms may act independently to repress distinct members of the same miRNA family, as is the case with the MIR200 family.
miR-10a and miR-10b are expressed in a fibroblast-specific fashion and appear to be under dual epigenetic repression in the epithelial cells. Both MIR10A and MIR10B show significant H3K27me3 and DNA methylation in their promoter regions and both are found in HOX gene clusters, gene families well known for rich and complex epigenetic regulation in normal and cancer cells (Rauch et al. 2007). Interestingly, miR-10b has been reported to be aberrantly expressed in breast cancer cells and this expression is linked to an aggressive cancer phenotype (Ma et al. 2007; Baffa et al. 2009), although this conclusion has been questioned based on the analysis of primary breast cancer specimens that showed no correlation between miR-10b expression levels and clinical progression (Gee et al. 2008). Several molecular, cellular, and organismal facets may contribute to these two different conclusions. The data presented in this study strongly suggest that epigenetic control of cell type–specific expression is one important facet. The experimental biological studies used pure cancer cell line populations to first detect and then verify miR-10b's phenotypic effects. In contrast, it appears that the analysis of breast cancer clinical specimens was performed on heterogeneous tissue samples. Detecting aberrant expression of miR-10b in breast tumor cells from a complex tissue specimen is likely to be difficult, since mammary fibroblasts can constitute a significant portion of stromal cell content typically found in such specimens, and they express more than three orders of magnitude higher levels of miR-10b compared with mammary epithelial cells. This very high expression of miR-10b in the stromal cells of the tissue specimens could obscure any significant changes within the breast cancer cells themselves.
Indeed, this cautionary note extends to all the cell type–specific miRNAs identified in this study. The large magnitude differences seen in the expression levels and epigenetic states of these miRNAs between HMEC and HMF limit the ability to easily determine over- or underexpression or epigenetic status from the analysis of heterogenous tissue samples. Precise analysis of these miRNAs will require approaches that assess the expression or epigenetic state of these miRNAs in specific cell types within a tissue specimen. This is especially important since many of the epigenetically regulated cell type–specific miRNAs identified in this study have already been associated with or functionally linked to human carcinogenesis. The tissue-specific miRNAs that have been found deregulated in cancer are summarized in Supplemental Table 2.
In summary, our study has revealed a significant level of cell type–specific miRNA expression associated with human mammary epithelial and fibroblast cells. We also showed that epigenetic modifications play an important role in this cell type–specific miRNA regulation. The two epigenetic pathways responsible for deployment of repressive epigenetic marks—DNA methylation and H3K27me3—act largely independently. H3K27me3, a hallmark of polycomb repression, appears to play the major role in the normal cell type–specific repression of miRNA genes. Examples of polycomb targets are the MIR200B/200A/429 and MIR183/96/182 genes. Some highly cell type–specific miRNA genes like MIR200C/141 and MIR199A2/214, however, are not polycomb targets and their differential expression is determined predominantly by cell type–specific DNA methylation at the promoter region, often in collaboration with the H3K9me2 histone mark in nonexpressing cells. A common feature of these miRNA genes repressed by DNA methylation is the lack of CpG islands. Different genes forming miRNA families are repressed by different epigenetic marks, indicating that they are targeted by different epigenetic mechanisms. Some miRNA genes, including MIR205, MIR10A, and MIR10B, are under dual epigenetic repression by both DNA methylation and H3K27me3. The miRNAs found to be cell type–specific and repressed by epigenetic marks in nonexpressing cells are often deregulated in cancer, indicating that the disruption of normal epigenetic regulation of cell type–specific miRNA expression can be involved in carcinogenesis. Knowing which epigenetic marks are involved in repression of individual miRNA genes in normal cells and the locations of relevant regulatory regions thus contributes to our better understanding of these processes.
Methods
Cell lines and cell culture
Finite lifespan prestasis HMEC from specimens 184 (batch D), 48 (batch RT), and 240L (batch B), were derived from reduction mammoplasty tissue of women aged 21, 16, and 19, respectively (Garbe et al. 2009). Cells were initiated as organoids in primary culture in serum-containing M85 medium supplemented with oxytocin (Bachem) at 0.1 nM and maintained in M87A medium supplemented with oxytocin and cholera toxin at 0.5 ng/mL (Garbe et al. 2009). Fibroblasts from specimens 184, 48, and 240L were obtained by growing primary reduction mammoplasty cells in DMEM/F12 with 10% FBS and 10 μg/mL insulin, and further propagated in DMEM/F12 with 10% FBS, as previously described (Garbe et al. 2009). The cells used in this study were within cell culture passages 4–9 from primary tissue. Our previous study shows that these cells do not acquire epigenetic changes as late as passage 14. It is only following the emergence from the stasis proliferation barrier (Garbe et al. 2009) that HMEC first show the acquisition of significant epigenetic changes (Novak et al. 2009).
Small RNA library preparation and sequencing
Total RNA from epithelial cells or fibroblasts was extracted using the TRIzol method (Invitrogen). The small RNA fraction (18–35 nt) was purified on a 15% denaturing polyacrylamide gel. A preadenylated adaptor (rAppCTGTAGGCACCATCAAT3ddC) was ligated to the 3′ end of a small RNA using truncated T4 RNA ligase 2 (New England Biolabs), followed by purification of the ligation product on a 15% denaturing polyacrylamide gel. An Illumina-specific 5′ adaptor (GTTCAGAGTTCTACAGTCCGAcgauc; uppercase: DNA, lowercase: RNA) was ligated using T4 RNA ligase 1 (New England Biolabs) and the product was purified on a 10% denaturing polyacrylamide gel. Small RNAs with ligated adaptors were reverse transcribed into DNA using Superscript III reverse transcriptase (Invitrogen) and a primer with a Illumina-specific extension on its 5′ end (GACATCCGTGGTAGTTAGCATACGGCAGAAGACGAAC). The cDNA was then amplified by 15 cycles of PCR using Phusion DNA polymerase (Finnzymes) and Illumina-specific primers (AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA and GACATCCGTGGTAGTTAGCATACGGCAGAAGACGAAC). The resulting ∼110-bp PCR products were separated and purified from a 3% agarose gel, and submitted for Illumina sequencing to NCGR. The data were deposited in SRA archive, accession number SRP001530.
Data analysis
Results from the Illumina Genome Analyzer were received in the fastq format. The reads were mapped to the hg18 human genome assembly using the program Novoalign (www.novocraft.com). Output from Novoalign was further analyzed in R (R_Development_Core_Team 2011). First, data were converted to bed format and peaks of reads were found for pooled data from all samples. The peak regions were used for counts and annotation of reads in individual samples. Counts were normalized for the total number of reads in individual libraries (see Filtered reads in Supplementary Table 1). The miRNAs with at least 10 reads across the libraries represented in at least two libraries were considered expressed and further analyzed. Package edgeR was used for the differential expression calculation.
miRNA gene tiling microarray design
Human miRNA-coding sequence positions were downloaded from miRBase (ver 13) (http://microrna.sanger.ac.uk/sequences/). This data set contains information about the positions of 718 miRNA-coding regions in the human genome. A total of 454 out of 718 miRNA-coding regions have their promoter predicted (Marson et al. 2008). Another 149 of the miRNA-coding regions are part of known protein-coding genes. For these miRNAs the TSS of the host gene was assumed as TSS for miRNA and the position of this region was obtained as the 5′ end of the known gene from the UCSC Genome Browser. The entire region from 10 kb upstream of the predicted promoter (TSS) region down to 5 kb downstream from the miRNA hairpin coding region was tiled for all of these 603 miRNA-coding regions. For the remaining 115 miRNA-coding regions, where there was no prediction of TSS nor do they lie within protein-coding genes, the whole region from 70 kb upstream of the hairpin coding region down to 5 kb downstream was tiled. Additionally, about 100 protein-coding genes, including controls like GAPDH and ACTB, were added. The probes used to tile the specified regions were from the Agilent whole genome tiling microarray set and the Agilent promoter 2 microarray set. The resulting number of probes totals over 99,000, with approximately five probes per kilobase pair. The microarrays were manufactured by Agilent (Agilent Technologies) using their 2 × 105 k platform. Agilent design ID is 024305.
Methyl cytosine DNA and chromatin immunoprecipitation
Methyl cytosine DNA immunoprecipitation (MeDIP) was performed using 5-methylcytosine-specific monoclonal antibody as described (Weber et al. 2005).
Chromatin immoprecipitation (ChIP) was performed as described previously (Oshiro et al. 2003; Vrba et al. 2008) using antibodies specific for histone H3 trimethylated at lysine 4 (#05-745, Upstate), histone H3 trimethylated at lysine 27 (#07-449, Millipore), acetylated histone H3 (#06-599, Millipore), and histone H3 dimethylated at lysine 9 (CS200587, Millipore).
Sample labeling and microarray hybridization
MeDIP DNA or ChIP DNA samples were amplified using the random primed approach and 2 ug of amplified DNA was labeled as described (Vrba et al. 2008). Cy5 was used for input samples, Cy3 for immunoprecipitated samples. After labeling and purification, Cy3 and Cy5 labeled samples were pooled and vacuum concentrated to a volume of 91.5 μL. A total of 12.5 μL of human Cot-1 DNA (1 μg/μL, Invitrogen Cat. No. 15279-011), 26 μL of Agilent blocking agent (10×), and 130 μL of Agilent hybridization buffer (2×) were added. Samples were heated for 3 min at 95°C, transferred to 37°C, incubated for 30 min, and then used for microarray hybridization for 28h at 65°C. After hybridization, slides were washed in Agilent Oligo aCGH/ChIP-on-Chip Wash Buffer 1 for 5 min at room temperature, then in Agilent Oligo aCGH/ChIP-on-Chip Wash Buffer 2 for 5 min at 37°C, washed in acetronitrile for 10 sec at room temperature, and finally in stabilization and drying solution for 30 sec at room temperature. The scanning was performed using an Axon GenePix 4000B microarray scanner (Axon Instruments) and GenePix 6.0 software at 5 μm resolution and PMT settings 750 (635 nm) and 600 (532 nm).
Microarray data analysis
Output from GenePix (*.gpr files) were imported to R using the limma package. Individual channels were first spatially normalized within arrays using ma2D function from the package marray and then loess normalized between arrays using the function normalize.loess from package affy. The RG object was transformed to an MA object and M values were again loess normalized between arrays. M values (log2 ratios of input to immunoprecipitated channel) were used for further analysis as a measure of enrichment of a region centered on individual probes. Differences in histone modification were determined in a 2-kb region centered on a TSS region or miRNA hairpin region. The paired t-test was used to analyze data from all probes in each 2-kb region. Differentially enriched regions were defined as regions where the average difference of ratio was at least 1.5-fold and the P-value was ≤0.05.
Transcription start regions prediction
We predicted miRNA TSS regions based on the fact that H3K4me3 is present in TSS regions, reaching maximum enrichment at ∼0.5 kb downstream from TSS (Guenther et al. 2007). To reduce the amount of false predictions, we used the most proximal major peak of H3K4me3 enrichment, upstream of the miRNA hairpin region, which was at least twofold enriched over input. Further, we limited the predictions only for miRNAs that are expressed (at least 100 reads in libraries). For differentially expressed miRNAs, the pooled H3K4me3 data from expressing samples (either HMEC or HMF) were used, for other miRNAs, the pooled H3K4me3 data from all six samples were used. Once the peak of H3K4me3 enrichment was identified, the area between the peak and nearby Switch gear TSS or EST 5′ end was considered a TSS region. Switch gear TSS and EST positions were downloaded from the UCSC browser. For miRNAs where there was no Switch gear TSS or EST end in the vicinity (2.5 kb upstream, 0.5 kb downstream) of the H3K4me3 peak, the region from H3K4me3 peak 1 kb upstream was considered TSS region.
DNA methylation analysis by MassARRAY
DNA methylation analysis by MassARRAY was performed as described (Novak et al. 2009). Primer sequences are listed in Supplemental Table 3.
All oligonucleotides used in this study were ordered from Integrated DNA Technologies.
Data access
The sequence data from this study have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP001530. The microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE28380.
Acknowledgments
This work was supported by grants CA-65662 and 1U01CA153086-01, and by the Margaret E. and Fenton L. Maynard Endowment for Breast Cancer Research. J.C.G. and M.R.S. were supported by Department of Defense grant BCRP BC060444 carried out at Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123935.111.
References
- Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, et al. 2009. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 219: 214–221 [DOI] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13: 496–506 [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS, Wyrobek AJ, Stampfer MR 2009. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res 69: 7557–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M 1987. CpG islands in vertebrate genomes. J Mol Biol 196: 261–282 [DOI] [PubMed] [Google Scholar]
- Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM, Ragoussis J, Harris AL 2008. MicroRNA-10b and breast cancer metastasis. Nature 455: E8–E9 [DOI] [PubMed] [Google Scholar]
- Greene SB, Herschkowitz JI, Rosen JM 2010. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol 7: 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ 2008a. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ 2008b. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7: 3112–3118 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Menard S, Croce CM, Tagliabue E 2009. microRNA-205 regulates HER3 in human breast cancer. Cancer Res 69: 2195–2200 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Piovan C, Croce CM 2010. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta 1799: 694–701 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J 2008. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res 68: 2094–2105 [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci 101: 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. 2008. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci 105: 13556–13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH 2010. miR-10 in development and cancer. Cell Death Differ 17: 209–214 [DOI] [PubMed] [Google Scholar]
- Ma L, Weinberg RA 2008. MicroRNAs in malignant progression. Cell Cycle 7: 570–572 [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Jones PA 2007. DNA methylation: the nuts and bolts of repression. J Cell Physiol 213: 384–390 [DOI] [PubMed] [Google Scholar]
- Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW 2009. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res 69: 5251–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Domann FE, Futscher BW 2003. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene 22: 3624–3634 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev 22: 3172–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME 2009. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Development_Core_Team 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP 2007. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci 104: 5527–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA 2011. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res 71: 3087–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z 2010. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem 285: 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrba L, Junk DJ, Novak P, Futscher BW 2008. p53 induces distinct epigenetic states at its direct target promoters. BMC Genomics 9: 486 doi: 10.1186/1471-2164-9-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW 2010. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE 5: e8697 doi: 10.1371/journal.pone.0008697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]