Abstract
We have determined methylation state differences in the epigenomes of uncultured cells purified from human, chimpanzee, and orangutan, using digestion with a methylation-sensitive enzyme, deep sequencing, and computational analysis of the sequence data. The methylomes show a high degree of conservation, but the methylation states of ∼10% of CpG island-like regions differ significantly between human and chimp. The differences are not associated with changes in CG content and recapitulate the known phylogenetic relationship of the three species, indicating that they are stably maintained within each species. Inferences about the relationship between somatic and germline methylation states can be made by an analysis of CG decay, derived from methylation and sequence data. This indicates that somatic methylation states are highly related to germline states and that the methylation differences between human and chimp have occurred in the germline. These results provide evidence for epigenetic changes that occur in the germline and distinguish closely related species and suggest that germline epigenetic states might constrain somatic states.
The epigenome is a complex assortment of proteins and chemical modifications that are associated with DNA, control its transcription (Brink 1960; Bernstein et al. 2007), and mediate stable phenotypic states as exemplified by cell differentiation. The genome and the epigenome are associated in the chromosomes and are inherited together, but the degree to which the epigenome is encoded by the genome is not known. Furthermore, it is not clear to what extent the epigenome is maintained in the germline and transmitted between generations (Feng et al. 2010). It might be reset in each generation using genetically encoded information, but persistence of epigenetic states in the germline creates the potential for semi-independent inheritance of epigenetic information (Rakyan and Beck 2006; Richards 2006). Finally, it is not clear if epigenetic states that are present in the germline influence somatic epigenotypes, or conversely, if somatic epigenetic states are generated during cell differentiation using genetically encoded information.
We have explored the use of comparative epigenomic analysis (“phyloepigenomics”) to obtain insights into changes in the epigenome in human evolution. Epigenetic differences provide a means to modulate the regulatory activity of noncoding regions. Functionally significant changes may be more readily identified in the epigenome than in the genome: Sequence change is not always associated with functional change (Boffelli et al. 2004), but the epigenome mediates genome function by controlling transcription (Brink 1960; Bernstein et al. 2007), and so changes in it are more likely to reflect functional changes. Epigenomic comparison might thus complement the evidence of potentially adaptive genomic changes identified with multiple sequence-based approaches (Pollard et al. 2006; Prabhakar et al. 2006; Grossman et al. 2010; Yi et al. 2010).
This study focuses on one component of the epigenome, cytosine methylation, a covalent modification of DNA that acts as a focal point in mechanisms that suppress transcription initiation (Klose and Bird 2006). We have compared the methylomes of human and chimpanzee in a homogeneous somatic cell type, the neutrophil, using the orangutan as an outgroup. Our comparison uses a “methyltyping” assay based on digestion with the methylation-sensitive restriction enzyme HpaII and deep sequencing (MethylSeq) (Ball et al. 2009; Brunner et al. 2009). MethylSeq data was analyzed with MetMap (Singer et al. 2010), a computational method that we developed to correct bias in MethylSeq data and infer the true methylation status of all HpaII sites within the scope of the experiment. The combination of MethylSeq and MetMap obtains very deep sequence data focused on regions of the genome that are not methylated and can reliably detect methylation changes in these regions (Singer et al. 2010). Compared to methods that use methylation-independent restriction enzymes, MethylSeq retrieves information spanning more of the genome because of a more favorable profile of fragment sizes (Singer et al. 2010).
We find that, while the methylomes of human and chimp are similar, a set of ∼1500 stable differences in CpG island-like regions distinguishes human from chimp; these differences identify regions that may have diverged in gene regulatory function. The methylation states can be used to build a tree that recapitulates the phylogenetic relationship of the three species. Analysis of CG substitution patterns in CpG island-like regions that have conserved their methylated state in human, chimp, and orang indicates that methylation in the neutrophil reflects germline methylation. Our findings demonstrate that methyltyping can identify differences that distinguish human from chimp and that these differences exist in the germline. This raises the question of whether a germline epigenome is transmitted along with the genome, and if so, whether it is completely determined by genome sequence.
Results
A comparative epigenomic study should use cells of a single homogeneous type because different cell types have distinct epigenomes (Meissner et al. 2008; Lister et al. 2009), and cells should be uncultured because the epigenome can be distorted by cell culture (Meissner et al. 2008). Neutrophils are abundant circulating cells that are morphologically indistinguishable in humans and chimps and can readily be isolated as a pure population without culturing. Since neutrophils in their circulating form are accessible and relatively homogenous, they are a suitable cell type for an interspecies comparison. We isolated neutrophils to >99% purity from the peripheral blood of four young adult male humans (age 20–25 yr old) and four age-equivalent male chimpanzees (age 12–16 yr old, which is young adult, after accounting for differences in age of maturity [Fleagle 1999]). To further attempt to control environmental variation, we selected individuals who were healthy, well-nourished, afebrile, and not part of any study of infectious agents or other treatments.
DNA from neutrophils was digested with HpaII, 50–300-bp fragments isolated from an agarose gel, Illumina sequencing libraries constructed, and sequencing carried out on an Illumina sequencer (Supplemental Table S1). Single-end reads were quality filtered and aligned to their respective genomes with Bowtie (Langmead et al. 2009), which produced stacks of reads at digested HpaII sites. Using MethylSeq data and a reference genome sequence, MetMap assigns to each HpaII site within the scope of the experiment a probability of being unmethylated p(U) and a probability of being part of an unmethylated region p(I) (Singer et al. 2010). (A HpaII site is within the scope of the experiment if it lies on a fragment that has HpaII sites at its ends and is of a length that allows it to pass the size selection step used in the construction of the sequencing libraries.) MetMap produces a reduced representation survey of the methylome: Of the 28,163,863 CGs in the human genome (the sites that can be methylated), 2,292,175 fall within a HpaII site; and of these, 1,349,376 are within the scope of this experiment; similarly, there are 26,602,442 CGs in the chimpanzee genome; 2,122,178 fall within a HpaII site, of which 1,197,715 are within the scope of this experiment. Only sites within the scope of the experiment receive a methylation probability p(U) and are considered in this analysis.
MetMap annotates CpG island-like regions, called strongly unmethylated islands (SUMIs), based on experimental evidence of their unmethylated state. Although CpG islands are typically identified by high CG content, their key feature is hypomethylation, which is associated with transcriptional function (Illingworth and Bird 2009) (for a detailed discussion of the similarities and differences between SUMIs and CpG islands, see Singer et al. 2010). The human neutrophil methylome contains 20,986 SUMIs that are present in at least one individual (Supplemental Table S2); they largely overlap with the reference CpG island annotation (20), but 4651 have not previously been annotated as CpG islands (Supplemental Table S2). We obtained similar results for the chimp methylome (Supplemental Table S2). Our comparison of the human and chimp methylomes used the 14,316 SUMIs that could be unambiguously mapped between the genomes of the two species.
Methylation probabilities calculated by MetMap were used to compare methylation states between human and chimp, revealing a high degree of similarity between the methylomes but also significant differences. Methylation states are more conserved at sites outside SUMIs than within them. At the 606,496 orthologous human and chimp HpaII sites that were not in SUMIs, methylation probabilities were highly correlated (r2 = 0.74, P < 10−3) (Fig. 1A); 87% of these orthologous sites had p(U) < 0.2, consistent with observations that CGs outside CG islands are usually methylated (Meissner et al. 2008; Lister et al. 2009). The 122,878 methylation probabilities at orthologous HpaII sites within SUMIs show a lower degree of correlation (r2 = 0.61, P < 10−3) (Fig. 1B). However, we observed a higher interspecies correlation when using the average p(U) calculated over all HpaII sites in each SUMI (r2 = 0.65, P < 10−3) (Fig. 1C). The higher conservation of the methylation state of whole SUMIs, relative to the state of individual CGs within a SUMI, suggests that the average methylation of SUMIs is more informative in a comparative study.
Figure 1.
Comparison of the human and chimp neutrophil methylomes. Methylation probabilities p(U) are plotted, with human on the x-axis and chimp on the y-axis. Low p(U) indicates that a site is likely to be methylated, high p(U) indicates that a site is likely to be unmethylated. Individual sites are plotted; grayscale intensity is proportional to the number of sites at each position. Sites deviating from the diagonal have differential methylation in the two species. (A) Methylation probabilities for 606,496 orthologous human and chimp HpaII sites outside 14,316 orthologous human and chimp SUMIs. The sites are highly correlated (r2 = 0.74, P < 10−3 by permutation analysis) indicating conservation of methylation states between the species. (B) Methylation probabilities for 122,878 orthologous human and chimp HpaII sites within the 14,316 orthologous SUMIs. The sites are less correlated than sites outside SUMIs (r2 = 0.61, P < 10−3). (C) Mean methylation probabilities of the 14,316 orthologous human and chimp SUMIs (r2 = 0.65, P < 10−3). The distribution of methylation probabilities along the diagonal appears to be bimodal, with a cluster at p(U) < 0.2 and a cluster at 0.3 < p(U) < 0.9; 15% of HpaII sites within SUMIs are methylated in at least one species.
We used permutation analysis to empirically define thresholds and identify SUMIs whose average methylation differs significantly between human and chimp. Among the 14,316 orthologous human and chimp SUMIs, we identified 1525 that were significantly different at P < 0.01. The differentially methylated SUMIs have a length distribution and CG content very similar to SUMIs in general (Supplemental Fig. S1); SUMIs have many similarities with computationally defined CG islands, as we have discussed elsewhere (Singer et al. 2010). Differential methylation of SUMIs between human and chimp is unlikely to be due to substitutions or polymorphisms at CGs in their genomes: Analysis of CG dinucleotide content in these SUMIs indicates that ∼20% of differentially methylated SUMIs, including some of the most strongly differentially methylated, have no difference in CG content (Supplemental Fig. S2); restriction of the analysis to HpaII sites, whose presence in both species could be confirmed by MspI digestion, identifies essentially the same SUMIs (Supplemental Fig. S3); similarly, restriction of the analysis to HpaII sites that are not polymorphic in dbSNP produced the same results (Supplemental Fig. S4).
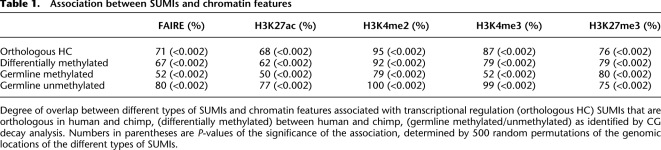
The differentially methylated SUMIs are associated with epigenetic features, including open chromatin (from FAIRE data [Myers et al. 2011]) and histone tail modifications, that are consistent with these SUMIs’ involvement in gene regulation (Table 1). Differentially methylated SUMIs are also modestly but significantly (P = 0.005) more likely to be associated with genes that were found to be differentially expressed in liver, heart, or kidney of human and chimp (Blekhman et al. 2008). Differentially methylated SUMIs are often found near transcription start sites, but not as frequently as SUMIs in general (Supplemental Fig. S5). Taken together, these data suggest that SUMIs in general— and differentially methylated SUMIs in particular—participate in transcriptional regulation, as might be expected since they are essentially CG islands (Singer et al. 2010).
Table 1.
Association between SUMIs and chromatin features
The orangutan methylome provides an outgroup to infer the ancestral state of the methylation differences noted between human and chimp SUMIs. Determination of the methylome of a single young adult male orangutan with the same procedure used for human and chimp identified 11,718 orthologous human-chimp-orang SUMIs. To determine if characteristic methylation states identify a species, we constructed a phylogenetic tree based on mean methylation probabilities of all 11,718 SUMIs that are orthologous in human, chimp, and orang, using each of the four humans, four chimps, and one orang as an independent operational taxonomic unit. To assign a SUMI to either a methylated or an unmethylated state, methylation probabilities were made binary using a stringent threshold of p(U) = 0.2. We calculated a distance matrix using Jukes Cantor-corrected Hamming distances and built a tree using Neighbor Joining. The tree recapitulates the established phylogeny of the three species, with orang as the outgroup and all human individuals clustering together on a branch that is separate from the chimpanzee cluster (Fig. 2). Bootstrap analysis indicates that this topology is highly significant; the tree also indicates that methylation of SUMIs has changed more frequently in human than in chimp, relative to the ancestral state (see also Supplemental Fig. S6). We obtained similar trees using the subset of SUMIs that have the same numbers of HpaII sites in human and chimp (Supplemental Fig. S7A), the subset of HpaII sites whose presence is confirmed by MspI digestion and deep sequencing (Supplemental Fig. S7B), and the subset of HpaII sites that do not have sequence polymorphisms reported in dbSNP (Supplemental Fig. S7C). The similar structures of these trees indicate that the tree structure in Figure 2 is not an artifact due to sequence changes in one of the species. A tree built from the methylation states of all orthologous HpaII sites, irrespective of their location within a SUMI or not, also recapitulates the phylogeny of the three species (Supplemental Fig. S7D).
Figure 2.
Phylogenetic tree built from mean methylation probabilities of the 11,718 orthologous human-chimp-orang SUMIs. The separate clustering of the human and chimp specimen indicates that certain methylation states are stably inherited within each species. The bootstrap values (1000 permutations) are shown next to each branch. The scale bar indicates the number of substitutions per site.
The mechanisms leading to the methylation differences between species are unknown. The separate clustering of humans and chimps is consistent with the stable inheritance of methylation states within the two species; however, it does not demonstrate that those changes were driven by selection or establish their functional significance, and it is also possible that at least some of the differences we observe are caused by factors in the separate environments of the humans and chimps in this study (Carone et al. 2010). The apparent heritability of methylation states could simply reflect the determination of neutrophil methylation states by genome sequence. However, it could also stem from stable maintenance of methylation states, or other epigenetic marks that determine methylation states, in the germline (i.e., pure epigenetic inheritance) (Richards 2006). Taken together with the tree structure in Figure 2, evidence of a correspondence between somatic and germline epigenetic states would support epigenetic inheritance, although it could not prove it because of the possible dependence of epigenetic states on genome sequence or environmental factors.
The possibility that methylation states are heritable raises the question of whether the neutrophil methylation states are related to germline methylation states. If they are, then SUMIs identified as methylated in the neutrophil should have a higher rate of CG decay than SUMIs that are unmethylated in the neutrophil. Methylated CGs undergo mutation to TG (but not to AG or GG) much more frequently than unmethylated CGs (Coulondre et al. 1978). The mutation is heritable if it occurs in the germline; this is the basis for the underrepresentation of the CG dinucleotide in vertebrate genomes (Sved and Bird 1990). We identified the subsets of orthologous SUMIs that had consistent methylation levels in human, chimp, and orangutan, varying from highly methylated to highly unmethylated, retrieved their sequences, and used Ambiore (Hwang and Green 2004) to determine rates of the different substitution types involving the C in a CG dinucleotide. The rate for CG to TG transition was proportional to the probability that a SUMI is methylated (Fig. 3). In contrast, rates of CG transversion to either AG or GG were independent of the neutrophil methylation state of a SUMI. These results are consistent with the hypothesis that neutrophil methylation states are related to germline methylation states. These inferences of germline methylation based on neutrophil methylation are in good correlation with the published methylome of an embryonic stem (ES) cell line determined by whole-genome bisulfite sequencing (corr = 0.43) (Supplemental Fig. S8A). These results reveal a relationship between the neutrophil methylome and the methylome of germ cells but do not mean that methylation is maintained at all stages of germ cell differentiation; germ cells undergo multiple stages of differentiation, and the pattern of CG decay indicates only that methylation is present in at least some of those stages.
Figure 3.
CG decay rates in orthologous human, chimp, and orang SUMIs. Substitution rates were calculated for all different types of substitutions involving the C in a CG dinucleotide ([diamonds] CG→TG, [squares] CG→AG, [triangles] CG→GG) in orthologous human, chimp, and orang SUMIs that have consistent methylation levels in the three species. Only the CG to TG substitution rate is expected to be affected by germline methylation. SUMIs in each p(U) bin have neutrophil methylation probabilities within the indicated bin boundaries in human, chimp, and orang (i.e., SUMIs considered in this analysis did not change neutrophil methylation state during these species’ evolution). Rates of CG to TG, but not to AG or GG, substitution vary as a function of methylation, in a manner consistent with neutrophil methylation states reflecting germline methylation states.
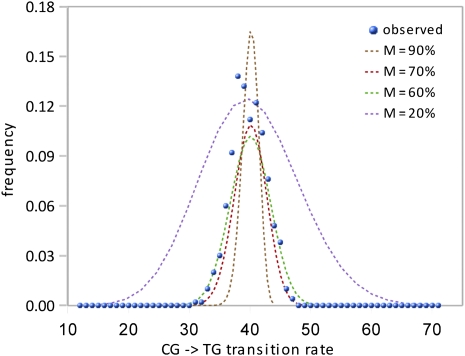
The CG decay result does not indicate how many of the SUMIs that are consistently methylated in the neutrophil are also methylated in the germline. To estimate the fraction of germline-methylated SUMIs, we calculated CG decay rates in a large number of subsets sampled from the 761 SUMIs that are consistently methylated in human, chimp, and orang: The variance of CG decay rates of the subsets reflects the ratio of methylated to unmethylated SUMIs in the whole set. The resulting distribution of CG decay rates is best fit by a simulated distribution with a fraction of methylated SUMIs of ∼0.66 (Fig. 4). This analysis also allows us to estimate the CG to TG transition rate in methylated CGs. The CG to TG rate in the entire set of neutrophil methylated SUMIs is scored at 40 arbitrary units. Adjusting this figure for the proportion of germline-methylated SUMIs derived from the subset analysis (0.66) gives an estimated rate of the CG to TG transition rate in methylated CGs of 60 arbitrary units. This is 13.5-fold higher than the transversion rate for CGs (CG to AG or GG), a figure that is consistent with other measurements of the CG to TG substitution rate (see Discussion).
Figure 4.
Estimation of the proportion of germline-methylated SUMIs. To estimate the proportion of germline-methylated SUMIs in the 761 SUMIs that are methylated in human, chimp, and orang neutrophils, we calculated CG decay rates in 500 random subsets of these SUMIs. The observed distribution of decay rates of the subsets is shown by the blue circles. Simulated distributions calculated for different proportions of methylated SUMIs are shown by the dotted lines. The variance of the distribution is a measure of the proportion of methylated and unmethylated SUMIs in the 761 SUMIs: A small variance indicates that most of the SUMIs are methylated (curve for M = 90%); a large variance indicates that most of the SUMIs are unmethylated (curve for M = 20%). The value of M that gives the best visual fit to the distribution of observed rates is between 60% and 70%.
The SUMIs that are overrepresented in the subsets that generate the highest CG decay rates in Figure 4 are the ones more likely to be methylated in the germline. Gene Ontology (GO) term analysis of these SUMIs shows that they are more frequently associated with genes involved in biological regulation, relative to all orthologous SUMIs (Supplemental Table S3). A similar analysis of the 775 SUMIs that are consistently unmethylated in human, chimp, and orang indicates that they are almost all unmethylated in the germline as well (Supplemental Fig. S9); however, they are not associated with any specific GO term, relative to all orthologous SUMIs. While germline-methylated SUMIs show a slight preference for promoter regions, they are often found at considerable distance from promoters; in contrast, SUMIs that are unmethylated in the germline are largely found near promoters (Supplemental Fig. S5). The fraction of SUMIs predicted to be methylated or unmethylated in the germline by our subset analysis is in very good agreement with the fraction of SUMIs that are methylated or unmethylated in the human embryonic stem cell line H1 (Supplemental Fig. S8B).
We asked if the correlation between methylation states in the neutrophil and the germline is also valid for SUMIs whose methylation state has diverged in human, since these are responsible for the clustering of human and chimp on the tree in Figure 2. If the correlation is valid, we should observe an excess of C/T polymorphism in human SUMIs that have become methylated. We compared the frequency of C/T polymorphism obtained from dbSNP130 in 312 human SUMIs that became methylated and 438 SUMIs that became unmethylated, in human relative to the ancestral state inferred using the orang methylome. In regions that became methylated, we counted 71 CG to TG polymorphisms out of 10,611 total CG sites; in regions that became unmethylated, there were 42 CG to TG polymorphisms out of 13,966 total CG sites. The difference in polymorphism counts is highly significant (P = 3.6 × 10−05, binomial 2-sample proportion test). Although the observed excess of C/T polymorphism is lower than the CG transition/transversion ratio discussed above, changes in methylation state may have occurred too recently for C/T polymorphisms to accumulate; this data indicates only that some human-specific changes in neutrophil methylation reflect changes in germline methylation states.
Discussion
We have carried out a comparison of methylation states in multiple primates. Our comparison of neutrophils in humans and chimps reveals differences that occur more frequently within CpG island-like regions. Humans and chimps segregate on separate branches of a tree built from the neutrophil methylation data, and the CG decay and C/T polymorphism rates indicate that neutrophil methylation is related to germline methylation states. The CG decay analysis is made possible for the first time by the availability of our multispecies methylation data (to determine subsets of SUMIs with the same methylation state in the species analyzed) combined with the availability of genome sequences (to calculate substitution rates). It reveals regions whose methylation state is functionally constrained in human, chimp, and orang.
The choice of cell type for our comparative study was dictated by our requirement for an accessible and homogeneous cell. Somatic tissues are composed of multiple differentiated cell types that have different epigenotypes; differences in the proportions of these cells can potentially have a dramatic impact on the apparent epigenotype of a tissue. Blood is the most accessible tissue, but nucleated blood cells are made up of several cell types in proportions that can vary widely among individuals or even at different times in a single individual. Furthermore, some types of blood cells, such as B and T lymphocytes, are made up of multiple subtypes. Neutrophils are among the most abundant nucleated blood cells, and they are the most homogeneous. They mature in the bone marrow and circulate briefly (∼12 h) as mature cells. Immature forms, principally the band form, make up only a small proportion in healthy individuals (and were <5% in our subjects). Neutrophils form part of a system of nonspecific immunity: They engulf and destroy microorganisms (Nathan 2006) in vertebrates and other animal species using mechanisms that do not require antigen-specific interactions (Segal 2005). This deeply conserved function is unlikely to have changed much in human evolution.
The SUMIs identified by MetMap are defined by criteria that include both CG content and methylation status, so that they are functional equivalents of CG islands (Singer et al. 2010). Our data indicate that ∼2/3 of the SUMIs that are consistently methylated in the neutrophils of all three species are also methylated in the germline. The validity of this analysis is supported by comparison of the ratio of CG transition to transversion rates as inferred by our analysis (13.5-fold; see Results) with ratios calculated in other studies. A study based on Ambiore analysis of 1.7 Mb of sequence containing the cystic fibrosis transmembrane receptor gene found an ∼10-fold excess of CG transitions over transversions (Hwang and Green 2004); this study is probably the most pertinent because it analyzed a large region of DNA with the same computational model used in our study. Other studies have found transition-transversion ratios varying between 11 and 40 (Nachman and Crowell 2000; Arndt et al. 2003; Kondrashov 2003; Zhang et al. 2007); the highest ratio was found in Arndt's study of repetitive elements, and Hwang and Green have suggested that this figure is an overestimate . Thus the rate of CG to TG substitution that we infer for the germline methylated SUMIs is consistent with similar estimates obtained with a variety of methods and experimental data.
The number of SUMIs (i.e., CG islands) that are methylated in the germline may be larger than established by our study, for two reasons. First, because we wanted to compare CG decay rates in methylated vs. unmethylated SUMIs, we restricted our analysis to SUMIs that were consistently methylated or unmethylated in all three species. It is possible, and even likely, that some SUMIs are methylated in the germline of one of the species but not the others, but our analysis cannot address this. Second, there may be SUMIs that are methylated in the germline but unmethylated in neutrophils. A full definition of the set of SUMIs that are methylated in the germline would require analysis of many cell types. However, our analysis (Supplemental Fig. S5) indicates that the great preponderance of SUMIs that are unmethylated in the neutrophil are also unmethylated in the germline.
The findings that somatic methylation states are related to germline methylation states, and that somatic methylation states recapitulate the phylogeny of human, chimp, and orang, raises some intriguing points.
First, the phylogenetic trees built from methylation states show that epigenetic states can be maintained as characters that are predictably transmitted within a species. The ability to reconstruct phylogenies is not unique to CG methylation states: Many phenotypic characters can be used for this purpose (Felsenstein 2004). However, these characters are not independently heritable but reflect genomic sequences that encode the characters, i.e., the character merely acts as a surrogate for information encoded in the genome. This is not necessarily the case with CG methylation: While it may be determined by underlying genotype, it differs from other characters because it is a covalent modification of DNA itself. Thus, it is intriguing to consider that the ability to reconstruct phylogenies using methylation states need not be a simple reflection of the inheritance of DNA sequence but may instead reflect heritable epigenetic information carried by the methylation states themselves. It is, nevertheless, difficult to rule out genetic control of, or contribution to, the epigenetic states we observe. The relationship between methylation state and genotype is complex: Examples exist in which a methylation state is completely determined by the DNA sequence on which it resides (obligate), is influenced but not determined by the DNA sequence (facilitated), or is purely epigenetic and can change without any change in DNA (Richards 2006) . Our data are consistent with any or all of these scenarios.
Furthermore, the evidence for germline methylation states raises the possibility that epigenetic states are inherited directly but does not demonstrate that methylation states per se are maintained and inherited in the germline. Germ cells go through a number of phases of differentiation, in some of which methylation is largely removed from the genome and subsequently replaced (Reik 2007; Popp et al. 2010). The evidence for epigenetic resetting in germ cells implies that if methylation states are heritable, they must be so either because they are determined by DNA sequence or because methylation is faithfully reestablished due to the retention of other components of the epigenome. piRNAs have been implicated in the setting of germline methylation states and are good candidates for such a role (Aravin et al. 2008).
Second, CG decay and C/T polymorphism analyses indicate that somatic methylation of SUMIs often reflects the presence of methylation at some stage in the germline. This observation is made possible for the first time by the availability of comparative methyltyping data, which has allowed us to obtain substitution rates in sequences with the same known methylation state. It raises the possibility that there are previously unsuspected constraints by germline methylation states on somatic states, i.e., that epigenetic information in the germline determines to some extent the epigenetic state of somatic cells. Phenotypic differences mediated by epigenetic inheritance would necessitate this type of control; however, as discussed above, our finding does not demonstrate or require that germline epigenetic states be independent of genotype.
Regardless of the means (genetic or epigenetic) by which inheritance of the methylation state of a regulatory element is mediated, deviation from the inferred ancestral methylation state implies a change in its functional potential. King and Wilson suggested that mutations in transcriptional regulatory sequences would account for the phenotypic divergence of human and chimp (King and Wilson 1975). A change in methylation state can accomplish the same thing as a regulatory mutation, without sequence change. In particular, loss of a methylated state provides a simple mode by which regulatory activity could be expanded without requiring gain-of-function through changes in DNA sequence. We speculate that a SUMI that is methylated in the germline remains methylated in most somatic cell types but is active in a restricted set of cell types in which it is demethylated; germline transition to a demethylated state might broaden the spectrum of somatic cell types in which such a SUMI is active, thus in effect creating a regulatory sequence in the cell types that gain the new activity. Thus, germline changes in methylation states could readily have functional and potentially adaptive consequences. This model of regulatory evolution is simpler than one requiring sequence change to create one or more transcription factor binding sites, but it remains to be seen if methylation states change without associated sequence change. Epigenetic differences, such as those we identified between the human and chimp methylomes, may be a novel source of variation to explain interspecies phenotypic divergence and possibly phenotypic variation within a species.
Methods
Sample collection and isolation
Animal samples were collected with IACUC approval. Human samples were collected with IRB approval after obtaining informed consent. We obtained blood samples from four young adult male humans (age 20–25 yr old) and four age-equivalent male chimpanzees (age 12–16 yr old, which is young adult, after accounting for differences in age of maturity [Fleagle 1999]). Young adults are fully developed but have not yet undergone age-related changes. To further attempt to control environmental variation, we selected individuals who were healthy, well-nourished, afebrile, and not part of any study of infectious agents or other treatments. Immediately after phlebotomy, leukocytes were isolated by Ficoll centrifugation. Neutrophils were isolated from the leukocyte fraction with CD-16 microbeads (Miltenyi). An aliquot of each specimen was Wright-Giemsa-stained and examined microscopically; all specimens contained >99% neutrophils.
Generation of MethylSeq libraries
DNA was extracted by standard methods and digested overnight with HpaII (NEB). HpaII cuts the sequence CCGG; methylation of the central cytosine on one or both strands protects the sequence from digestion with HpaII (Harland 1982). HpaII fragments 50–300 bp in length were isolated on an agarose gel. Single-read sequencing libraries were constructed from human and chimp samples using the standard Illumina kit, and sequenced on an Illumina GA to collect reads of 32 bases. A paired-end sequencing library was constructed from the orangutan sample and sequenced on an Illumina GAII to collect paired reads of 36 bases; only the first read of the paired-end sequencing reaction was analyzed in this study. As a control for the HpaII digestion, the DNA of human sample 1 and chimp sample 1 was digested with MspI, a methylation-insensitive isoschizomer of HpaII; MspI single-read libraries were generated and sequenced as described for the HpaII libraries. Sequencing data were processed by the Illumina Pipeline for base calling and quality filtering. The first three sequencing cycles (corresponding to the “CGG” sequence from the digested HpaII sites) of the orang sample were skipped to facilitate cluster calling. Only reads passing the Illumina quality filter (chastity filter = 0.6) were further processed. Supplemental Table S1 shows the number of reads collected for each sample.
Generation of methylation maps from sequencing data
For a detailed description and explanation of the computational pipeline for analysis of MethylSeq data, see Singer (Singer et al. 2010). Quality-filtered reads were aligned, using Bowtie v0.9.9.2 (Langmead et al. 2009), to their respective reference genomes, which were retrieved from the UCSC Genome Browser (Rhead et al. 2010): human genome (hg18, March 2006), chimpanzee genome (panTro2, March 2006), orangutan genome (ponAbe2, July 2007). We used an alignment policy that allows up to two mismatches in the first 28 bases and reports only reads that align with a single best match (parameters: -all -m 1; in Bowtie v 0.9.9.2, hits are “stratified” by default). Reads whose 5′ end aligned to a CGG corresponding to a HpaII site (Supplemental Table S1) were analyzed with MetMap to assign to each HpaII site within the scope of the experiment a probability of being unmethylated p(U) and a probability of being part of an unmethylated region p(I) (Singer et al. 2010). For each of these sites, a species p(U) was determined by averaging the p(U) values of the four individuals (human and chimp) of that species at that site. For orang, we used the p(U) values from the single individual analyzed. MetMap is available for download at www.cs.berkeley.edu/∼meromit/MetMap.html.
Annotation of SUMIs
MetMap annotates strongly unmethylated islands (SUMIs) as regions in which all HpaII sites have a p(I) greater than 0.1 and at least two HpaII fragments within the region are represented in the MethylSeq data, or by setting a 600-bp interval around each HpaII site that had a p(I) value smaller than 0.1 and a p(U) higher than the prior probability of being unmethylated outside of an unmethylated island (Singer et al. 2010). We then concatenated all overlapping windows and considered as SUMIs those regions in which at least 30% of the HpaII sites had a p(U) larger than the prior-set threshold (0.175) and in which at least two fragments within the region are present in the MethylSeq data. SUMIs share properties with CpG islands (Singer et al. 2010), but because they are defined by experimental data, they are specific to a data set; the process of annotating SUMIs is most similar to the original definition of CpG islands as HTF (HpaII tiny fragment) islands (Bird 1986). While a SUMI is annotated based on the presence of unmethylated HpaII sites in some specimen, it can be scored as methylated if the majority of the HpaII sites within it are methylated. Additionally, since SUMIs are experimentally defined, a region identified as a SUMI in one individual can be methylated in another (Supplemental Table S2).
Cross-genome mapping of SUMIs and HpaII sites
We define as orthologous a HpaII site or a SUMI that passes the following LiftOver procedure: A site or SUMI is mapped from the chimp or the orang genome to the human genome using LiftOver (hgdownload.cse.ucsc.edu/admin/exe/) and then mapped back to the first genome to ensure that it has a unique correspondent in both genomes. Furthermore, all SUMIs whose sequence contained a stretch of more than 10 contiguous “N” in at least one species was excluded from the analysis.
Comparison of human and chimp methylomes
The human and chimp methylomes were compared using scatter plots of the p(U) values of the 729,374 orthologous HpaII sites within the scope of the experiment or of the mean p(U) values of the 14,316 orthologous human-chimp SUMIs. The significance of the correlation values of each scatter plot was determined by permutation analysis (1000 permutations). To determine the significance of differences in the methylation state of human and chimp SUMIs, we generated a null mean-p(U) value distribution assuming that there is no significant difference in methylation between the four human and the four chimp samples. We generated groups for the null distribution by considering all divisions of the human and chimp individuals into two groups, such that in each group there were two humans and two chimps. The distribution of the absolute differences in mean-p(U) values was used to set the thresholds at P = 0.01 (p(U) difference = 0.19). Of the 14,316 SUMIs considered, 1525 had differences with P < 0.01.
Overlap between SUMIs and chromatin features
Human genome (hg18) annotation of chromatin features was obtained from the UCSC Genome Browser. FAIRE data were obtained from http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=219820407&c=chrX&g=wgEncodeChromatinMap, using the union of the data for the following cell lines: GM12878, H1hESC, HUVEC, NHEK, Panislets. Histone tail modification data were obtained from http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=219820407&c=chrX&g=wgEncodeBroadChipSeq, using the union of the data for the following cell lines: GM12878, H1hESC, HMEC, HSMM, HUVEC, NHEK, NHLF (H1hESC data was not available for H3K27ac). A SUMI and a chromatin feature were scored as overlapping if they shared at least one base. To evaluate the significance of the overlap between SUMIs and chromatin features, the location of the SUMIs was randomized 500 times using shuffleBed (Quinlan and Hall 2010). For each randomization, we computed the overlap between randomized SUMIs and chromatin features; P-values are reported as the frequency of the number of times a degree of overlap of a given chromatin feature with randomized SUMIs was equal or greater than that with the original SUMI data.
Association between differentially methylated SUMIs and differentially expressed genes
We considered only SUMIs that have exactly one transcription start site (TSS), as defined by the refGene table of the UCSC Genome Browser, within ±2000 bp from the SUMI boundary. Out of 14,316 orthologous human-chimp SUMIs (HC SUMIs), there are 6457 such cases; out of the 1525 differentially methylated SUMIs (diffSUMIs), there are 507 such cases. Each of these SUMIs was associated with the its proximal RefSeq gene. Blekhman et al. generated human–chimp differential gene expression data from three tissues for 17,231 genes (Blekhman et al. 2008). The intersection of the sets of SUMIs proximal to a TSS with the genes studied by Blekhman et al. identified 5277 HC SUMIs and 384 diffSUMIs for which we had gene expression data. Of the 384 diffSUMIs, 180 (46.9%) matched a gene that was differentially expressed, as determined by Blekhman, in at least one of the three tissues tested. In contrast, of the 5277 HC SUMIs, 2139 (40.5%) matched a gene that was differentially expressed in at least one of the tissues they tested. To evaluate the significance of this difference, we randomized the association between SUMIs and genes and generated a P-value after 1000 iterations— in each iteration, 384 SUMIs were picked at random from the 5277 SUMI set, and the number of those SUMIs for which the associated gene was determined as differentially expressed in one of the tissues was counted. In five of the 1000 cases, that number was larger or equal to 180, resulting in a P-value of 0.005.
Inference of methylation state in the last common ancestor
We used the orangutan methylome as the outgroup to infer the ancestral methylation state of human and chimp SUMIs. Out of the 14,316 orthologous human-chimp SUMIs, we identified 11,718 that had an orthologous orang SUMI meeting the same criteria described above in the section on cross-genome mapping. Only one unrooted tree topology is possible for any three species; for each orthologous human, chimp, and orang SUMI, we calculated the branch length of the unrooted tree from mean p(U) values of the three species using the REML method for continuous traits (Felsenstein 2004). Given the branch lengths, the methylation state of the common ancestor for each SUMI was inferred using squared-change parsimony. We calculated the amount of change in methylation state as the difference for each SUMI between the p(U) values of the common ancestor and the extant species. The value is positive if the extant species is less methylated than the common ancestor, and negative if the extant species is more methylated.
Construction of a phylogenetic tree based on methylation states
We built a phylogenetic tree based on the average methylation states of the 11,718 orthologous human-chimp-orang SUMIs. Each SUMI was assigned a p(U) value of 1 if its mean p(U) score was larger than 0.2, and 0 otherwise (this conservative threshold of 0.2 for calling a SUMI “methylated” is consistent with the CG decay analysis—see Fig. 3). The Jukes-Cantor distances (for binary characters) were calculated for each pair of individuals to obtain a distance matrix. We used the SplitsTree program (Huson and Bryant 2006) to construct a phylogenetic tree using the Neighbor-Joining algorithm and to bootstrap the resulting tree (1000 permutations) (Felsenstein 2004).
CG decay analysis
From the set of the 11,718 orthologous human, chimp, and orang SUMIs, we determined the subsets of SUMIs in which all the three species had mean p(U) within defined thresholds, computed multiple sequence alignments using ClustalW (Larkin et al. 2007), and concatenated the alignments of all SUMIs within each subset. The concatenated multiple sequence alignments were submitted to Ambiore (Hwang and Green 2004) to calculate the substitution rates of all possible substitution types involving the C of a CG dinucleotide.
To analyze CG decay in SUMIs that had changed methylation state in the human lineage, we identified human SUMIs whose methylation difference from the inferred last common ancestor was p(U) < −0.19 (identifying SUMIs that have become more methylated), or p(U) > 0.20 (identifying SUMIs that have become less methylated). For each SUMI, C/T polymorphisms mapping to a CG dinucleotide were retrieved from dbSNP build 130; only polymorphisms validated as “by-hapmap” and “1000genome” were used.
Estimation of the proportion of germline methylated/unmethylated SUMIs from CG decay analysis
The 761 SUMIs that are consistently methylated in human, chimp, and orang neutrophils will contain m SUMIs that are methylated in the germline, and u SUMIs that are unmethylated in the germline. We sampled 500 random subsets of size 70 (without replacement) from these 761 SUMIs, calculated the C→T transition rate for each subset, and obtained the distribution of the frequencies of the transition rates in the subsets (“observed” line in Fig. 4). To estimate the number of methylated SUMIs (m) within the set of 761 SUMIs, we assumed that the observed transition rate follows the simple model:
where obs_rate is the observed transition rate, m is the number of methylated SUMIs, u the number of unmethylated SUMIs, Tm is the transition rate C→T for methylated sequences, and Tu the transition rate C→T for unmethylated sequences. We set obs_rate = 40; this is the value calculated by Ambiore for the set of 761 SUMIs (Fig. 3, 0–0.2 bin). Tu can be estimated from either the C→A and C→G transition rates at Tu = 4.4 (Fig. 3), or from C→T in the set of SUMIs that are consistently unmethylated in human, chimp, and orang at Tu = 9.3 (Fig. 3); the analysis described below was performed for both values, and the results obtained were similar (difference < 5%). For different values of m and u = 761−m, we calculated Tm using the equation above and the expected distribution of the fraction of m and u in the subsets of size 70 using the appropriate hypergeometric distribution. We then calculated the simulated obs_rate for the different subsets using the assumed model; the expected obs_rate distributions are plotted in Figure 4. Under the simplifying assumptions that Tm and Tu are represented by single values and do not depend on SUMI length and that Ambiore and the simplified model used for the simulations have the same variance, we can estimate the number of methylated SUMIs present in the data set by the value of m for which a simulated distribution is closest (in variance) to the distribution observed by computing transition rates for the subsets. From this analysis, we expect that ∼500 SUMIs (i.e., 2/3 of 761) are methylated in the germline; the identity of the germline-methylated SUMIs is inferred by determining which SUMIs are overrepresented in the subsets that generate the highest C→T rates. We carried out a similar subset analysis on the 775 SUMIs found in the methylation bins 50–60, 60–70, 70–80, and 80–100 of Figure 3.
GO term analysis
We used GREAT (McLean et al. 2010) for all GO term analyses described in this study. We used the following parameters to associate genomic regions with genes: Proximal regulatory domain = ± 5kb; distal regulatory domain = 500kb. As a background set, we used the 11,718 orthologous human, chimp, and orang SUMIs. As the test set for SUMIs with differential methylation in human and chimp, we used the 458 SUMIs with either p(U) < 0.2 in human and p(U) > 0.3 in chimp and orang, p(U) > 0.3 in human and p(U) < 0.2 in chimp and orang, p(U) < 0.2 in chimp and p(U) > 0.3 in human and orang, p(U) > 0.3 in chimp and p(U) < 0.2 in human and orang. The subset analysis of CG decay rates shows that ∼500 of the 761 SUMIs that are consistently methylated [p(U) < 0.2] in human, chimp, and orang neutrophils are also methylated in the germline; as the test set for germline methylated SUMIs, we used the 493 SUMIs that were most represented four or more times among the subset of SUMIs with C→T rate > 43 in the subset analysis of the 761 SUMIs described in the “CG decay analysis section”. As the test set for germline-unmethylated SUMIs, we used the 775 SUMIs found in the methylation bins 50–-60, 60–70, 70–80, and 80–100 of Figure 3, which are shown to be unmethylated in the germline by the subset analysis of CG decay rates.
Data access
The sequence data used in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE22376.
Acknowledgments
This work was supported by NIH grants HL084474 (D.B.), ES016581 (D.I.K.M.), and CA115768 (D.I.K.M.). J.D. was supported by the California Institute of Regenerative Medicine. This study used biological materials obtained from the Southwest National Primate Research Center, which is supported by NIH-NCRR grant P51 RR013986. We thank Cole Trapnell for help with Bowtie alignments, and Cath Suter for helpful comments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122721.111.
References
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt PF, Petrov DA, Hwa T 2003. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol Biol Evol 20: 1887–1896 [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES 2007. The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Bird AP 1986. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213 [DOI] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet 4: e1000271 doi: 10.1371/journal.pgen.1000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM 2004. Comparative genomics at the vertebrate extremes. Nat Rev Genet 5: 456–465 [DOI] [PubMed] [Google Scholar]
- Brink RA 1960. Paramutation and chromosome organization. Q Rev Biol 35: 120–137 [DOI] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. 2009. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res 19: 1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 2004. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- Feng S, Jacobsen SE, Reik W 2010. Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleagle JG 1999. Primate adaptation and evolution, p. 257. Academic Press, San Diego, CA [Google Scholar]
- Grossman SR, Shylakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, Zuk O, et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886 [DOI] [PubMed] [Google Scholar]
- Harland RM 1982. Inheritance of DNA methylation in microinjected eggs of Xenopus laevis. Proc Natl Acad Sci 79: 2323–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Hwang DG, Green P 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci 101: 13994–14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands—“a rough guide.” FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kondrashov AS 2003. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 21: 12–27 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C 2006. Neutrophils and immunity: Challenges and opportunities. Nat Rev Immunol 6: 173–182 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168 doi: 10.1371/journal.pgen.0020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM 2006. Accelerated evolution of conserved noncoding sequences in humans. Science 314: 786 doi: 10.1126/science.1130738 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Beck S 2006. Epigenetic variation and inheritance in mammals. Curr Opin Genet Dev 16: 573–577 [DOI] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ 2006. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet 7: 395–401 [DOI] [PubMed] [Google Scholar]
- Segal AW 2005. How neutrophils kill microbes. Annu Rev Immunol 23: 197–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Boffelli D, Dhahbi J, Schoenhuth A, Schroth GP, Martin DIK, Pachter L 2010. MetMap enables genome-scale Methyltyping for determining methylation states in populations. PLoS Comput Biol 6: e1000888 doi: 10.1371/journal.pcbi.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci 87: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bouffard GG, Wallace SS, Bond JP 2007. Estimation of DNA sequence context-dependent mutation rates using primate genomic sequences. J Mol Evol 65: 207–214 [DOI] [PubMed] [Google Scholar]