Abstract
Drug development efforts against cancer are often hampered by the complex properties of signaling networks. Here we combined the results of an RNAi screen targeting the cellular signaling machinery, with graph theoretical analysis to extract the core modules that process both mitogenic and oncogenic signals to drive cell cycle progression. These modules encapsulated mechanisms for coordinating seamless transition of cells through the individual cell cycle stages and, importantly, were functionally conserved across different cancer cell types. Further analysis also enabled extraction of the core signaling axes that progressively guide commitment of cells to the division cycle. Importantly, pharmacological targeting of the least redundant nodes in these axes yielded a synergistic disruption of the cell cycle in a tissue-type-independent manner. Thus, the core elements that regulate temporally distinct stages of the cell cycle provide attractive targets for the development of multi-module-based chemotherapeutic strategies.
Current approaches aimed at anti-cancer drug development primarily focus on identifying those signaling intermediates where mutations have led to constitutive activity in a given cancer. The aim then is to develop inhibitors against these targets (Evan and Vousden 2001; Vermeulen et al. 2003). The prevailing systems view, however, describes the signaling machinery as being organized into a complex network (Barabasi and Oltvai 2004; Zhu et al. 2007) that exhibits a nonlinear response behavior (Alon 2007). This implies that the effects of inhibiting an intermediate need not necessarily be the inverse of that which is obtained upon its constitutive activation. In this context, at least one factor influencing the outcome would be the level of functional redundancy exhibited by this intermediate (Tononi et al. 1999; Edelman and Gally 2001). That is, although activation of mitogenic pathways may derive from an oncogenic mutation in a given signaling intermediate, inhibition of this intermediate may, however, have only a minimal effect if its functional role is compensated for through contributions from other intermediates.
The above possibility is especially relevant for cancer cells where mutations in more than one signaling molecule are often the norm (Vogelstein and Kinzler 2004). In other words, the greater degree of plasticity associated with oncogenic pathway activation, relative to its suppression, also indicates that the ideal targets for pathway inhibition need not necessarily coincide with those that are involved in its activation. Such increments to our understanding of the complex properties of biological systems illuminate that drug development efforts will be significantly aided by a better resolution of the signaling circuitry that controls the cell cycle, as well as a description of the least redundant nodes (i.e., functionally least replaceable nodes) that participate in this process.
Despite accumulation of information on mitogen-activated signaling cascades, a clear picture of how they integrate to modulate the cell cycle program is, however, still lacking (Papin et al. 2005). Indeed, the current challenge is to develop approaches that can distill the available information and provide a coherent view of the core pathways involved (Papin et al. 2005; Del Sol et al. 2010). In the present study, we combined the results of a siRNA screen targeting the signaling machinery with graph theoretical analysis to extract the core modules that processed mitogenic signal in the context of the individual phases of the cell cycle. We demonstrate that these modules, indeed, constitute functionally conserved features of mitogen-dependent signaling networks, and that the least redundant nodes present in them provided effective targets for chemotherapy. Thus, in addition to providing new insights into mechanisms regulating cell cycle progression, our results also highlight that multi-module targeting—wherein the temporal dimensions of a biological process are also taken into account—may provide a useful refinement to current drug development efforts.
Results
An RNAi screen targeting cellular kinases and phosphatases identifies regulators of the cell cycle
To identify signaling molecules that regulate cell cycle progression, we performed a siRNA screen against all known kinases (758 proteins) and phosphatases (294 proteins) in cycling cells of the murine B lymphoma cell line CH1. This cell line has been previously used as a model system to study signaling events regulating the cell cycle (Aflakian et al. 2009; Jamal et al. 2010). Changes in cell cycle phase distribution at 72 h after siRNA transfection were monitored by flow cytometry (Methods). Since our aim was to decipher processes regulating mitogen-dependent cell cycle progression, only those effects that perturbed the cell cycle phases without significantly affecting cell viability were considered. A primary screen followed by a validation exercise (Fig. 1A; Supplemental Experimental Procedures) identified 38 kinases and five phosphatases whose silencing yielded significant effects (Supplemental Table S1). A microarray analysis subsequently confirmed that the genes coding for all of these target proteins were, indeed, expressed in CH1 cells (Supplemental Table S2; Aflakian et al. 2009). A subsequent analysis using the Novoseek gene–disease relationship scores identified nearly half of these target molecules to exhibit a strong association with either one or more forms of cancer (Supplemental Fig. S3), thus emphasizing the functional relevance of the hits identified by the screen.
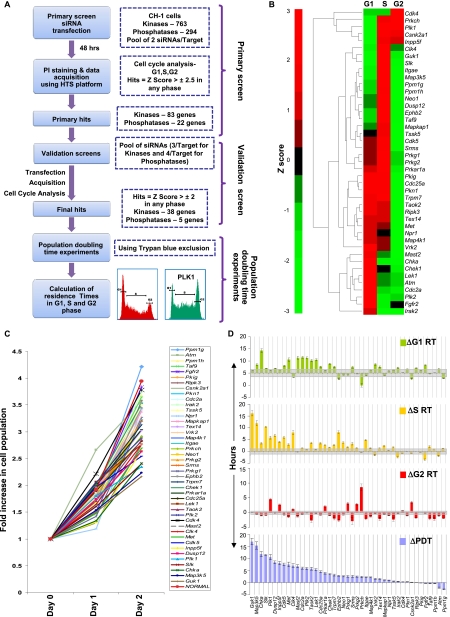
Figure 1.
Design of the siRNA screen and the cell cycle phase–specific effects of the identified targets. (A) Schematic of the strategy used to identify genes involved in cell cycle regulation. The overall z-factor obtained for our screen was 0.56, confirming its high overall quality. Details of the design, assay validation, and target verification are provided in the text, Supplemental Experimental Procedures, and Supplemental Figures S1 and S2. (B) The hierarchical clustering of the siRNA hits according to the observed phenotype on cell cycle. The dendogram represents the various clusters of genes identified by our screen for the observed cell cycle distribution pattern. This pattern is based on the normalized z-scores of each of the siRNA hits in the respective cell cycle phases. (C) The range of observed changes in proliferation rate. The population growth curves of CH1 cells obtained in response to transfection with either nonsilencing siRNA (NORMAL), or with siRNA against each of the validated hits are plotted. Here, cells were plated at 48 h after siRNA transfection (Day 0), and results are plotted in terms of the fold increase in cell number at the indicated time points. Values are the mean of three separate determinations. The subset highlighted by the bracket indicates those target-specific siRNAs that produced a significant increase in the population doubling times (P-value < 0.05, as indicated by the asterisk). (D) A further analysis wherein the results in panels B and C were combined to determine the effects of siRNA-mediated target silencing on residence time (RT) of the individual cell cycle phases. Changes (Δ), relative to the corresponding values in GFP-silenced cells, in PDT and RT of the individual phases are shown here as the mean (±SD, gray bar) of three experiments. Details of their calculation are provided in Supplemental Figure S3.
A hierarchical clustering analysis of the nature of the effect caused by depletion of these proteins revealed that the most common one was that of a simultaneous increase in the pool size of cells resident in the G1 and S phases (Fig. 1B). In addition, there were also a significant number of cases in which siRNA treatment specifically increased either the G1 (12), or the S (nine) phase populations, whereas only a limited number of siRNAs had any detectable affect on the G2 phase (Fig. 1B).
Silencing of signaling intermediates induces differential effects on cell doubling times
Examination of the effects of target-specific depletion on cell population doubling times (PDTs) yielded a diverse spectrum ranging from negligible to a marked reduction in the proliferation rate (Fig. 1C). While the PDT of control cells (i.e., transfected with nonsilencing siRNA) was 26 ± 3 h, 16 of the 43 siRNAs tested produced a significant extension with PDTs of 30 h or more. The individual PDT values were then used to calculate the residence time (RT) of cells in G1, S, and G2 phases under each of the knockdown conditions (Supplemental Fig. S3; Supplemental Experimental Procedures), and these results are compared to the control value in Figure 1D. It is evident that each siRNA pool caused specific perturbations in one or more of the individual phases of the cell cycle, although the effects differed widely between the various knockdowns. For example, the G1 phase RT varied from about twofold lower (in five cases) to about twofold greater (in 22 cases) than that in control cells (Fig. 1D). No significant effect was observed for the remaining 16 siRNAs (Fig. 1D). A similar degree of variation was also observed for the S phase, whereas the G2 phase was only sensitive to 19 of the siRNAs tested, of which eight caused an increase in its RT (Fig. 1D).
To simplify our subsequent analysis, we categorized the target-specific siRNAs solely on the basis of the phase-specific extensions in RT that they induced (Fig. 1D). That is, although in some cases extension in RT of a given phase was compensated for by shortening of another phase, the latter effect was not taken into account. Accordingly, the largest group of siRNAs (15) corresponded to those inducing an extension of both the G1 and the S phases, whereas that of only the G1 phase was observed for an additional group of 12 cases (Fig. 1D). Another set of nine siRNAs specifically extended the S phase alone. Perturbations prolonging the G2 phase were seen in response to four siRNAs, and the remaining three target-specific siRNAs affected either the S and G2 (two), or the G1 and G2 phases (Fig. 1D). These results are consistent with the known biological roles of these target proteins in the literature (Supplemental Table S3).
Since the role of mitogenic signals is to move cells through the G1 phase and then ensure their entry into the S phase (Jones and Kazlauskas 2000; Sears and Nevins 2002), our subsequent analysis concentrated on only those targets where depletion resulted in an extension in RT of either specifically the G1, the S, or both of these phases simultaneously. This latter group of targets was termed as those extending RT of the “G1S phase,” and available literature suggests that it likely represents a window extending from the later stages of G1 to initiation of the S phase (Jones and Kazlauskas 2000).
Defining the source and target relationships for phase-specific regulation of the cell cycle
An examination of results from two previously reported genome-wide screens performed in the human cell lines HeLa and U2OS (Mukherji et al. 2006; Kittler et al. 2007) revealed that there was a <5% overlap between them at the level of the signaling molecules that were identified. Relative to this, the overlap between the signaling molecules identified here and those in either of these two previous reports was between 5% and 10%. This poor concordance exemplifies the emerging limitations of RNAi-based screening approaches where—in addition to the preponderance of false negatives—differences in cell type and the experimental conditions used, also significantly influence the outcome (Goff 2008; Bushman et al. 2009). It was, therefore, imperative to explore whether a further interrogation of our results would yield more meaningful, and consistent, insights into signal-mediated regulation of the cell cycle.
The trade-off between sensitivity and robustness constrains the architecture of the signaling network into a bow tie—or hourglass—structure, which represents the convergence of diverse and redundant input processes onto a conserved core module or set of proteins (Csete and Doyle 2004; Supper et al. 2009). Such core elements function as the key regulators of plasticity in the cellular response, by calibrating a range of output processes (Kitano and Oda 2006; Del Sol et al. 2010). Thus, mitogen-activated signaling networks also likely incorporate such conserved regulatory elements that process input signals and translate them into the output response of cell cycle progression. We next attempted to reconstruct the network of pathways through which the effects of the identified targets were likely enforced.
Consequently, we defined the hits identified by our siRNA screen as “source nodes” (Fig. 2A, nodes in dark red) to indicate that these signaling proteins constitute sources for inducing perturbations in the cell cycle. Furthermore, since the effects of siRNA-mediated depletion of each of these source proteins was likely to be eventually enforced through modulations in activity of one or more constituents of the core cell cycle regulatory machinery, we considered this latter group of proteins as target nodes (Fig. 2A, light red nodes). The objective then was to trace the network of pathways connecting the sources to the targets and search for any associated topological features (shown as green nodes linked through dotted edges) that may be central to the regulation of G1- and S-phase progression. While highlighting regulatory pathways, we also anticipated that this strategy would illuminate any other key regulators that may have been missed by the screen.
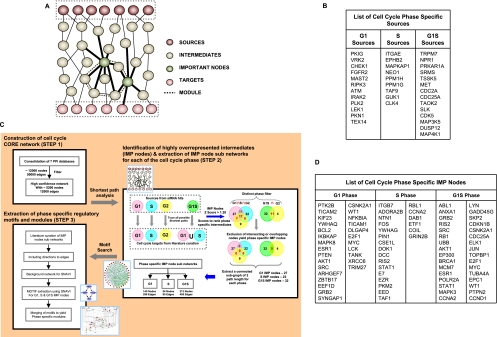
Figure 2.
A systems approach to the analysis of cell cycle regulation and the identification of phase-specific IMP nodes. (A) The rationale underlying the examination of source to target subnetworks in order to identify key intermediate effectors of cell cycle perturbation. Here the sources denote the hits identified by the siRNA, while the targets denote the key molecules involved directly in progression of cell cycle and growth. (B) The “source” characterization of the hits obtained in the siRNA screen, based on the phase-specific effects obtained. The genes described here are the human orthologs of the murine counterparts (see text). The flowchart in panel C provides a stepwise summary of the in silico methodologies used for eventual identification of the phase-specific regulatory modules. STEP 1 describes the stages involved in delineation of the core network, whereas STEP 2 illustrates the subsequent analysis of this core network to extract the phase-specific IMP node subnetworks. Finally, STEP 3 depicts the stages through which an analysis of the IMP node subnetworks eventually yielded the phase-specific modules. These individual steps are described in detail in the Supplemental Experimental Procedures. (D) The IMP nodes eventually identified through this network analysis for regulation of the G1, S, and G1S phases.
Proteins whose depletion resulted in an extension in RT either of the G1, the S, or the G1S phases were labeled as source nodes specific for the respective phases (Fig. 2B). This was then followed by an attempt to define the putative end targets for each of these source node groups, that is, those constituents of the core cell cycle machinery that directly influence progression of cells through the G1 or the S phases (Murray 2004). Combining literature survey with KEGG, Reactome, NCI, and INOH pathway databases (Kanehisa and Goto 2000; Matthews et al. 2009; Schaefer et al. 2009) identified 74 such proteins, which could then be grouped either as G1- or S-phase-specific “targets” based on their known functional roles. Supplemental Table S4 lists these targets, along with links that describe evidence in support of their categorization. Due to the contiguous nature of cell cycle progression, however, it was not possible to distinguish G1S-specific targets from those involved either in the G1 or the S phases. Therefore, the list of G1- and S-phase targets were combined together and taken as targets for the G1S phase.
In silico network construction and identification of important intermediates using shortest path analysis
Our next goal was to map the network of pathways linking the source and target nodes in each of the relevant phases. The approach used is outlined in Figure 2C. As the first step, we consolidated the mammalian protein–protein interactions (PPI) from BIND, IntAct, HPRD, BioGRID, MINT, Reactome, and NetworKIN databases to build an undirected high-confidence network consisting of 5200 nodes and 12,000 interactions (or, edges) (see Supplemental Table S5; Supplemental Materials). To identify those molecular intermediates that were enriched in a cell cycle phase–specific manner (IMP nodes), we next traced all possible shortest paths from each of the classified phase-specific sources (G1, S, or G1S) (see Fig. 2B) to all of the corresponding phase-specific targets described in Supplemental Table S4. Shortest paths were traced from each G1source to all G1target, from each Ssource to all Starget, and also from all G1Ssource to all G1Starget (STEP 2, Fig. 2C).
Counting the number of times an intermediate occurred in the ensemble of shortest paths for each phase (Supplemental Table S6), and calculating the corresponding Z-scores, enabled short-listing of the high scoring phase-specific IMP nodes; while normalizing for differences in the number of source and target nodes between the different cell cycle phases (STEP 2, Fig. 2C; Supplemental Table S6; Supplemental Experimental Procedures). Here, our inclusion of a G1S phase ensured that any relevant node that may potentially be involved in regulating both the G1 and S phases was not missed. This process identified 27, 22, and 32 IMP nodes from the G1, S, and G1S phase-specific shortest paths, respectively, and these are listed in Figure 2D.
Delineation of IMP node-dependent regulatory modules
While the IMP nodes identified represented molecule sets that were enriched in pathways regulating the distinct cell cycle phases, it was, however, important to delineate the temporal/topological regulatory features of mitogen-induced progression from G1 to S. We first took the IMP nodes from each cell cycle phase as the seed nodes and extracted connected subnetworks of two-path length cutoff from the core network as described in Supplemental Table S5 (STEP 3 of Fig. 2C). The resulting G1 IMP node network was composed of 145 nodes and 296 edges, whereas the S IMP node network contained 54 nodes and 85 edges, with 215 nodes and 495 edges describing the G1S IMP node network (Fig. 3A–C). These subnetworks likely encapsulate the specific elements responsible for regulating the respective phases of the cell cycle.
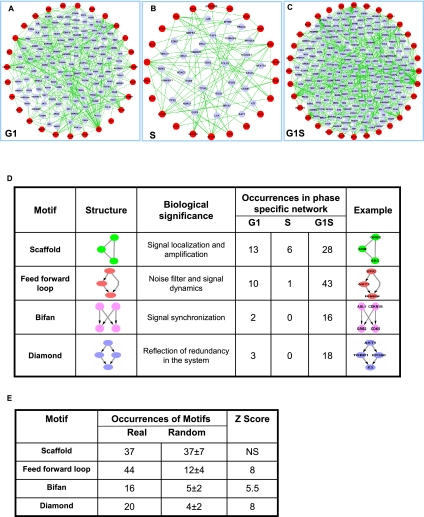
Figure 3.
Extraction of phase-specific subnetworks and their constituent regulatory motifs. (A–C) The phase-specific IMP node subnetworks extracted from our cell cycle core network for G1, S, and G1S phase regulation. The IMP nodes are defined in red, while the blue nodes denote the intermediate node linking any two or more IMP nodes. (D) A compilation of the different functional motifs identified in each of the phases, along with a brief description of their biological implications. In addition, a typical topological representation of each kind of motif is also shown. Here both three- and four-node motifs in the FFL categories have been grouped together for the sake of convenience. (E) Comparison of the number of different motifs obtained for the integrated IMP node network (Real) versus that in a hundred randomized networks (Random, values given as mean ± SD). In these randomizations both the nodes and the number of links were kept at a constant. The statistical significance of the difference in numbers between the Real and Random cases is indicated by the corresponding Z-scores, which were calculated as (<Real> − <Random>)/SD). Here NS indicates that there was no significant change in the case of scaffolds. This is not surprising given that scaffolds are composed of undirected edges.
To next delineate regulatory elements present in these IMP node networks, we adopted the approach outlined in STEP 3 of Figure 2C. First, a survey of the literature assisted in clarifying the nature of the biochemical reaction represented by each of the edges in the IMP node subnetworks (compiled in Supplemental Table S7). The resulting classification of these edges into functional categories of activation, inhibition (both directed), or neutral (undirected) facilitated conversion of the individual phase-specific IMP node subnetworks into mixed networks that consisted of directed and undirected edges. Taking each of these, we then probed for any embedded motifs that also included the respective IMP nodes. Here, we used SNAVI (Ma'ayan et al. 2009), and Figure 3D lists the various types of motifs that could be identified and the number of times that these occurred in each of the individual phase-specific IMP node subnetworks. Integrating the phase-specific IMP node networks and randomizing the edges while keeping the degree distribution constant over a hundred iterations led to a marked reduction in the number of motifs involving directed edges (i.e., excluding the scaffolds) (Fig. 3E). This confirmed that the phase-specific motifs identified were, indeed, statistically significant. Furthermore, our identification that the relative enrichment of signal-dependent regulatory motifs follows the order of G1S > G1 > S is consistent with fact that the G1/S checkpoint represents the step that is most tightly regulated by growth factor-mediated signaling (Zetterberg et al. 1995; Sears and Nevins 2002).
Extracting cell cycle regulatory modules and the identification of vulnerable nodes
Motifs in a network do not function in isolation but, rather, integrate with one another to form discrete clusters (Dobrin et al. 2004). In our case, such motif clusters can be considered to represent the core units that process mitogenic signals to drive the cell cycle. Therefore, we merged all motifs detected in the G1-specific IMP node network to generate the G1-phase motif cluster. Similarly, motifs present in the G1S- or the S-specific IMP node networks were also independently merged to generate motif clusters defining the regulation of corresponding phases. Importantly, these motif clusters exhibited a high degree of edge density when compared with random subnetworks extracted from IMP node networks (see Supplemental Materials). Thus, these phase-specific motif clusters likely define regulatory modules that specifically function during the cell cycle phases from which they were derived. A compilation of the nodes present in these modules, along with a classification of the functional groups to which they belong, is provided in Supplemental Table S8.
Having extracted the cell cycle phase–specific regulatory modules, we next sought to identify those constituent nodes that were most critical for their functioning. For this we used the centrality measures of “stress” and “betweenness.” Stress and betweenness represent complementary indices that together describe the functional importance of a node in a regulatory module (Newman 2005; Manimaran et al. 2009). We calculated the stress and betweenness measures for each node in each phase-specific module, and the resulting plot of these values for the G1- and G1S-specific modules is shown in Figure 4A. Here, as expected, the S-specific module was composed of only a few nodes (see Supplemental Table S8), with just two giving any measurable value for stress and betweenness. The modules for the G1, G1S, and S phases are shown in Figures 4B–D, where the nodes are color-coded according to their stress and betweenness values. The larger the size of the node, the higher is the respective value of stress, whereas the betweenness value is indicated by the shade of the color for that node.
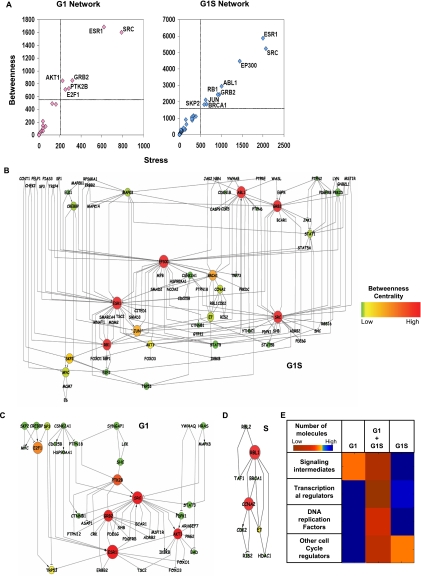
Figure 4.
Delineating phase-specific regulatory modules and defining the vulnerable nodes. (A) Summary of the results of our analysis of centrality measures of the nodes in each of the phase-specific regulatory modules. This is depicted as a Stress-versus-Betweenness plot for all of the molecules in the G1 and G1S cell cycle modules. Only those nodes that are above the cutoff of 2.5 times that of the mean value (indicated by the dotted line; see text) are identified here. (B–D) The IMP node-based regulatory modules obtained for each of the individual phases. These represent phase-specific modules extracted by merging the motifs identified for the G1, G1S and S phases, respectively. The nodes are color-coded according to their betweenness centrality measure. The size of the nodes represents the stress parameter, where the size of the nodes increases with increasing stress value. The gradation of the betweenness values is defined by the color bar. (E) The stage-dependent modulation in biochemical activities of the core cell cycle regulatory network spanning from early G1 to completion of the S phase. This is presented as a heat map that defines the percent contribution of signaling, transcription regulatory, DNA replicative, and cell cycle regulatory molecules, in each of the indicated windows.
We next segregated the functionally most significant nodes as those with stress and betweenness values that were both greater than 2.5-fold that of the mean for all nodes in the corresponding module (Supplemental Experimental Procedures; Fig. 4A). The composition of the resulting shortlist was then compared across the individual phases. Intriguingly, the high-stress and high-betweenness nodes identified for the G1, G1S, and S windows of the cell cycle did not constitute distinct sets of molecules. Rather, the group of vulnerable nodes identified for a given window overlapped significantly with that for the adjacent window. For example, while PTK2B was exclusively present in the G1 module ESR1, SRC, and GRB2 commonly occurred as high-stress and high-betweenness nodes in both G1 and G1S modules (Fig. 4A). Interestingly, although AKT1 and E2F1 were also present in both modules (Supplemental Table S8), these proteins exhibited properties of high stress and betweenness only in the context of the G1 module (see Fig. 4A–C). The nodes exclusively present in the G1S module were ABL1, JUN, BRCA1, EP300, and RB1 (Fig. 4A). In the S module, only CCNA2 and RBL1 showed functional nonredundancy (i.e., high stress and betweenness) (Fig. 4D). The observed overlap in structure between modules describing the adjacent windows of the cell cycle is consistent with the fact that individual stages of this process do not represent discrete events, but rather reflect windows that capture temporally defined stages of a contiguously evolving biochemical cascade.
Interestingly the majority of the G1-unique nodes represented signaling molecules, whereas nodes specific for the G1S IMP node network were distributed between signaling molecules, transcription regulators, and proteins involved in DNA replication (Supplemental Table S8). The nodes enriched in the S-phase network included CDK2 and CCNA2, which form a complex that drives cells through the S phase (Arias and Walter 2007). Other such enriched nodes included here were the damage repair protein BRCA1, the DNA replication factor RIS2, and RBL1—a key regulator of entry into cell division (Harper and Elledge 2007). Figure 4E summarizes the distinct classes of biochemical activities described by each of the IMP node subnetworks in the form of a heat map. The observed layering of the initial activation of signaling events (G1 and G1S) over the overlapping phase involving the induction of transcriptional processes (G1S), and subsequent recruitment of the DNA replication machinery (G1S and S) accurately recapitulates the broad sequence of events that govern cell cycle progression.
These collective results, therefore, substantiate the information-rich nature of the IMP node modules shown in Figure 4. They also support that these modules together capture the dynamic transitions in biochemical activities that enforce commitment of cells to the division cycle.
Verifying phase specificity of vulnerable nodes
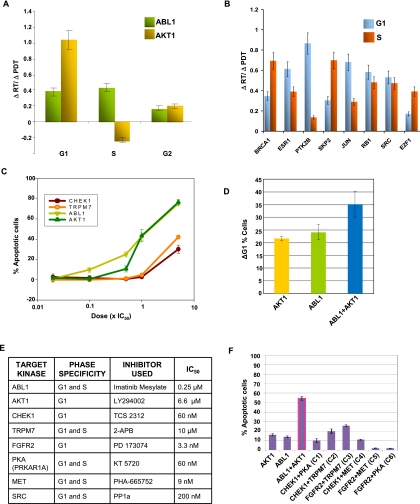
To further validate relevance of the phase-specific modules, we experimentally verified both the selection of high-stress, high-betweenness nodes and the characterization of their phase specificity. We first compared the effects obtained upon perturbation either of AKT or ABL. Our IMP node modules had suggested that the functional significance of AKT1 was restricted to the G1-specific module, whereas ABL1 was specific to the G1S module. Therefore, we treated CH1 cells with pharmacological agents that either inhibited AKT phosphorylation (LY294002) or ABL activity (imatinib mesylate). The concentration of both inhibitors was adjusted such that a similar PDT was obtained in both cases, thereby enabling a direct comparison between the RT of the individual phases. Inhibition of AKT resulted in a specific extension of the G1 phase, with a marginal reduction in RT of the S phase of the cycle (Fig. 5A). In contrast, the increased PDT obtained upon ABL inhibition involved contributions from both the G1 and S phases (Fig. 5A). These results therefore verify the distinctions in phase specificity that were earlier suggested for these two nodes, on the basis of centrality index estimations.
Figure 5.
Vulnerable nodes provide targets for synergistic disruption of the cell cycle. (A) The effects of inhibition of either AKT or ABL on RTs of the individual phases. The increase in RT (ΔRT) of a given phase, relative to untreated cells, is expressed as a ratio of the corresponding increase—again relative to untreated cells—in PDT (ΔPDT). Here, LY294002 and imatinib mesylate were used at concentrations that were half of their respective IC50 values, and the increase in PDT obtained over that of untreated cells was 3 h. The effects of perturbation of the remaining high-stress, high-betweenness nodes identified in A are similarly shown in B. With the exception of SRC, where PP1a was used for inhibition, perturbation was achieved through siRNA-mediated depletion in all cases. Accordingly, the control cells used for the purposes of comparison were those treated with nonsilencing (i.e., GFP-specific) siRNA. The results for GRB2 and EP300 silencing are not included because high levels (>80%) of cell death were obtained in these cases. Values in both panels are the mean (±SD) of three independent experiments. No significant effect on RT of the G2 phase was noted in any of the cases. See also Supplemental Figure S4. (C) The dose-response profile for apoptosis in CH1 cells treated with pharmacological inhibitors of the indicated kinases. Doses for each inhibitor used were in multiples of their corresponding IC50 values as noted. Values (mean ± SD of three experiments) are expressed as the percent of apoptotic cells obtained 72 h later, after normalizing for spontaneous apoptosis. (D) The bar graph shows the corresponding accumulation of CH1 cells in the G1 phase obtained at 18 h with the 5 × IC50 concentration of inhibitors. Values (mean ± SD of three experiments) are expressed as the increase in percent of the G1 population, over that in untreated cells. (E) The target kinases examined, their phase specificity, the pharmacological inhibitors used for inhibition of these kinases, and the corresponding IC50 values of these inhibitors. (F) CH1 cells were treated with inhibitors against the indicated kinases (or kinase combinations), and the consequent effect on cellular apoptosis was determined 48 h later. The concentration of the relevant inhibitor used was fivefold greater than its corresponding IC50 value in all cases, and results are the mean (±SD) of three experiments. For panels C, D, and F, cells were treated with a single addition of the inhibitor or inhibitor combination.
We also tested the effects of perturbing the remaining high-stress, high-betweenness nodes that were identified (Fig. 5B). As earlier discussed, the large majority of high-stress, high-betweenness nodes identified were those that were either unique to G1S or shared between the G1 and G1S modules. Consistent with this, perturbation at any of these nodes resulted in a simultaneous increase in RT of both the G1 and the S phase (Fig. 5B). However, RT of the G2 phase was unaffected. Furthermore, in line with our identification of this protein as a G1-specific regulatory factor, silencing of PTK2B led to a biased extension of the G1 phase (Fig. 5B). The only discrepant result obtained was for E2F1, whose silencing perturbed both the G1 and S phases, although this protein was characterized as a node whose vulnerability was restricted to the G1 phase (Fig. 4A). This, however, may not be surprising given that the downstream products of E2F-dependent transcriptional regulation (e.g., cyclins A, B, and E) drive both the later stages of G1, as well as the S phase of the cycle.
Finally, with the exception of CDK5, silencing of nodes with low values for stress and betweenness (i.e., <1.5-fold of the mean value) had no significant effect on phase-specific RTs. Although CDK5 is suspected to play a role in cell cycle regulation, its properties and function have been less explored (Dhavan and Tsai 2001). This may account for the lesser number of associations known for CDK5 and, thereby, the resulting low values for stress and betweenness. Thus, the results in Figure 5, A and B, provide experimental support for the functional description of nodes, on the basis of centrality indices, from the corresponding regulatory modules. However, they also underscore the possibility that some additional critical nodes may have been missed due either to insufficient information or noise in the curated PPI databases.
High-stress, high-betweenness nodes represent vulnerable constituents of phase-specific regulatory modules
The identification of critical nodes on the basis of centrality measures also implied that these were the functionally least redundant, or most vulnerable, nodes within the respective cell cycle phase–specific modules. Consequently, inactivation of one or more of these nodes should then exert a profound influence on the cell cycle. Consistent with this, treatment of CH1 cells with an inhibitor of either AKT1 activation or that of ABL1 activity resulted in a dose-dependent increase in the frequency of apoptotic cells (Fig. 5C). Importantly, this apoptotic response was preceded by an accumulation of cells in the G1 phase (Fig. 5D), confirming that cell death derived from an arrest in the cycle. Here, for the purposes of comparison, we also examined the effects of inhibition of five representative target proteins that had been identified by our siRNA screen (Fig. 1). Of these, two (CHEK1 and FGFR2) represented G1-specific source nodes, and the remaining three (TRPM7, PRKAR1A, and MET) were G1S source nodes (see Fig. 2B). Whereas inhibitors of CHEK1 and TRPM7 were significantly less potent than that of AKT1 and ABL1, only marginal effects were obtained upon either PRKAR1A, FGFR2, or MET inhibition (Fig. 5C). The inhibitors used in these experiments, along with the IC50 values for their respective targets, are listed in Figure 5E.
Our interpretation that the vulnerable character of AKT1 and ABL1 emerged in temporally distinct windows of the cell cycle regulatory module also implied that simultaneous inhibition of both AKT1 and ABL1 should produce a cooperative effect on cell cycle arrest, and the consequent apoptosis. This expectation was indeed borne out in a subsequent experiment (Fig. 5F), and the Loewe Combination Index (CI) obtained was 0.56 ± 0.08. Here, a CI of <1 is indicative of a synergistic response (Chou and Talalay 1981). In contrast, apoptosis induction by combinations of inhibitors of either CHEK1 or FGFR2 (G1-specific source nodes) with that of TRPM7, PRKAR1A, or MET (G1S-specific source nodes) was poor (Fig. 5F). Thus, the cumulative results in Figure 5, C and F, confirm that both AKT and ABL exhibit a degree of vulnerability that is significantly greater than at least some of the targets identified in our siRNA screen. This, however, does not rule out the possibility that there may be other combinations not tested in this study, which may also be effective. Furthermore, the cooperative effects of combined inhibition observed also support our inferred distinction in phase-specific involvement (G1 vs. G1S) of at least two of the nodes (AKT1 and ABL1) identified on the basis of centrality indices.
IMP node modules define the signaling axes that govern G1 and G1S windows of the cell cycle
The extraction of the G1 and G1S IMP node modules also helped to resolve the nature of receptor-proximal signals that govern commitment of cells to the division cycle. In the G1 IMP node module, four molecules likely represent key constituents of the signaling cascade that governs the early G1. While AKT1 and PTK2B play a more specific role in this process, SRC and the adaptor molecule GRB2 represent the least redundant constituents of both G1 and G1S modules (Fig. 4). SRC likely serves as a common sensor that bridges AKT1 activation with both RTKs and mitogenic GPCRs. Whereas the SRC/GRB2 complex represents a constituent of the signalosome complex recruited by RTKs (Luttrell and Luttrell 2004), PTK2B mediates SRC activation by GPCRs (Litvak et al. 2000). This may then explain earlier observations that distinct mitogenic signals activate a common signaling cascade in the early G1 response (Jones and Kazlauskas 2001).
ABL1 was the unique kinase present as a high-stress, high-betweenness node in the G1S IMP node network (Fig. 4). While its inferred functional significance was experimentally verified (Fig. 5A), earlier studies have also demonstrated that depletion in levels of ABL1 significantly delays entry of cells into the S phase (Furstoss et al. 2002). Similar to AKT1, activation of ABL1 is also primarily dependent on the upstream SRC, which occurs both through direct and indirect mechanisms (Plattner et al. 1999; Sirvent et al. 2008). Thus, the early G1 is likely regulated through the complement of pathways initiated by the SRC–AKT1 signaling axis. Continuance through late G1, and then entry into the S phase, however, appears to be contingent on a switch in this axis where ABL1 now replaces AKT1 as the nonredundant substrate of SRC (see Fig. 6, inset).
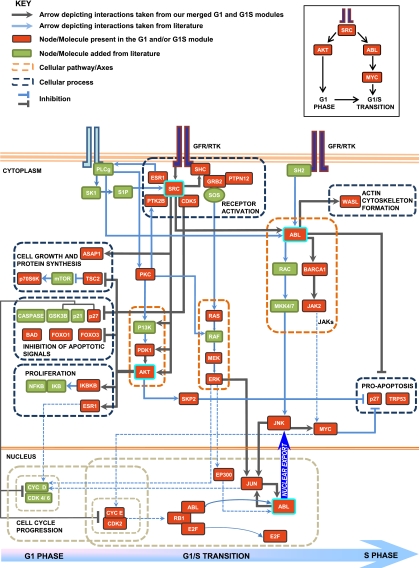
Figure 6.
Network of core pathways that mediate commitment of cells to the division cycle. In the pathway model, the nodes in red are proteins that are present in the G1 and/or G1S modules. Nodes present in green are derived from the literature (they include intermediates or effectors of cellular processes captured by our merged modules). The gray arrows depict interactions taken from the merged G1 and G1S modules. The blue arrows are links added from the literature. The dotted orange boxes highlight cellular signaling pathways, and the dotted gray boxes group molecules involved in regulating similar cellular processes. The dashed blue arrows indicate a known role of the source node in regulating the target node, derived from the literature. AKT and PTK2B are G1-specific high-stress and betweenness nodes. SRC and GRB2 show high stress and betweenness in both modules. ABL and JUN are present as high-stress betweenness nodes only in the G1S modules. A detailed description of the pathways captured is given in the Supplemental Material.
IMP node modules capture the network of core pathways that mediate commitment of cells to the division cycle
To translate our delineation of the distinct receptor-proximal signals into a more coherent view of G1 and G1S phase regulation, we merged the corresponding IMP node modules and extracted the motifs incorporating the combined list of high-stress, high-betweenness nodes. By then enriching the resulting links with information from the literature, we transformed these into the pathway map shown in Figure 6. It is evident that this map represents a synopsis of the multiplicity of biochemical events that cumulatively drive commitment of cells to the division cycle. Importantly, it also elaborates the individual modes of functioning of the G1- and G1S-specific signaling cascades, as well as the links between them that coordinate their respective activities.
The analysis reiterates that AKT1-dependent pathways, indeed, initiate cell cycle progression, by a concomitant inhibition of pro-apoptotic pathways being coordinated with mechanisms regulating cell growth and proliferation (Fig. 6). Similarly, this figure also rationalizes the key role ascribed to ABL1 in regulation of late G1 and the subsequent G1/S transition. As shown, pathways emanating from the cytoplasmic pool of ABL1 influence both cell cycle progression and also suppress pro-apoptotic mechanisms (Fig. 6). A prominent downstream consequence of cytoplasmic ABL1 activation is the enhancement in levels of cellular MYC (Boureux et al. 2005), the central regulator that links external signals to the cell cycle machinery (Obaya et al. 1999). Importantly, the cell cycle regulatory effects of MYC predominantly impinge on the later stages of G1 and facilitate entry of cells into the S phase (Heikkila et al. 1987; Santoni-Rugiu et al. 2000).
Release of the nuclear-localized ABL1 from its inhibitory complex with RB, which occurs as cells approach the G1/S boundary (Welch and Wang 1995), also contributes toward regulating MYC levels. The resulting convergence in signaling pathways, initiated from both the cytoplasmic and nuclear pools of ABL1, may well explain the long-standing question of how cellular MYC levels are temporally modulated during cell cycle progression. While expression of this protein is induced in the early G1 phase, these levels are further enhanced as cells prepare for S-phase entry (Jones and Kazlauskas 2001). Interestingly, consistent with our proposal, the timing of this second phase of MYC enhancement coincides with that of dissociation of the RB–ABL complex (Welch and Wang 1993). Synergism between the cytoplasmic and nuclear pools of ABL may therefore provide a plausible mechanism that enables ABL1 to replace AKT1 as the dominant signaling node in the G1S phase. This effect is likely augmented by the fact that, subsequent to its release from RB, a fraction of the nuclear ABL also partitions into the cytoplasmic compartment (di Bari et al. 2006). The resulting increase in the pool of cytoplasmic ABL1 would then further intensify signals generated from this subset of the kinase. Significantly, our proposal that ABL1-dependent signaling is markedly amplified as cells approach the G1/S boundary is well supported by recent findings that cellular levels of phosphorylated ABL1 peak at the G1S phase (Olsen et al. 2010).
These cumulative results therefore rationalize our assignment of key, but distinct, regulatory roles to AKT1 and ABL1 in the early and late G1 stages, respectively. The MAP kinase pathway likely serves as a common link that facilitates transition between these two phases. Thus, in addition to delineating the key receptor-proximal molecules, the results of our graph theoretical analysis also facilitated synthesis of existing—albeit piecemeal—data into a more unified scheme. This scheme captures the temporal evolution of signaling cascades, as they propel cells through the mitogen-sensitive phase of the cycle.
A comparative analysis of independently conducted screens yields a common IMP node network
Since cell cycle regulation is common to all dividing cells, it would be reasonable to expect that the core modules identified here—and the IMP node network from which they were extracted—would represent a common feature of dividing cells, independent of their tissue type of origin. To verify this, we also analyzed results of the previously described RNAi-based screens for cell cycle regulators that were performed in HeLa and U2OS cells (Mukherji et al. 2006; Kittler et al. 2007). As noted earlier, the overlap in signaling molecules identified between these two screens was <5%.
From each of these reports, we separately shortlisted the signaling molecules described as “hits” for either the G1 or S phase (the G1S phase was not characterized in these studies), and categorized them as G1 + S source nodes. We next repeated the analysis described in Figure 2A, wherein shortest paths were traced from each of these source nodes, to each of the G1 + S target nodes described earlier. By then subtracting against the corresponding G2-specific source to target network, we extracted the IMP nodes as described in Figure 2C. A similar exercise was also performed with the results of our present screen, which then allowed us to compare the resulting IMP node list with that obtained from the HeLa and U2OS cell screens. Remarkably, in spite of the nominal degree of concordance at the level of the RNAi-defined hits, there was a >70% overlap in the IMP nodes derived from the three screens (Supplemental Fig. S5). Furthermore, when IMP node networks were independently generated from each of these three lists, the resulting overlap between them was >92% (Supplemental Fig. S5).
To rule out any potential bias that our procedure for identifying IMP nodes may only detect highly connected proteins, we performed validation exercises at two levels. First, three different random pairs of protein sources and targets were sampled from the core PPI network, and IMP nodes were identified with respect to this background. The resulting overlap between three random source-target nodes (<30%) (Supplemental Fig. S5) was significantly less than the 70% value observed in our comparison between IMP nodes from the three studies. Second, to assess for any bias resulting from the employment of a common set of target nodes, we sampled a random set of sources from the core PPI network while retaining the targets (i.e., cell cycle targets; Supplemental Table S4). The overlap in IMP nodes identified from this exercise was ∼40% (Supplemental Fig. S5). Thus, any random set of source when used with the same cell cycle targets does not identify a conserved set of IMP nodes. Finally, the overlap between our CH1 IMP nodes and the three sets of random IMP nodes was <38% (Supplemental Fig. S5), further confirming that the observed overlap in IMP nodes between the three independent screens obtained with the two other independent RNAi screen results was, indeed, significant and not a random or biased event.
These cumulative findings thus establish the significance of our methodology for further interrogating the siRNA screen results. In addition, they also support the likelihood that the IMP nodules identified from our data constitute conserved regulatory elements of the mitogen-dependent signaling network.
The G1 and G1S IMP node modules represent conserved elements of mitogen-activated signaling cascades
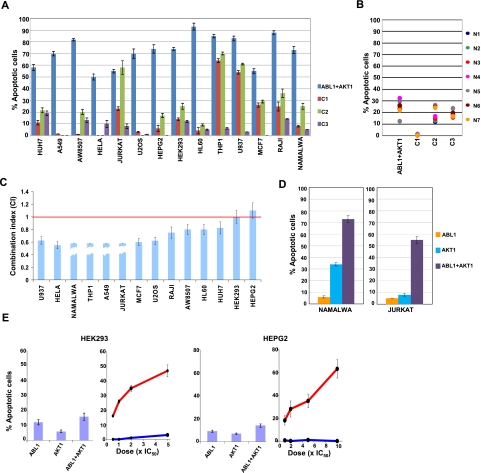
To experimentally verify our above inference that the delineated IMP node modules may constitute invariant features that are common to all dividing cells, we took a panel of 14 human cell lines derived from a diverse range of tissue types (see Supplemental Fig. S6B). These cells were treated with a single dose of either the combined inhibitors of AKT and ABL or, for purposes of comparison, combinations of inhibitors against the siRNA-identified targets (C1, C2, and C3 in Fig. 4F). The frequency of apoptotic cells obtained in each case was then determined 48 h later, and the results are presented in Figure 7A. The efficacy of the inhibitor combinations C1, C2, and C3 was highly restricted with the combination C2 inducing significant (i.e., >30%) apoptosis only in four of the cell lines tested. Furthermore, with the exception of Jurkat cells, effects on the remaining cell lines were predominantly due to CHEK1 inhibition with little or no contribution from PKA inhibition (Supplemental Fig. S6). This was also true of the effects of combination C1 on THP1 and U937 cells, whereas the combination C3 was virtually ineffective (Fig. 7A).
Figure 7.
Vulnerable nodes constitute regulatory elements that are conserved across a broad range of cell types. (A) The effects of combined inhibition of the indicated kinase combinations (at concentrations of 5 × IC50 values) on 14 different cancer cell lines, following a protocol similar to that described for Figure 5F; (B) the results of a similar experiment with indicated inhibitor combinations performed on normal B-lymphocytes purified from the peripheral blood of six different individuals. (C) The CI values for the combined inhibition of AKT and ABL obtained for the individual cell lines. CI values were calculated as previously described (Chou and Talalay 1981). For four cell lines, CI values could not be estimated since either one (for Namalwa and THP1) or both (for Jurkat and A549) inhibitors showed no effect when added individually (indicated as broken bars). The red line defines the cutoff (CI = 1) for interpreting responses as being synergistic, and values are the mean (±SD) of three determinations. (D) Representative results for the synergy between AKT and ABL inhibition where one (Namalwa) or both (Jurkat) inhibitors were ineffective when added individually. (E) The effects of the inclusion of PP1a, in addition to LY294002 and imatinib mesylate, on the apoptotic responses of HEK293 and HEPG2 cells. Results are shown in two subpanels for each cell line, where the subpanel on the left indicates the effects either of LY294002, imatinib mesylate alone (each at 4 × IC50), or a combination of both. The subpanel on the right gives the results of the addition of increasing concentrations of PP1a either alone (blue line), or in the presence of the combined inhibitors for AKT and ABL (each at 4 × IC50, red line). Values again are the mean (±SD) of three determinations.
In contrast to these limited effects, simultaneous inhibition of AKT and ABL induced significant levels of apoptosis in all the cell lines tested, which also included HeLa and U2OS cells (Fig. 7A). This effect was further enhanced by the 72-h time point, where the number of apoptotic cells was >80% in all cases. These findings, therefore, further support the functional distinction between at least the hits from the siRNA screen tested here, and the high vulnerability nodes identified from a graph theoretical derivation of the phase-specific regulatory modules. That is, in comparison with the former group, the latter represents nodes that are involved in a less redundant manner, during cell cycle regulation of a broader range of cancer cell types. Notably, none of the inhibitor combinations examined had any significant effect on viability of normal B-lymphocytes purified from the peripheral blood of six different individuals (Fig. 7B). This confirms that the functional relevance of AKT1 and ABL1 is only expressed in actively cycling cells.
To assess whether responses to the combined inhibition of ABL and AKT in Figure 7A were synergistic, we determined the CI values from dose-response curves either to LY294002, imatinib mesylate, or the combination of both in each instance. Figure 7C shows clear evidence for a synergistic effect in 12 of the 14 cell lines. Importantly, this also included examples in which either one (imatinib mesylate in Namalwa and THP1 cells) or both inhibitors (Jurkat and A549 cells) had no significant effect on cell survival when administered individually (Fig. 7D), further emphasizing the extent of cooperativity achieved by the inhibitor combination in these cells. For the remaining two cell lines, however (HEK293 and HEPG2), combined inhibition of both ABL and AKT yielded an outcome that was only additive in nature. We reasoned that this absence of synergy for the inhibitor combination in these latter two cell lines could derive from subtle, cell-type-inherent, differences in the architecture of the IMP node modules. This could then influence the degree of vulnerability exhibited by AKT1 and ABL1. Consequently, we probed this latter group further by also including SRC inhibitor PP1a, along with those for ABL and AKT. As earlier discussed, SRC principally functions by mediating either AKT1 activation in the G1 phase or ABL1 activation in the G1S phase.
Figure 7E shows that although SRC inhibition alone had no significant effect on either of the cell lines tested, a strong synergistic effect was nonetheless obtained when it was combined with the inhibitors of both AKT and ABL. Thus, by combining SRC inhibition along with that for AKT1 and ABL1, it seems possible to also guard against any cell-type-specific variations in topology of the G1 and G1S IMP node modules. Important here is that in all cases involving inhibition of vulnerable nodes—or their combinations—in Figure 7, increased apoptosis was always preceded by an accumulation of cells in the G1 phase, supporting that cell death was, indeed, mediated through an arrest in the cell cycle. These cumulative results, therefore, confirm that the G1 and G1S IMP node modules described in Figure 4, indeed, represent signal processing elements that commonly mediate cell cycle progression in at least a wide range of cell types. Furthermore, the vulnerable nodes that they incorporate provide sensitive targets for achieving an efficient disruption of the cell cycle regulatory network.
Discussion
RNAi-based screens are now being commonly used to probe a wide variety of biological processes. Recent results, however, have begun to highlight limitations inherent to this approach. Prominent among these are the preponderance of false negatives and the highly cell-type-restricted nature of the results obtained (Bushman et al. 2009). Consequently, the poor convergence between results from different laboratories often hampers the extraction of any meaningful interpretation. It was for this reason that we were prompted to explore graph theoretical approaches for unraveling the mechanistic implications of the hits identified in our screen. Previous studies support the utility of analyzing network interactions in receptor-specific pathways, to generate insights into signaling mechanisms. Furthermore, they also emphasized the potential of this approach for identifying new drug targets (Tu et al. 2009; Astsaturov et al. 2010; Bandyopadhyay et al. 2010).
Central to our analysis was the identification of phase-specific IMP nodes, which were selected on the basis of their unique over-representation in each of the phase-specific regulatory subnetworks. These IMP nodes likely combine the traits of minimal functional redundancy and increased specificity of action, in the context of regulation of the respective window of the cell cycle. By extension then, the modules defined by the phase-specific group of IMP nodes also likely constitute the key regulatory elements that process mitogenic (and oncogenic?) signals for the corresponding cell cycle phases. Here, a reexamination of data from the two previously described screens validated both the relevance and significance of our approach. The virtually similar IMP node network that was extracted in all cases also lent preliminary support to our proposition of the existence of a conserved set of core elements that regulate mitogenic signals.
Examination of the incorporated links revealed novel details on the mode of functioning of these modules. For instance, the G1 and G1S modules both participate in the critical function of coordinating cell cycle progression with cell growth, in a setting in which pro-apoptotic pathways are inhibited. The shared activities between the individual modules maintains contiguity during progression of cells from early G1 up to the S phase, whereas integration between the three modules promotes temporal evolution of the spectrum of biochemical processes that drive commitment of cells to the cycle. Particularly significant here was our discovery that the G1 and G1S phases were controlled by distinct signaling axes that were composed of SRC-AKT and SRC-ABL, respectively. Furthermore, we also subsequently elucidated that it was the switch between these two axes that guided transition of cells from early to late G1/G1S.
In addition to defining the receptor-proximal signals, however, our cumulative results further established that the IMP node modules, in fact, also capture the key regulatory elements that translate the effector consequences of mitogenic/oncogenic signaling to the respective stages of the cell cycle. In this connection, our experiments demonstrating that these cumulative functional properties of the G1 and G1S modules were conserved across a diverse panel cell lines is particularly significant. That is, these phase-specific modules presumably describe core information-processing units of the mitogen-activated signaling network, which function—in at least a broad range of cell types—to ensure irreversible commitment to the cycle. Future studies on the systems properties of these modules may provide important clues on mechanisms underlying cell transformation, and also on factors that govern properties related to cellular growth rate such as tumor aggressiveness and metastasis.
The identification of IMP node modules and delineation of their vulnerable constituents also facilitated the exploration of a potentially more effective strategy for chemotherapy. In view of the pathway redundancies that are inherent to signaling networks, current emphasis is on approaches that involve a concerted pharmacological intervention at multiple key targets (Lehar et al. 2008b). The challenge, however, is to identify the most appropriate combination of nodes to be targeted, and present approaches in this direction are limited by the vast space of high-order combinations that are theoretically possible (Lehar et al. 2008a). Here, our present strategy of extracting vulnerable nodes from the core elements that process input signals in a context-specific manner alone serves to reduce the dimensions of combination space to be explored by several orders of magnitude. Furthermore, as we have demonstrated, the relative functional conservation of these regulatory elements also implies that the benefits of targeting such nodes would be at least less cell-type-restricted.
From a broader point of view, the above results also support that targeting nodes derived from modules that regulate temporally distinct stages provide for a more effective means for breaching the fragility barrier of the cell cycle regulatory network. That is, these findings emphasize the potential of exploiting the temporal dimensions of a biological process, an aspect that is presently ignored in ongoing drug development exercises. At one level, targeting combinations of nodes whose functional vulnerabilities are conserved can potentially override current limitations faced due to problems of cell- and tissue-type heterogeneity. In addition, the observed synergism between AKT1 and ABL1 inhibition also entails a broader therapeutic window, thus providing an advantage over several of the chemotherapeutic agents currently in use (Kaelin 2005). Our proposal is particularly well supported by emerging evidence that a role for constitutive ABL activity is not restricted to CML alone but, rather, is now implicated in a broader spectrum of cancers (Sirvent et al. 2008). This would be consistent with our present description of ABL1 as the key regulatory signaling molecule that drives the G1S phase. Also to be noted in this context is the possibility of exploiting the other vulnerable nodes that were identified in the individual IMP node modules, through the development of specific inhibitors.
Thus, in summary, our present report describes an integration of results from a siRNA screen with graph theoretical analysis, to delineate the core signaling modules that regulate the early phases of the cell cycle. Importantly, these modules could be demonstrated to represent elements that were conserved across a broad range of cell types and could be exploited for elaborating novel mechanistic features pertaining to signal-mediated regulation of the cell cycle. Notable among these was the delineation of the distinct core signaling axes that governed early and late G1 phases, and subsequent elucidation of the seamless manner in which the switch between them was integrated. Furthermore, successful resolution of the least redundant nodes in each of the regulatory modules also permitted the development of a strategy for multi-module targeting in order to achieve synergistic disruption of the cell cycle. These latter findings thus pave the way for overcoming the daunting challenge presently faced, in identifying target combinations that can yield synthetic lethal effects in mammalian cells (Kaelin 2005, 2009).
Methods
Materials
All cell lines used in this study were originally procured from the American Tissue Culture Collection with the exception of AW8507, which was obtained from Shubha Chiplunkar (ACTREC, Navi Mumbai). Normal B-lymphocytes were purified from the blood of donors by using the B-cell negative selection kit from Dynal and following the protocol recommended by the manufacturer. Purity of these cells were >90% as assessed by CD19 staining. Antibodies were purchased from Cell Signaling Technologies. For the primary screen, siRNAs were obtained from QIAGEN, whereas for the validation screen they were obtained either from Sigma Chemical Co. (for kinases) or from Dharmacon Inc. (for phosphatases). Further details of these are provided in the Supplemental Experimental Procedures. All chemical inhibitors used here were purchased from Tocris Biosciences except for imatinib mesylate, which was obtained as a gift from Sundeep Dugar (Sphaera Pharma).
siRNA screen
Details of the standardization of conditions for the siRNA screen, the protocol adopted, and the data analysis procedures are provided in the Supplemental Experimental Procedures. Briefly, CH1 cells were seeded in wells of a 96-well plate at 2 × 104/well and transfected, in duplicate, either with control or target-specific siRNA. At 72 h later these cells were stained with propidium iodide and analyzed for the DNA content by flow cytometry. An analysis of the results using the FlowJo software yielded the DNA histograms for quantification of the sub-G1, G1, S, and G2 populations. The Z-scores for each parameter were then calculated by using the corresponding mean values, and the standard deviations, of the five negative controls in each plate as described in the Supplemental Experimental Procedures. Short-listed hits were then taken for further validation as described in the text. By using various criteria, we also confirmed that our screen was robust, reproducible, and sensitive with an overall z-factor of 0.56 (see the Supplemental Experimental Procedures). Determination of the effect of siRNA treatment on the population doubling time and the consequent estimation of phase-specific RTs is also described in detail in the Supplemental Experimental Procedures.
Network analyses
An outline of the approach is described in Figure 2C. A step-by-step description of the graph theoretical approaches for arriving at the IMP node networks and—eventually—the vulnerable nodes is provided in the Supplemental Experimental Procedures.
Data access
Publicly available Protein–Protein interaction databases used in this study have been mentioned and also have been cited at the appropriate places in the text. Novoseek disease relationship scores were obtained from http://www.novoseek.com/Welcome.action. The Supplemental Material includes six additional figures and eight tables.
Acknowledgments
This work was supported by a grant (to K.V.S.R.) from the Department of Biotechnology, Government of India. N.J. and S.R. are recipients of a Senior Research Fellowship from the CSIR, and S.R.H. received a Senior Research Fellowship from the UGC. We thank Shubha Chiplunkar (ACTREC) and Ramesh Bamezai (JNU) for gifts of the AW8507 and MCF7 cells, respectively.
Authors' contributions: N.J and K.V.S.R. managed the project. S.R., S.R.H., S.C.M., and N.J. performed the network and screen analysis. N.J. and Z.S. performed the experiments. N.J., S.R., and K.V.S.R. interpreted results and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116145.110.
References
- Aflakian N, Ravichandran S, Jamal MS, Jarvenpaa H, Lahesmaa R, Rao KV 2009. Integration of signals from the B-cell antigen receptor and the IL-4 receptor leads to a cooperative shift in the cellular response axis. Mol Biosyst 5: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Alon U 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, Devarajan K, Silverman JS, Tikhmyanova N, Skobeleva N, et al. 2010. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal 3: ra67 doi: 10.1126/scisignal.2001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, et al. 2010. A human MAP kinase interactome. Nat Methods 7: 801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN 2004. Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Boureux A, Furstoss O, Simon V, Roche S 2005. Abl tyrosine kinase regulates a Rac/JNK and a Rac/Nox pathway for DNA synthesis and Myc expression induced by growth factors. J Cell Sci 118: 3717–3726 [DOI] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, et al. 2009. Host cell factors in HIV replication: Meta-analysis of genome-wide studies. PLoS Pathog 5: e1000437 doi: 10.1371/journal.ppat.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P 1981. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur J Biochem 115: 207–216 [DOI] [PubMed] [Google Scholar]
- Csete M, Doyle J 2004. Bow ties, metabolism and disease. Trends Biotechnol 22: 446–450 [DOI] [PubMed] [Google Scholar]
- Del Sol A, Balling R, Hood L, Galas D 2010. Diseases as network perturbations. Curr Opin Biotechnol 21: 566–571 [DOI] [PubMed] [Google Scholar]
- Dhavan R, Tsai LH 2001. A decade of CDK5. Nat Rev Mol Cell Biol 2: 749–759 [DOI] [PubMed] [Google Scholar]
- di Bari MG, Ciuffini L, Mingardi M, Testi R, Soddu S, Barila D 2006. c-Abl acetylation by histone acetyltransferases regulates its nuclear-cytoplasmic localization. EMBO Rep 7: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrin R, Beg QK, Barabási AL, Oltvai ZN 2004. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics 5: 10 doi: 10.1186/1471-2105-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman GM, Gally JA 2001. Degeneracy and complexity in biological systems. Proc Natl Acad Sci 98: 13763–13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Vousden KH 2001. Proliferation, cell cycle and apoptosis in cancer. Nature 411: 342–348 [DOI] [PubMed] [Google Scholar]
- Furstoss O, Dorey K, Simon V, Barila D, Superti-Furga G, Roche S 2002. c-Abl is an effector of Src for growth factor-induced c-myc expression and DNA synthesis. EMBO J 21: 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP 2008. Knockdown screens to knockout HIV-1. Cell 135: 417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ 2007. The DNA damage response: Ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM 1987. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature 328: 445–449 [DOI] [PubMed] [Google Scholar]
- Jamal MS, Ravichandran S, Jailkhani N, Chatterjee S, Dua R, Rao KV 2010. Defining the antigen receptor-dependent regulatory network that induces arrest of cycling immature B-lymphocytes. BMC Syst Biol 4: 169 doi: 10.1186/1752-0509-4-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Kazlauskas A 2000. Connecting signaling and cell cycle progression in growth factor-stimulated cells. Oncogene 19: 5558–5567 [DOI] [PubMed] [Google Scholar]
- Jones SM, Kazlauskas A 2001. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat Cell Biol 3: 165–172 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr 2005. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 5: 689–698 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr 2009. Synthetic lethality: a framework for the development of wiser cancer therapeutics. Genome Med 1: 99 doi: 10.1186/gm99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H, Oda K 2006. Robustness trade-offs and host-microbial symbiosis in the immune system. Mol Syst Biol 2: 2006.0022. doi: 10.1038/msb4100039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol 9: 1401–1412 [DOI] [PubMed] [Google Scholar]
- Lehar J, Krueger A, Zimmermann G, Borisy A 2008a. High-order combination effects and biological robustness. Mol Syst Biol 4: 215 doi: 10.1038/msb.2008.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Stockwell BR, Giaever G, Nislow C 2008b. Combination chemical genetics. Nat Chem Biol 4: 674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Tian D, Shaul YD, Lev S 2000. Targeting of PYK2 to focal adhesions as a cellular mechanism for convergence between integrins and G protein-coupled receptor signaling cascades. J Biol Chem 275: 32736–32746 [DOI] [PubMed] [Google Scholar]
- Luttrell DK, Luttrell LM 2004. Not so strange bedfellows: G-protein-coupled receptors and Src family kinases. Oncogene 23: 7969–7978 [DOI] [PubMed] [Google Scholar]
- Ma'ayan A, Jenkins SL, Webb RL, Berger SI, Purushothaman SP, Abul-Husn NS, Posner JM, Flores T, Iyengar R 2009. SNAVI: Desktop application for analysis and visualization of large-scale signaling networks. BMC Syst Biol 3: 10 doi: 10.1186/1752-0509-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manimaran P, Hegde SR, Mande SC 2009. Prediction of conditional gene essentiality through graph theoretical analysis of genome-wide functional linkages. Mol Biosyst 5: 1936–1942 [DOI] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. 2009. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 37: D619–D622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, et al. 2006. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci 103: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW 2004. Recycling the cell cycle: Cyclins revisited. Cell 116: 221–234 [DOI] [PubMed] [Google Scholar]
- Newman MEJ 2005. A measure of betweenness centrality based on random walks. Soc Networks 27: 39–54 [Google Scholar]
- Obaya AJ, Mateyak MK, Sedivy JM 1999. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene 18: 2934–2941 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3 doi: 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Papin JA, Hunter T, Palsson BO, Subramaniam S 2005. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol 6: 99–111 [DOI] [PubMed] [Google Scholar]
- Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM 1999. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev 13: 2400–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J 2000. Involvement of Myc activity in a G1/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol 20: 3497–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH 2009. PID: the Pathway Interaction Database. Nucleic Acids Res 37: D674–D679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC, Nevins JR 2002. Signaling networks that link cell proliferation and cell fate. J Biol Chem 277: 11617–11620 [DOI] [PubMed] [Google Scholar]
- Sirvent A, Benistant C, Roche S 2008. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell 100: 617–631 [DOI] [PubMed] [Google Scholar]
- Supper J, Spangenberg L, Planatscher H, Drager A, Schroder A, Zell A 2009. BowTieBuilder: modeling signal transduction pathways. BMC Syst Biol 3: 67.. doi: 10.1186/1752-0509-3-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM 1999. Measures of degeneracy and redundancy in biological networks. Proc Natl Acad Sci 96: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Argmann C, Wong KK, Mitnaul LJ, Edwards S, Sach IC, Zhu J, Schadt EE 2009. Integrating siRNA and protein–protein interaction data to identify an expanded insulin signaling network. Genome Res 19: 1057–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen K, Van Bockstaele DR, Berneman ZN 2003. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 36: 131–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 2004. Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JY 1993. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell 75: 779–790 [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JY 1995. Abrogation of retinoblastoma protein function by c-Abl through tyrosine kinase-dependent and -independent mechanisms. Mol Cell Biol 15: 5542–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg A, Larsson O, Wiman KG 1995. What is the restriction point? Curr Opin Cell Biol 7: 835–842 [DOI] [PubMed] [Google Scholar]
- Zhu X, Gerstein M, Snyder M 2007. Getting connected: analysis and principles of biological networks. Genes Dev 21: 1010–1024 [DOI] [PubMed] [Google Scholar]