Abstract
Gene duplication via retrotransposition has been shown to be an important mechanism in evolution, affecting gene dosage and allowing for the acquisition of new gene functions. Although fixed retrotransposed genes have been found in a variety of species, very little effort has been made to identify retrogene polymorphisms. Here, we examine 37 Illumina-sequenced North American Drosophila melanogaster inbred lines and present the first ever data set and analysis of polymorphic retrogenes in Drosophila. We show that this type of polymorphism is quite common, with any two gametes in the North American population differing in the presence or absence of six retrogenes, accounting for ∼13% of gene copy-number heterozygosity. These retrogenes were identified by a straightforward method that can be applied using any type of DNA sequencing data. We also use a variant of this method to conduct a genome-wide scan for intron presence/absence polymorphisms, and show that any two chromosomes in the population likely differ in the presence of multiple introns. We show that these polymorphisms are all in fact deletions rather than intron gain events present in the reference genome. Finally, by leveraging the known location of the parental genes that give rise to the retrogene polymorphisms, we provide direct evidence that natural selection is responsible for the excess of fixations of retrogenes moving off of the X chromosome in Drosophila. Further efforts to identify retrogene and intron presence/absence polymorphisms will undoubtedly improve our understanding of the evolution of gene copy number and gene structure.
Recent studies have revealed a large number of cases in which changes in gene copy-number rather than nucleotide substitutions have contributed to adaptive evolution (for review, see Demuth and Hahn 2009). In Drosophila in particular, it has become clear that natural selection can favor both gene gains (Long and Langley 1993) and gene losses (Greenberg et al. 2006). Because all adaptive differences in gene copy-number between species must first arise as polymorphisms, recent genome-wide efforts have focused on describing the number and type of copy-number variants (CNVs) within populations of Drosophila melanogaster (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010; CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.). These studies have collectively identified thousands of CNVs, including newly duplicated genes segregating at high frequency that may be influenced by adaptive natural selection.
Most methods used to identify CNVs only detect long stretches of either duplicated or deleted nucleotides (Dopman and Hartl 2007; Emerson et al. 2008) or only confidently detect duplications that lie in tandem to the original locus (Cridland and Thornton 2010). However, single genes can also be duplicated by retrotransposition (referred to as “retrogenes” when functional), in which a gene is transcribed into mRNA, reverse transcribed into cDNA, and then reinserted into a new genomic position (Hollis et al. 1982; Karin and Richards 1982; Ueda et al. 1982). These polymorphic retrogenes (“retroCNVs”) will only have signatures of duplication in exons and may be inserted anywhere in the genome; they are therefore likely to have been missed by previous studies of copy-number variation.
Notwithstanding challenges in detection, previous studies of copy number variation may have ignored retroCNVs because they rarely have regulatory DNA copied along with them (with exceptions described by Okamura and Nakai 2008) and are therefore most often present as dead-on-arrival pseudogenes. However, comparative genomics has shown that between 0.5 (Drosophila) (Bai et al. 2007) and two (mammals) (Vinckenbosch et al. 2006) new functional retrogenes are fixed per million years. Several of these new retrogenes have been found to evolve adaptively shortly after duplication (e.g., Long and Langley 1993; Betrán and Long 2003; Burki and Kaessmann 2004), and they are therefore likely to make an important contribution to organismal adaptation. Despite the wealth of data on retrogenes provided by comparative genomics, we still know little about the rate at which they arise and the evolutionary forces that determine their trajectories through populations.
One of the most interesting patterns to arise from studies of retrotransposition is the excess number of retrogenes that move from the X chromosome to the autosomes in Drosophila (Betrán et al. 2002; Meisel et al. 2009) and the mosquito Anopheles gambiae (Toups and Hahn 2010) and both onto and off of the X in human and mouse (Emerson et al. 2004). In Drosophila, recent studies have shown a similar bias for DNA-based gene duplication events, at least in some species (Meisel et al. 2009; Vibranovski et al. 2009b). In the case of retrogenes, the movement between chromosomes can be polarized because both the parental gene (with introns) and the daughter retrogene (without introns) have known locations in the genome. Two main explanations have been given for the excess of retrotransposition involving the X chromosome: escape from meiotic sex chromosome inactivation (Betrán et al. 2002; Vibranovski et al. 2009a) and sexually antagonistic selection against male-favorable genes (Ranz et al. 2003; Wu and Xu 2003; Vicoso and Charlesworth 2006). In addition, nonadaptive explanations—such as biased integration of reverse-transcribed cDNAs onto autosomes (Metta and Schlotterer 2010)—have been put forward, though studies of the movement of pseudogenized retrogenes in both Drosophila and mammals have not found any biased integration (Emerson et al. 2004; Potrzebowski et al. 2008; Meisel 2009), nor has such a bias been observed with respect to transposable elements (Fontanillas et al. 2007). Much of the evidence for selection driving retrogenes off the X has been correlative: Such retrogenes often have testis-biased or even testis-specific gene expression, a pattern consistent with the advantage of autosomal copies that are not precociously silenced during spermatogenesis (Betrán et al. 2002; Vibranovski et al. 2009a). Due to the lack of direct evidence in support of adaptive explanations of this X-to-autosome bias in Drosophila, the forces that drive the genomic movement of retrogenes over evolutionary time-scales remain unknown.
The retrogenes identified in previous studies of Drosophila are likely to have fixed in the population long ago; these fixed retrogenes originated as individual mutations and were then fixed by either directional selection or genetic drift. Because polymorphism data can be used to distinguish between selective and neutral forces (e.g., McDonald and Kreitman 1991), studying retrogene polymorphisms should be of use in identifying the evolutionary forces leading to the migration of genes off the X chromosome. Thus far, very little effort has been devoted to the study of copy number-variant retrogenes, or retroCNVs. To our knowledge, the only study that has detected retroCNVs on a genome-wide scale is a recent microarray-based study of CNVs in humans (Conrad et al. 2010); no detailed analysis of these variants was reported. Here we present the first in-depth, genome-wide analysis of retroCNVs in any species to date. We use a novel, highly accurate method to detect these variants using the next-generation sequencing data generated as part of the Drosophila Population Genomics Project (DPGP) (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.), though in principle, data generated by any sequencing technology can be used. We use these data to show conclusively that natural selection drives the fixation of retrogenes moving from the X chromosome to the autosomes in D. melanogaster. Finally, we use a variant of our method to describe the first genome-wide set of intron presence/absence polymorphisms in Drosophila.
Results and Discussion
Number and frequency of retroCNVs in the Raleigh population
In order to detect retrogenes present in one or more of 37 Illumina-sequenced D. melanogaster inbred lines obtained from Raleigh, North Carolina (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.), but not the reference genome, we first examined read-depth at all annotated genes. We predict that retroCNVs will show excess read-depth only in exons because only exonic sequence is duplicated by retrotransposition (Fig. 1). We recorded the average and standard deviation of read-depth for each intron and the two flanking exons (with depth measured every 36 bp), as well as the ratio of exonic to intronic read-depth. This resulted in 224 genes with at least three out of four introns having an exon:intron read-depth ratio of 1.5 or greater.
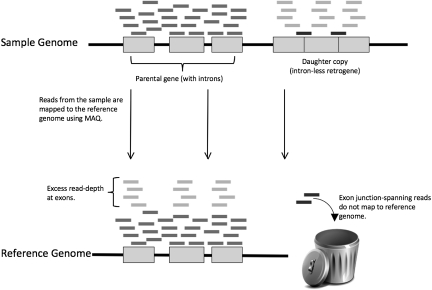
Figure 1.
Mapping reads from a genome containing a polymorphic retrocopy to a reference genome. The black line at the top represents a chromosome in a sample genome. Gray boxes represent exons within a gene, with the spaces in between representing introns. The gray boxes on the right with no introns in between them represent a retrogene derived from a parental gene (located downstream in this example). The short bars appearing above the two gene copies represent reads derived from the sample chromosome. The black line at the bottom represents the same chromosome in the reference genome to which these reads are mapped. Note that reads derived from the parental copy of the gene are mapped to the proper location, while reads from the retrocopy (light gray) are mapped to the exons of the parental copy, resulting in elevated read-depth. Also note that the reads crossing exon–exon boundaries in the retrocopy (dark gray) are not mapped to the reference genome. Our method to detect retroCNVs involves finding these reads by searching all unmapped reads against a database of exon–exon junctions.
Because read-depth is highly variable across the genome—and is dependent on both GC-content and the level of nucleotide polymorphism (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.)—we expected that relying on read-depth alone would result in a large number of false positives. Therefore, as a second step, we used reads that could not be mapped to the reference genome to search against a database of exon–exon junction sequences present in mRNAs annotated in FlyBase, regardless of read-depth. We expect reads mapping across such an exon–exon junction to be found in an individual containing a retrocopy of the gene in which the junction resides (Fig. 1); such reads should also be found in an individual missing an intron that is present in the reference genome (Fig. 2).
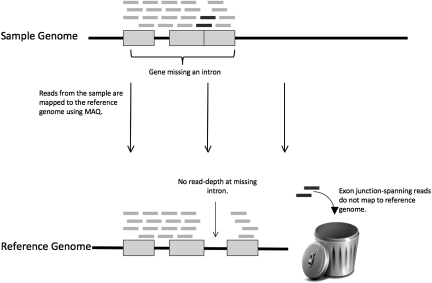
Figure 2.
Mapping reads from a genome containing a polymorphic intron deletion to a reference genome. As in Figure 1, the black line at the top represents a chromosome in a sample genome, the bottom line represents the chromosome in the reference genome, gray boxes represent exons, and short bars represent reads. Note that no reads are mapped to the intron that is deleted in the sample chromosome but present in the reference genome. Also note that the reads crossing the single exon–exon boundary in the sample chromosome are not mapped to the reference genome.
In total, we found 197 exon–exon junctions spanned by at least one read. To distinguish between spanned exon–exon junctions due to retroCNVs versus deleted introns, we examined the depth of reads mapped to the introns between skipped exons. We expect intronic read-depth to be at normal levels (∼10×) in genes that are retrotransposed but at low levels (less than 1×) in deleted introns. Three of the spanned exon–exon junctions were found to correspond to missing introns and are discussed further below; the remaining 194 cases were inferred to correspond to retroCNVs. These 194 exon–exon junctions correspond to 181 genes. The majority (155) of these genes had only one intron skipped by only a single read in a single line. Although these cases could represent true retrocopies located within poorly covered heterochromatic regions of the genome, we conservatively assumed that they are false positives and removed them from further analysis. Therefore, only the 34 retroCNVs having at least one exon–exon junction spanned by multiple reads were included in the remainder of our analysis. Although read-depth within exon–exon junctions could be used to infer copy numbers higher than two, we observed no cases suggestive of multiple polymorphic retrocopies originating from the same gene, and we conservatively assume that each of these 34 cases represents a single duplication resulting in only one additional copy of the gene.
To assess the accuracy of our retroCNV predictions, we attempted to confirm via PCR 18 exon–exon junctions that were spanned by overlapping reads, corresponding to seven different retroCNVs. Thirteen of these spanned junctions gave two bands of the size predicted by the presence of both a retroCNV and an intron-containing parental copy (see Methods), and each of the seven retroCNVs that we attempted to validate had at least one confirmed exon–exon junction. There were also 14 such junctions predicted in two lines for which low-coverage paired-end data has been collected (see Methods). By using the paired-end data, we were able to validate 10 of the 14 (71%) junctions corresponding to retroCNVs in these lines. Due to the low coverage of paired-end sequence data in these two lines, the true-positive rate may be significantly higher than this estimate, as not all retroCNVs present in these low-coverage sequenced lines will be covered by a pair of reads. For example, large deletions detected as part of the DPGP analysis only had adequate paired-end coverage to be validated ∼75% of the time (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.). The results of these independent methods of validation suggest that the vast majority of our 34 retroCNVs are true polymorphisms and not false positives. We also estimate that on average only 7.2% of exon–exon junctions corresponding to retroCNVs (or missing introns) are missed by this method (Methods). Information on each retroCNV is given in Table 1, and the lines containing each retroCNV and the transcripts inferred to be retrotransposed are listed in Supplemental Table 1.
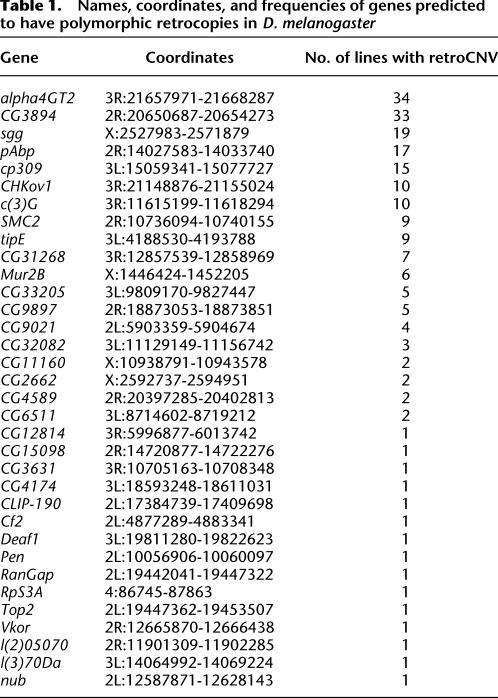
Table 1.
Names, coordinates, and frequencies of genes predicted to have polymorphic retrocopies in D. melanogaster
We examined the general characteristics of detected retroCNVs, finding that genes giving rise to retroCNVs are not significantly biased away from certain chromosomes, nor are they significantly clustered along chromosomes. We also examined the average coding sequence length of retrotransposed genes, finding that retrotransposed genes have slightly longer coding sequences on average than nonretrotransposed genes (P = 0.02; Wilcoxon rank-sum test). We also find that the majority of retrotransposed genes are expressed in the germline according to FlyAtlas (74% are present in the testis and 65% are present in the ovaries) (Chintapalli et al. 2007). This result is unsurprising, as mutations must occur in the germline in order to be inherited by offspring, and a gene must be expressed in order to be retrotransposed. Further work relying on larger sample sizes and data from additional species will further elucidate the mutational patterns of retroCNVs.
We searched the Drosophila simulans genome and found no retrogenes corresponding to our retroCNVs. We therefore conclude that the majority of our 34 retroCNVs are derived duplications (though some retrocopies may be present in unassembled regions of the D. simulans genome and therefore represent deletions of previously duplicated genes). In order to genotype these 34 retroCNVs among all lines, we simply inferred that any sequenced line having an exon–exon junction-spanning read has the retrocopy, and any line not having such a read does not have the retrocopy. The derived allele frequency spectrum of these retroCNVs is shown in Figure 3. Based on the allele frequencies of all observed variants, we calculate that any two chromosomes in the Raleigh population differ by the presence of 6.1 retroCNVs on average, accounting for ∼13% of gene copy number heterozygosity in the population (based on CNV data from CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.). It should be noted that retroCNVs derived from single-exon genes cannot be detected by this analysis, so this number may underestimate the amount of retrogene heterozygosity. This analysis also fails to detect any retroCNVs present in the reference genome. However, a recent study detected only one retrogene in the D. melanogaster genome that is not shared with D. simulans (Bai et al. 2007), so it is unlikely that this causes us to miss many retroCNVs. We also compared the derived allele frequency spectrum of these retroCNVs to the spectrum expected under neutrality in order to assess the strength of selection acting on these polymorphisms (see CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.). The resulting large confidence intervals imply that this analysis lacked adequate power to reject neutrality and that a larger data set will be required to confidently detect whether any selective forces may be acting on retroCNVs.
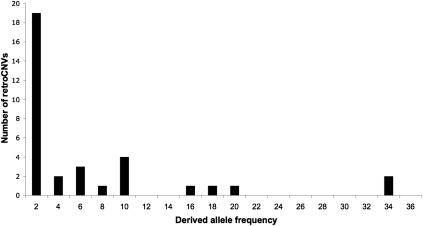
Figure 3.
Derived allele frequency spectrum of retroCNVs. The derived allele frequency is given as the number of sequenced lines containing a retrocopy.
Thousands of CNVs have previously been found in D. melanogaster (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010; CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.) and many other species (for review, Schrider and Hahn 2010). However, most methods used to detect CNVs rely on long stretches of increased or decreased read-depth, and the crenellated patterns of increased read-depth associated with retroCNVs (Fig. 1) are not likely to be detected unless exons are especially long. Even in the event that single exons are identified as CNVs, they may not be connected to the mechanism of retrotransposition. A previous microarray study in humans was able to detect several retroCNVs by querying retrogenes present in the reference genome, as well as the hybridization signal of exons relative to their intervening introns (Conrad et al. 2010). Several polymorphisms detected by this approach were deletions in sample individuals relative to the reference genome, though the investigators did not polarize them as evolutionary gains or losses. Clone-based paired-end methods offer the opportunity to detect possible retroCNVs if the DNA insert can be subsequently sequenced (e.g., Kidd et al. 2008); if not, the source of the insertion sequence remains unknown. While the results presented here suggest several ways that next-generation paired-end sequencing could be used to detect retroCNVs (not just to validate them), the method we use offers a straightforward and accurate way to identify these important polymorphisms.
Intron presence/absence polymorphisms
As discussed above, our method for detecting retroCNVs—increased exon:intron read-depth combined with reads that span exon–exon junctions—also identified three introns missing from sequenced lines. Because intron gain or loss polymorphisms could be useful for elucidating the forces driving the evolution of intron density, we decided to look for more intron copy-number polymorphisms. Upon further examination, we found several more introns in our data set with average depth less than one read per base pair (less than 1× coverage) that were not confirmed by reads spanning the exon–exon junction. Visual inspection of the read-depth in these introns suggested that the breakpoints of some intronic deletions (relative to the reference) did not exactly match the exon–intron boundaries. In other words, some introns may be “imperfectly” deleted relative to the exon–intron boundary, removing some exonic sequence in addition to the intron; in these cases, MAQ will not map any reads to the exon–exon junction. (There can also be imperfect deletions that do not remove the entire intron, but we are less likely to detect these because the average coverage of introns may be greater than 1×.) In order to detect imperfect intron deletions, we used the program BWA (Li and Durbin 2009) to look for reads “skipping” the portions of these genes with zero read-depth. Because BWA allows mapping with small indels, this method does not rely on identifying the exact breakpoints of any deletions beforehand. This analysis detected 11 additional intron deletions relative to the reference genome, though we still may have missed some deletions in cases where actual breakpoints do not closely match the boundaries of regions with zero depth. Reads supporting these imperfect intron deletions were then searched against the reference genome to verify that their best alignment was consistent with an intron deletion. In one case, all reads were found to have alignments inconsistent with an intron deletion, instead supporting a small intronic indel. This case was removed from the remainder of the analysis.
We were able to validate one of the “perfect” missing introns by PCR, and we also had adequate paired-end coverage to confirm an additional imperfect intron deletion. In order to ensure that all remaining introns missing in at least one line are in fact spliced out of mature mRNAs, we examined cDNA, EST, and RNA-seq data collected for the modENCODE project (Celniker et al. 2009). Four introns were found to reside in genes with little to no expression evidence in the modENCODE data. Although these genes may be expressed at low levels or in tissues/stages not queried by these experiments, the missing introns residing in these genes were conservatively removed from the remainder of the analysis. Our final data set consists of nine missing introns (three perfect and six imperfect), all of which have expression evidence supporting the annotated intron.
Since an apparent deletion in a sequenced line relative to the reference may correspond to either a deletion or a novel insertion allele present in the reference, the nine missing introns in our set may represent intron gains. To determine whether they are evolutionary gains or losses, we examined alignments of the flanking exonic sequence with D. simulans using the UCSC Genome Browser (Kent et al. 2002). In each case, the intronic sequence present in the D. melanogaster reference genome was also present in the D. simulans reference genome (with small indels in some cases). These results imply that the missing introns are all recent intron deletions in D. melanogaster.
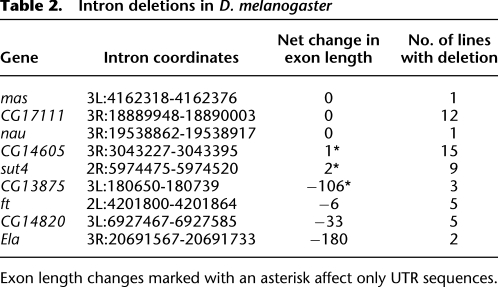
To determine the effect of imperfect intron deletions on coding sequences, we counted the number of exonic bases removed or added by these deletions, assuming any remaining intronic sequence is not spliced. Three imperfect intron deletions only remove sequence within UTRs, while the remaining three deletions result in a net loss in coding length of 6, 33, and 180 bases, respectively. Notably, none of the deletions appear to result in frameshifts. The fact that six of the intron deletions do not match the exon–intron boundaries strongly suggest that they have been removed by a DNA-based event and not conversion by cDNA, as is known to occur in fungi (Fink 1987; Derr and Strathern 1993; Goffeau et al. 1996; Stajich and Dietrich 2006). Though it seems highly unlikely that a random genomic deletion would perfectly remove an intron, we do observe three such events, which is suggestive of cDNA conversion. However, given that even the imperfect deletions preserve the reading-frame or only overlap UTRs, it is also possible that deletion mutations are occurring without respect to exon–intron boundaries, but that only those that do not significantly disrupt the protein sequence reach appreciable frequencies in the population.
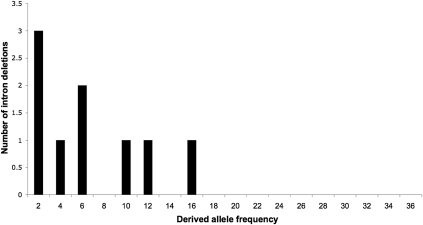
In order to genotype each of these nine intron deletions with more sensitivity, any sequenced DPGP line found to have less than 1.0 average depth in any of these introns was considered to contain the deletion allele as well, regardless of whether the deletion was confirmed by a read spanning the exon–exon junction in that particular line. A description of these intron deletion alleles and the genes in which they reside is given in Table 2. More detailed information, including the estimated deletion breakpoints and the sequenced lines containing the deletions, is listed in Supplemental Table 2; the derived allele frequency spectrum is shown in Figure 4. From these data, we estimate that, on average, any two chromosomes from the Raleigh population will differ in the presence of 2.3 introns, though this may be an underestimate due to false negatives. As with retroCNVs, we compared this allele frequency spectrum to the spectrum expected under neutrality, but were unable to reject the null hypothesis.
Table 2.
Intron deletions in D. melanogaster
Figure 4.
Derived allele frequency spectrum of intron deletions. The derived allele frequency is given as the number of sequenced lines lacking the intron.
Although most of the data on intron loss in Drosophila come from comparisons between species (e.g., Roy and Gilbert 2005; Coulombe-Huntington and Majewski 2007), one previous example of a polymorphic intron loss has been found in Drosophila teissieri (Llopart et al. 2002). Recently, a genome-wide analysis of two genomes from the species Daphnia pulex revealed 24 intron presence/absence polymorphisms (Li et al. 2009). These investigators found that 21 of the 24 polymorphisms in Daphnia were recent intron gains; in contrast, all of the polymorphisms we detect are losses. Our method does not allow us to detect introns present in resequenced lines but absent from the reference genome, so we can only detect polymorphic gains if the new intron is present in the reference. Given that no new introns were detected by a phylogenetic analysis on the branch leading to D. melanogaster since the split with Drosophila erecta (Farlow et al. 2010)—and only cases like these can be detected by our method—the fact that we find no intron gains should not be too surprising. Regardless of whether events are gains or losses, our results suggest that the growing number of population genomic studies will reveal intron presence/absence polymorphism in a large number of species and will improve our understanding of the evolutionary forces affecting changes in gene structure.
Natural selection drives retroCNVs off the X chromosome
As discussed in the Introduction, analyses of genes present in the reference genomes of multiple Drosophila species have revealed an excess of retrogene duplications moving from the X to the autosomes (e.g., Betrán et al. 2002; Dai et al. 2006; Bai et al. 2007; Meisel et al. 2009). The 34 retroCNVs described above represent the first opportunity to directly test the hypothesis that the excess of fixed retrogenes moving from the X to the autosomes in Drosophila is due to natural selection.
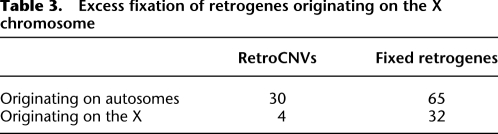
The approach we take to testing for directional selection follows the logic of the test laid out by McDonald and Kreitman (1991) for nucleotide data. If there is no positive selection acting on retrogenes arising on the X chromosome, then the ratio of polymorphic to fixed variants on the X should be approximately equal to the same ratio for variants on the autosomes. If, on the other hand, selection is driving the fixation of retrogenes moving off the X, then there will be an excess of fixed variants in this class relative to autosomes. Since we are not able to determine where our retroCNVs have inserted in the genome, we can only directly test whether or not there is an excess of fixed retrogenes originating on the X chromosome. To compare the number and location of retroCNVs we identified to an equivalent set of fixed retrogenes, we used data from Bai et al. (2007), which identified 97 retrogenes in D. melanogaster; 32 of these retrogenes originated on the X, with all but two moving to an autosome. For our analysis, we simply use the counts of parental genes on the X and autosomes from this data set.
As shown in Table 3, there is indeed an excess of fixed retrogenes originating on the X (P = 0.01). The proportion of retroCNVs on the X (i.e., the parental gene is on the X) is roughly equivalent to the proportion of total genes on the X chromosome (11.7% and 18.1%, respectively). However, the proportion of fixed retrogenes originating on the X (33%) is much higher than the genome-wide expectation. We find the same result when using fixed retrogene data from another recent study (Zhang et al. 2010). This result strongly rejects the hypothesis that mutational biases could be responsible for the excess of fixed retrogenes originating on the X chromosome and landing on the autosomes. Instead, our data provide direct support for the hypothesis that natural selection is driving the fixation of retrogene polymorphisms off the X.
Table 3.
Excess fixation of retrogenes originating on the X chromosome
If the movement of retrogenes off the X is indeed driven by positive selection, we would also expect there to be signatures of this selection in flanking nucleotide variation, as has been found previously for transposed duplicates (Yi and Charlesworth 2000). Unfortunately, such an analysis requires that we know the insertion location of the retroCNVs: Otherwise, we do not know what nucleotide variation to examine. Paired-end Illumina data offer the opportunity to map the location of inserts when one read comes from the retroCNV and the other comes from flanking DNA at the insertion site (cf. Lee et al. 2008). However, we could not confidently map any of the detected retroCNVs, perhaps due to a combination of low paired-end coverage and retroCNVs often being inserted into repetitive sequence, as observed for nontandem polymorphic duplications detected in these same sequenced lines (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.). Indeed, paired-end mapping suggests that ∼55% of retroCNVs are inserted into repetitive elements (data not shown).
While our analysis of polymorphic retrocopies and fixed retrogenes provides support for adaptive hypotheses of gene movement off the X, it does not distinguish between the two major selective explanations for this pattern. One hypothesis for the advantage of moving off the X is that the new, autosomal copy escapes from precocious silencing of the X chromosome (Betrán et al. 2002; Vibranovski et al. 2009a). The fact that most new retrogenes have testis-biased expression (Betrán et al. 2002; Meisel et al. 2009), especially in post-meiotic cells (Vibranovski et al. 2009a), has been taken as evidence that selection favors germline expression of these genes after the X chromosome has been inactivated. A second hypothesis for the advantage of moving off the X is that sexually antagonistic forces will favor male-beneficial/female-harmful alleles that are located on autosomes because the X chromosome is in females two-thirds of the time (Wu and Xu 2003; Vicoso and Charlesworth 2006). The testis-biased expression in retrogenes has also been used to support the idea that these genes are involved in sexual antagonism, though there is no direct evidence for such a role. Identifying a large number of retroCNVs in D. melanogaster offers the opportunity to test several competing predictions of these two models. For instance, if gene expression in the daughter retroCNV can be distinguished from expression at the parental locus, one can examine patterns of expression among polymorphic retrogenes. As with the comparison of polymorphic X- and autosome-linked retrocopies above, we can ask whether those retroCNVs with testis-biased expression are fixing at higher rates than those without. In order to test predictions of the sexual antagonism hypothesis, strains of D. melanogaster that demonstrate strong sexual antagonism (e.g., Rice et al. 2005) can be screened for the presence of retroCNVs that have moved off the X. Conversely, the lines genotyped here can be measured at antagonistic phenotypes, with obvious predictions based on the presence or absence of retroCNVs.
Regardless of the precise selective forces driving overall patterns of retrogene movement, our study demonstrates that a large number of these polymorphisms, along with intron presence/absence polymorphisms, are detectable by next-generation sequencing. Given the considerable amount of variation found in our study, we believe that more effort should put into the detection and analysis of these types of variants in organisms with assembled genomes, as methods like ours will surely detect many new polymorphisms in a variety of species and improve our understanding of the mutational and selective forces affecting the evolution of gene families.
Methods
Sequenced DPGP inbred lines
As part of the DPGP (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.), 37 inbred D. melanogaster lines from Raleigh, North Carolina, were sequenced with Illumina technology, yielding ∼10× average coverage of 36-bp single-end reads for each inbred line. (Read data are available on the NCBI Short Read Archive under project ID SRP000224: http://www.ncbi.nlm.nih.gov/sra/SRP000224.) The reads were mapped to release 5 of the D. melanogaster reference genome using the software package MAQ (Li et al. 2008; CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.), and read-depth was recorded at every position. Additionally, two of these lines, RAL-437 and RAL-765, were sequenced to ∼3.5× and ∼2.8× coverage, respectively, with 45-bp paired-end Illumina reads, each with a 250-bp insert. The sections below detail how these data were used to detect and validate retroCNVs and missing introns.
Database of unique exon–exon junctions
D. melanogaster gene annotations were acquired from FlyBase version 5.23 (Tweedie et al. 2009). For each intron in each FlyBase transcript, the 36 (or fewer in the case of smaller exons) bases flanking each side of the intron were extracted and concatenated together to yield the exon–exon junction sequence that would be present in any retrotransposed copy of the gene but not in the parental gene copy (Fig. 1). Such an exon junction would also be present in a gene missing an intron (Fig. 2). To prevent spurious matches, each of these exon–exon junctions was then searched against release 5 of the D. melanogaster genome using BLAST. Any junction for which a 20-bp stretch overlapping it (from the last 10 bases of the first exon to the first 10 bases of the second exon) was found in the reference genome was removed from the analysis. Although this step ensures that no false-positive retroCNVs are inferred due to fixed retrocopies present in the reference genome, another consequence is we are unable to detect retrocopies originating from genes already containing a retrocopy in the reference. The remaining 51,453 exon–exon junctions were then included in a database of unique exon–exon junctions.
Mapping reads to exon–exon junctions
We gathered all sequence reads that were not successfully mapped to the reference genome in each of the 37 lines and attempted to map them against the exon–exon junction database described above. Any exon–exon junction having at least two reads (either in the same or different lines) mapped to it such that at least 5 bp of the read maps across the exon boundaries with at most one mismatch on each side of the boundary was considered as either a putative retroCNV or a putative intron deletion (Supplemental Fig. 1). To differentiate between these two cases, we examined the read-depth of the intron located between the two spanned exons. In cases where the average intronic depth in any sequenced line was less than or equal to 1× (i.e., one read per base pair), the exon–exon junction was considered to correspond to an intron that is missing in one or more of the lines. In 34 cases the introns had read-depth well over 1× and the exon–exon junction was considered to belong to a gene having a retrocopy in one or more lines. In one case, an exon–exon junction shared by two genes (pex1 and btl) was spanned by multiple reads, and the gene with a higher ratio of exonic to intronic read-depth (pex1) was inferred to be the parental gene.
Sensitivity of retroCNV calls to changes in mapping parameters
We modified a number of cutoffs involved in the retroCNV calling procedure to determine the extent to which each affected the final number of retroCNV calls. First, we began by examining the effect of a more stringent requirement for exon–exon junctions to be considered unique and included in the search. When we require each exon–exon junction to have no hits spanning the region from 5 bp upstream of the junction to 5 bp downstream, rather than 10 bp in both directions, 33 of 34 retroCNVs remain in the final count. We then examined the effect of changing read mapping cutoffs, finding that requiring each exon–exon junction to be spanned by four reads (rather than two) to be considered part of a retroCNV reduced the number of calls to 26. Finally, requiring each read to cross 10 bp on either side of the exon–exon junction rather than just 5 bp reduced the number of retroCNV calls to 21.
Detecting imperfect intron deletions
In a number of cases where the read-depth was higher in the exons than introns and the introns had less than 1× average depth, we failed to detect reads spanning the exon–exon junctions. Therefore, in order to detect intron deletions that did not perfectly match annotated intron boundaries, we examined all introns with less than 1× average depth in any of the 37 lines. For each of these introns, we found stretches of sequence in the reference genome to which no reads from the corresponding strain could be mapped by MAQ. The beginnings and ends of these stretches were then inferred to be the breakpoints of “imperfect” intronic deletions relative to the reference genome (Supplemental Fig. 2a). In cases where this strategy yielded multiple sets of possible breakpoints for the same intron in different lines, all possibilities were considered. Similar to the creation of the exon–exon junction database described above, sequences flanking each set of possible deletion breakpoints were extracted from the reference genome, concatenated, and incorporated into a database to be searched against by reads not matching the reference genome. Since deletion breakpoints may not necessarily correspond exactly to regions of zero depth, we used the program BWA (Li and Durbin 2009) to map reads because it allows mapping with small indels. BWA was used to search all unmapped reads against this database of concatenated sequences flanking putative deletions (Supplemental Fig. 2b).
To validate cases where reads were mapped by BWA to entries in this database as true deletions relative to the reference, we used BLAT (Kent 2002) to map these reads to release 5 of the D. melanogaster reference genome. The resulting alignment was then examined to ensure that it was consistent with an imperfect intronic deletion. The sequence between the putative deletion breakpoints was then examined manually to ensure that no other reasonable alignment existed for any read suggestive of an imperfect intronic deletion. This procedure was also performed to validate read mappings supporting deletions perfectly removing annotated introns.
Experimental validation of retroCNVs and deleted introns
In order to validate our computational predictions, we made primers designed to span 19 exon–exon junctions corresponding to seven retroCNVs and one deleted intron. If a retrogene is present, then PCR in the appropriate inbred line should result in two amplified sequences: one long sequence containing the intron (from the parental copy), and one short sequence missing the intron (from the retrocopy). If an intron is deleted, only one PCR product (lacking an intron) should be produced. The design of this experiment is illustrated in Supplemental Figure 3a. An image of a gel showing a true positive and a false positive is shown in Supplemental Figure 3b.
We also used paired-end Illumina sequences to validate calls in two of the inbred lines, RAL-437 and RAL-765. Paired-end mapping data is useful for detecting deletion polymorphisms, as paired reads from an individual containing a deletion will appear further apart than expected when mapped to a reference genome not containing the deletion (e.g., Tuzun et al. 2005; Korbel et al. 2007; Kidd et al. 2008). If two exons are located adjacent to one another in the sample genome with no intervening sequence, either due to an intron deletion relative to the reference genome or because of the presence of a retrogene, paired-ends spanning the two exons would appear farther apart than expected (∼250 bp in our data) when mapped to the reference genome (Supplemental Fig. 4). Based on the distribution of paired-end distances calculated from all reads mapped to the same chromosome arm in the expected orientation, we expect <2% of all inserts to be >350 bp apart. Thus, any skipped exon junction (corresponding to a retroCNV or a missing intron) lying within a region spanned by an insert inferred to be >350 bp long was considered confirmed by the paired-end data. As shown by Langley and colleagues (CH Langley, K Stevens, C Cardeno, YCG Lee, DR Schrider, JE Pool, SA Langley, C Suarez, R Detig-Corbett, B Kolaczkowski, et al., in prep.), the 350-bp cutoff for this paired-end data set results in very few spurious called deletions.
Estimating false-negative rates
In order to estimate the false-negative rate of our method, we randomly drew 1000 genomic positions and determined whether these positions were covered using the same cutoffs required to call exon–exon junctions corresponding to missing introns or retroCNVs. In other words, in order to be considered covered, each position must have been spanned by at least two reads, with at least 5 bp of the read landing on either side of the positions, and the reads having no more than one mismatch on either side of the position. Each of the 1000 unique positions used was a randomly selected boundary between exons and introns, though similar false-negative rates are obtained if random genomic coordinates are used (data not shown). False-negative rates were calculated in three randomly selected lines: RAL-303 (9.8%), RAL-307 (6.8%), and RAL-732 (5.1%). We note that the true false-negative rate for retrocopies or missing introns found in poorly covered heterochromatic regions is likely significantly higher than these estimates.
Acknowledgments
We thank A. Kern for conducting the frequency spectrum analysis and also thank D. Begun, A. Tucker, and K. Pollard for very helpful suggestions. K.S., C.M.C., and C.H.L. are supported by National Institutes of Health grant HG02942. D.R.S. and M.W.H. are supported by National Science Foundation grant DBI-0845494.
Footnotes
[Supplemental material is available for this article.]
Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116434.110.
References
- Bai YS, Casola C, Feschotte C, Betran E 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol 8: R11 doi: 10.1186/gb-2007-8-1-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Long M 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164: 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Thornton K, Long M 2002. Retroposed new genes out of the X in Drosophila. Genome Res 12: 1854–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Kaessmann H 2004. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet 36: 1061–1063 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang YJ, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J 2007. Intron loss and gain in Drosophila. Mol Biol Evol 24: 2842–2850 [DOI] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol 2: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai HZ, Yoshimatsu TF, Long MY 2006. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 385: 96–102 [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW 2009. The life and death of gene families. Bioessays 31: 29–39 [DOI] [PubMed] [Google Scholar]
- Derr LK, Strathern JN 1993. A role for reverse transcripts in gene conversion. Nature 361: 170–173 [DOI] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long MY 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540 [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Farlow A, Meduri E, Dolezal M, Hua LS, Schlotterer C 2010. Nonsense-mediated decay enables intron gain in Drosophila. PLoS Genet 6: e1000819 doi: 10.1371/journal.pgen.1000819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR 1987. Pseudogenes in yeast? Cell 49: 5–6 [DOI] [PubMed] [Google Scholar]
- Fontanillas P, Hartl DL, Reuter M 2007. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet 3: e210 doi: 10.1371/journal.pgen.0030210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell BG., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. 1996. Life with 6000 genes. Science 274: 546–567 [DOI] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Fang S, Wu CI 2006. Adaptive loss of an old duplicated gene during incipient speciation. Mol Biol Evol 23: 401–410 [DOI] [PubMed] [Google Scholar]
- Hollis GF, Hieter PA, McBride OW, Swan D, Leder P 1982. Processed genes: a dispersed human-immunoglobulin gene bearing evidence of rna-type processing. Nature 296: 321–325 [DOI] [PubMed] [Google Scholar]
- Karin M, Richards RI 1982. Human metallothionein genes: primary structure of the metallothionein-II gene and a related processed gene. Nature 299: 797–802 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheran E, Brudno M 2008. A robust framework for detecting structural variations in a genome. Bioinformatics 24: I59–I67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WL, Tucker AE, Sung W, Thomas WK, Lynch M 2009. Extensive, recent intron gains in Daphnia populations. Science 326: 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A, Comeron JM, Brunet FG, Lachaise D, Long M 2002. Intron presence-absence polymorphism in Drosophila driven by positive Darwinian selection. Proc Natl Acad Sci 99: 8121–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MY, Langley CH 1993. Natural-selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91–95 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654 [DOI] [PubMed] [Google Scholar]
- Meisel RP 2009. Evolutionary dynamics of recently duplicated genes: selective constraints on diverging paralogs in the Drosophila pseudoobscura genome. J Mol Evol 69: 81–93 [DOI] [PubMed] [Google Scholar]
- Meisel RP, Han MV, Hahn MW 2009. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol Evol 1: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metta M, Schlotterer C 2010. Non-random genomic integration - an intrinsic property of retrogenes in Drosophila? BMC Evol Biol 10: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Nakai K 2008. Retrotransposition as a source of new promoters. Mol Biol Evol 25: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol 6: e80 doi: 10.1371/journal.pbio.0060080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745 [DOI] [PubMed] [Google Scholar]
- Rice WR, Linder JE, Friberg U, Lew TA, Morrow EH, Stewart AD 2005. Inter-locus antagonistic coevolution as an engine of speciation: Assessment with hemiclonal analysis. Proc Natl Acad Sci 102: 6527–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W 2005. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc Natl Acad Sci 102: 5773–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hahn MW 2010. Gene copy number polymorphism in nature. Proc Biol Sci doi: 10.1098/rspb.2010.1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS 2006. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 5: 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Hahn MW 2010. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics. doi: 10.1534/genetics.110.118794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Nakai S, Nishida Y, Hisajima H, Honjo T 1982. Long terminal repeat-like elements flank a human-immunoglobulin epsilon pseudogene that lacks introns. EMBO J 1: 1539–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M 2009a. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet 5: e1000731 doi: 10.1371/journal.pgen.1000731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long MY 2009b. General gene movement off the X chromosome in the Drosophila genus. Genome Res 19: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B 2006. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet 7: 645–653 [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci 103: 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Xu EY 2003. Sexual antagonism and X inactivation - the SAXI hypothesis. Trends Genet 19: 243–247 [DOI] [PubMed] [Google Scholar]
- Yi S, Charlesworth B 2000. A selective sweep associated with a recent gene transposition in Drosophila miranda. Genetics 156: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long MY 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res 20: 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]