Abstract
While translational stop codon readthrough is often used by viral genomes, it has been observed for only a handful of eukaryotic genes. We previously used comparative genomics evidence to recognize protein-coding regions in 12 species of Drosophila and showed that for 149 genes, the open reading frame following the stop codon has a protein-coding conservation signature, hinting that stop codon readthrough might be common in Drosophila. We return to this observation armed with deep RNA sequence data from the modENCODE project, an improved higher-resolution comparative genomics metric for detecting protein-coding regions, comparative sequence information from additional species, and directed experimental evidence. We report an expanded set of 283 readthrough candidates, including 16 double-readthrough candidates; these were manually curated to rule out alternatives such as A-to-I editing, alternative splicing, dicistronic translation, and selenocysteine incorporation. We report experimental evidence of translation using GFP tagging and mass spectrometry for several readthrough regions. We find that the set of readthrough candidates differs from other genes in length, composition, conservation, stop codon context, and in some cases, conserved stem–loops, providing clues about readthrough regulation and potential mechanisms. Lastly, we expand our studies beyond Drosophila and find evidence of abundant readthrough in several other insect species and one crustacean, and several readthrough candidates in nematode and human, suggesting that functionally important translational stop codon readthrough is significantly more prevalent in Metazoa than previously recognized.
While the three stop codons UAG, UGA, and UAA typically lead to termination of translation and ribosome detachment from the messenger RNA (mRNA) molecule, protein translation can sometimes continue through a stop codon, a mechanism known as “stop codon readthrough.” During readthrough, the ribosome can insert an amino acid and continue translation in the same reading frame until a subsequent stop codon, so that a fraction of the resulting proteins include additional peptides (Doronina and Brown 2006; Namy and Rousset 2010). The tRNA that inserts the amino acid can be a cognate of the stop codon (a stop suppressor tRNA). Alternatively, a selenocysteine tRNA can insert a selenocysteine amino acid for UGA stop codons, if a selenocysteine insertion sequence (SECIS element) is present in the 3′-untranslated region (UTR). Lastly, for certain “leaky” stop codon contexts (more frequently subject to readthrough), a near-cognate tRNA can insert its cognate amino acid instead (Bonetti et al. 1995; Poole et al. 1998), which can result in a readthrough level of >5% (Namy et al. 2001). Downstream translation could alternatively result from a ribosomal bypassing event (Wills 2010), whereby the ribosome continues translation in the same or a different reading frame, thus failing to recognize a stop codon, in which case, the stop codon is bypassed but not read through.
Readthrough provides a compelling regulatory mechanism for exposing additional C-terminal domains of a protein at a lower abundance, which in contrast to alternative splicing can be regulated at the translational level. Viruses use this mechanism not only to increase functional versatility in a compact genome but also to control the ratio of two protein isoforms (Beier and Grimm 2001; Namy and Rousset 2010). In eukaryotes the ratio between readthrough and non-readthrough proteins can be controlled for many genes simultaneously by regulation of release factor proteins, themselves sometimes regulated post-translationally (von der Haar and Tuite 2007), and can vary by tissue (Robinson and Cooley 1997) and development stage (Samson et al. 1995). Readthrough interacts with many other regulatory processes: On one hand, it is enhanced by factors that reduce translational elongation and fidelity; on the other hand, it can affect mRNA abundance by slowing down transcript deadenylation, reducing nonsense-mediated decay, and triggering no-go decay or non-stop decay (von der Haar and Tuite 2007). Readthrough has been proposed as an evolutionary catalyst in yeast, where it is epigenetically controlled via a prion protein state, thus enabling the adaptation of new domains translated at low rates during normal growth but at higher rates in periods of stress when they might provide a selective advantage (True and Lindquist 2000). Readthrough, which extends the C terminus of the protein, is complementary to leaky AUG recognition, a mechanism for protein diversification at the N terminus by starting translation at a subsequent start codon, which has been shown to create pairs of similar proteins with different localizations or biological functions (Touriol et al. 2003). Lastly, readthrough can be induced by small molecules or by introducing suppressor tRNAs, offering potential new therapeutic avenues for patients with nonsense mutations (Keeling and Bedwell 2010).
In eukaryotic genomes, readthrough has been thought to play only a minor role outside selenocysteine incorporation, experimentally observed for only six wild-type genes in three species: Drosophila melanogaster genes syn (Klagges et al. 1996), kel (Robinson and Cooley 1997), and hdc (Steneberg and Samakovlis 2001); Saccharomyces cerevisiae genes PDE2 and IMP3 (Namy et al. 2002; Namy et al. 2003); and the rabbit beta-globin gene (Chittum et al. 1998). In addition, two nonsense alleles of elav (Samson et al. 1995) and a nonsense allele of wg (Chao et al. 2003) in D. melanogaster are known to undergo readthrough, and two additional D. melanogaster candidates, Sxl and oaf, have been proposed based on long ORFs downstream from the stop codon (Samuels et al. 1991; Bergstrom et al. 1995). According to the Recode-2 database (Bekaert et al. 2010), the only other known cases of readthrough in eukaryotes are in transposable elements that could be endogenous retroviruses.
The story changed dramatically with the sequencing of 12 Drosophila genomes (Drosophila 12 Genomes Consortium et al. 2007; Stark et al. 2007), which enabled a search for readthrough genes through the evolutionary lens of comparative genomics analysis. While evolutionary signatures associated with protein-coding selection usually showed sharp boundaries coinciding with the exact boundaries of protein-coding genes, we found a surprising 149 readthrough candidates for which the protein-coding constraint extends past the stop codon until the next in-frame stop codon (Fig. 1; Lin et al. 2007), suggesting not only that translation does not always stop at the stop codon but also that the specific polypeptide sequence of the extended protein confers selective advantages at the protein level. At the time, we ruled out selenocysteine insertion, but postulated that perhaps adenosine-to-inosine (A-to-I) editing was responsible for the observed signature, by editing away stop codons into tryptophan codons, since readthrough candidates were enriched for nervous system genes where editing is most active. However, we acknowledged that alternative explanations were possible, such as precisely positioned alternative splicing, which would also explain the observed signatures without readthrough.
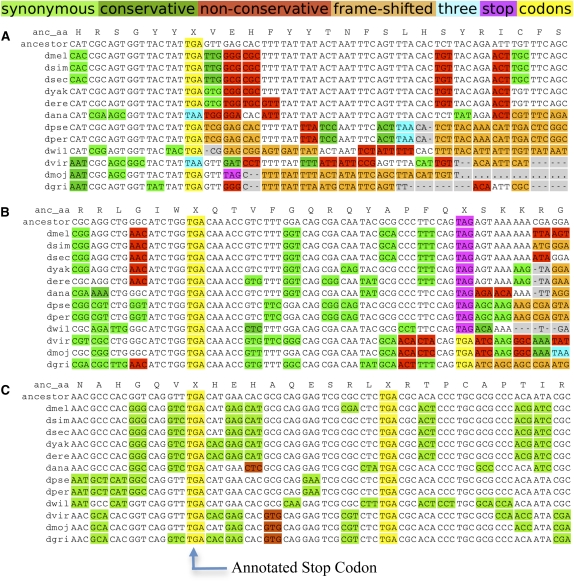
Figure 1.
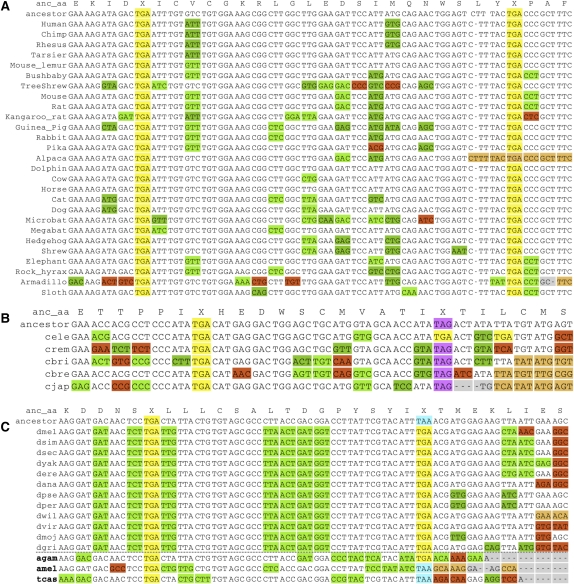
Protein-coding evolutionary signatures for typical, readthrough, and double-readthrough stop codons. Alignments surrounding the annotated stop codons of three genes for 12 Drosophila species and their inferred maximum-parsimony common ancestor. The color coding of substitutions and insertions/deletions (indels) relative to the common ancestor is a simplification for visualization purposes, as the actual PhyloCSF score sums over all possible ancestral sequences and weighs every codon substitution by its probability. Insertions in other species relative to D. melanogaster are not shown. (A) Alignment of a typical gene (bw), shows abundant synonymous and conservative substitutions (green) upstream of the stop codon, and many non-conservative substitutions (red), frameshifting indels (orange), and in-frame stop codons downstream from the stop codon. The stop codon locus shows several substitutions between different stop codons. (B) Alignment of CG17319, one of 283 readthrough candidates. The region between the annotated stop codon and the next in-frame stop codon shows mostly synonymous substitutions and lacks frameshifting indels, while the region downstream from the second stop shows non-conservative substitutions and indels typical of non-coding regions, providing evidence of continued protein-coding selection in the region between the two stop codons, and suggesting likely translational readthrough of the first stop codon. As is typical for readthrough candidates, the first stop codon is perfectly conserved, while the second stop codon shows substitutions between different stop codons. (C) Alignment of a double-readthrough candidate, Glu-RIB (one of 16 cases). Both the second ORF and the third ORF show protein-coding signatures, indicating that both stop codons are likely readthrough. Both readthrough stop codon positions show no substitutions.
For the three years since our initial findings, the phenomenon has remained a mystery, with no resolution about what underlying mechanism may enable translation of our candidate regions, no direct evidence of translation, and no evidence of how extensive readthrough is across the animal kingdom. To address these questions, we exploit the vast new transcriptional evidence provided by modENCODE data sets (Graveley et al. 2011) and new phylogenetic methods for detecting protein-coding selection (Lin et al. 2011). We manually curated a list of almost 300 genes that nearly doubles the previous number of candidates, and we show that translational readthrough is the only plausible explanation for most of these. We provide experimental evidence of downstream translation in several cases using GFP transgenic flies and mass spectrometry. We also investigate additional genomic properties of readthrough candidates that yield insights into their mechanisms of function. Lastly, we apply several genomic techniques to search for evidence of readthrough in additional genomes using both comparative and single-species evidence, revealing readthrough across the animal kingdom. The result is an expanded picture of abundant readthrough in Drosophila and related species, many new insights into the mechanism and evolution of readthrough, and a dramatically expanded list of species showing evidence of translational readthrough, including the human.
Results
Comparative evidence and list of candidates
We first sought to expand and substantiate our previous list of D. melanogaster readthrough candidates by generating an initial list of likely coding regions downstream from stop codons and then using newer computational methods and data sets followed by manual curation to eliminate those for which there is a plausible explanation other than translational stop codon readthrough (Fig. 2). For each annotated protein-coding transcript, we evaluated the coding potential of the region between the annotated stop codon (“first stop codon”) and the next in-frame stop codon (“second stop codon”), which we refer to as the “second ORF,” or, for readthrough candidates, as the “readthrough region.” We refer to the codon position aligned to the first or the second stop codon in each species as the first or second “stop locus,” respectively.
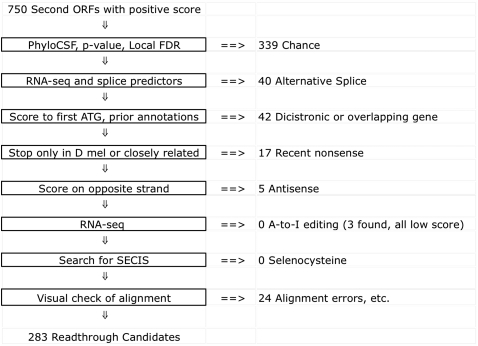
Figure 2.
Manual curation distinguishes 283 readthrough candidates. Steps of filtering method used to eliminate transcripts with other plausible explanations for the observed second-ORF protein-coding selection, leading to the final list of 283 unambiguous readthrough candidates.
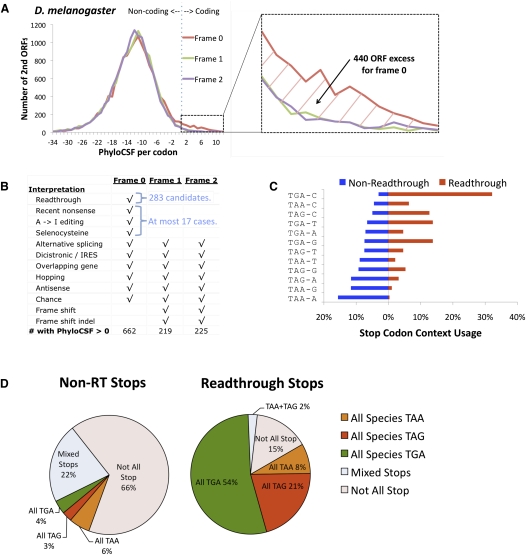
We scored the protein-coding potential of each second ORF using PhyloCSF (Lin et al. 2011), an improved, phylogenetic version of the Codon Substitution Frequency (CSF) score we had previously applied (Lin et al. 2008) to identify short, conserved coding regions. This new algorithm, based on a statistical comparison of phylogenetic codon models, provides higher resolution for detecting protein-coding evolutionary signatures in small regions. For example, the alignment of the second codon in Figure 1B (headed by AGG) is 1145 times more likely to occur in a coding region than in a non-coding region. The PhyloCSF score is a log-likelihood ratio, with a positive score indicating that the alignment is more likely to occur in a coding region than in a non-coding one. We found 750 transcripts with positive second-ORF PhyloCSF scores, of which 411 had sufficiently high scores that they were unlikely due to chance (see Methods).
We used next-generation deep transcript sequencing data (RNA-seq) to detect alternative splicing events or RNA editing that could potentially explain the observed protein-coding selection in the absence of readthrough. The modENCODE project has provided 2.3 billion reads across 30 stages of development, which recover 93% of known splice junctions across all of the genes we were evaluating. We used these data, supplemented with EST and mRNA evidence reported by the UCSC Genome Browser and with two genomic splice prediction algorithms, to look for possible 3′-splice acceptor sites shortly downstream from the first stop codon. This revealed likely splice sites in only 40 of our 411 candidates with strongly positive scores, suggesting that splicing is not a likely explanation for the observed widespread phenomenon, and leaving 371 candidates that are not explained by alternative splicing. We also used RNA-seq data to show that continued translation of the second ORF is not a result of RNA editing of adenosine to inosine (Aphasizhev 2007), which we had originally hypothesized would convert stop codons to UGG tryptophan codons (Lin et al. 2007). In fact, no readthrough stop codons among our candidates were found to be edited (Supplemental Text S1).
We next excluded 42 readthrough regions that overlap another gene in the same frame or a possible internal ribosome entry site (IRES) marking the start of a second cistron (an independent complete ORF in the 3′ UTR of a coding transcript), using existing annotations and scoring the region between the first stop and subsequent in-frame ATGs (as second cistrons would follow lower-scoring untranslated regions). While we cannot rule out the possibility that some of the remaining candidates are translated through a non-AUG IRES (Sugiharas et al. 1990; DeSimone and White 1993; Takahashia et al. 2005), we have used a similar analysis to show that this could not explain the evolutionary signatures for most candidates (Supplemental Text S2). We also excluded 17 possible recent nonsense mutations in D. melanogaster, five cases where the positive PhyloCSF score could be due to overlap with coding regions on the opposite strand (“antisense”), and 24 for other reasons such as potentially incorrect alignments. Additional details of the manual curation are provided in Supplemental Text S2.
We also ruled out both selenocysteine insertion and a related Drosophila readthrough mechanism that can read through TGA stop codons using the same SECIS element and proteins as selenocysteine insertion but without inserting selenocysteine (Hirosawa-Takamori et al. 2009). We looked for SECIS elements in the 3′ UTRs of the remaining transcripts using an existing program, SECISearch (Kryukov et al. 2003), and found none scoring above the recommended threshold. We also studied readthrough codon alignments in Drosophila willistoni, a Drosophila species that has lost most of the selenocysteine incorporation machinery and thus substitutes a cysteine codon for the TGA stop at all known selenocysteine insertion sites (Chapple and Guigo 2008). Indeed, D. willistoni retains the TGA stop and shows continued protein-coding selection in the second ORF for nearly all readthrough candidates with TGA first stop codons, suggesting that selenocysteine insertion does not explain more than a handful of our candidates, if any. Further evidence ruling out selenocysteine insertion is presented in Supplemental Text S2.
We were left with 283 transcripts (Supplemental Data 1), henceforth referred to as the “readthrough candidates,” for which the most plausible explanation of the observed comparative signature is functional and evolutionarily conserved stop codon readthrough. The readthrough regions vary in length from 4 to 788 codons, with a mean of 67 codons. We similarly curated the subsequent open reading frame (“third ORF”) of each readthrough candidate and found 16 double-readthrough candidates (Fig. 1C). We did not find any candidates for triple readthrough.
Experimental evidence of readthrough
We used two experimental lines of evidence to verify that readthrough occurs in several of our candidates.
Evidence of stop codon readthrough using GFP transgenic fly strains
We created transgenic flies in which the second stop codon of a readthrough candidate was replaced with a construct encoding GFP, so that GFP would be expressed if the first stop codon were read through (Fig. 3A), and tested the resulting strains for the presence of GFP in embryos and larvae.
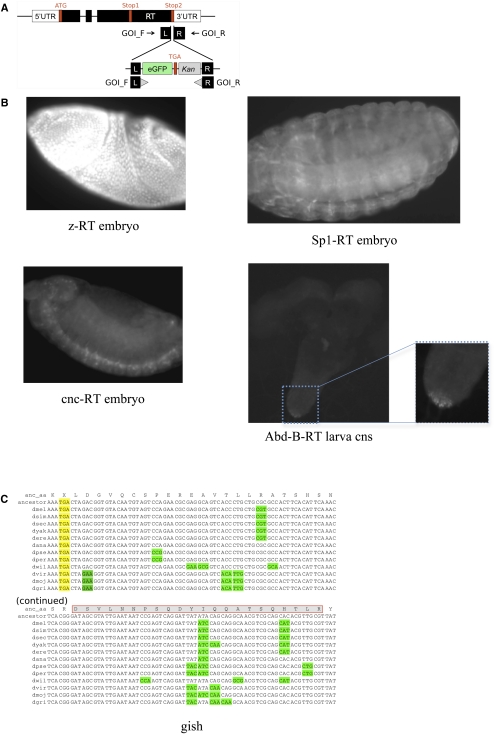
Figure 3.
Experimental validation of readthrough. (A) GFP insert construct replacing the second stop codon so that GFP is only observed after translation of the 3′ end of the second ORF and subsequent eGFP gene. GOI_F and GOI_R are 50-bp homology arms on the forward and reverse strands specific to each gene of interest (GOI). (B) Expression of GFP in transgenic constructs showing that translation continues through to the second stop codon for four of the readthrough (RT) candidates. Strains shown are z-RT, Sp1-RT, and cnc-RT in embryos, and Abd-B-RT in the central nervous system of a larva. No GFP expression was found in a wild-type strain used as a control (Supplemental Fig. S12). (C) Mass spectrometry evidence of readthrough. Example of readthrough region (gish) supported by a 22-amino-acid peptide match (red rectangle) to mass spectrometry Drosophila PeptideAtlas (one of nine cases). With no ATG codon between the stop codon and the peptide, and no observed alternative splicing events across thousands of RNA-seq reads overlapping this region, readthrough seems the only plausible explanation for translation of this peptide.
We created five transgenic fly strains labeled Abd-B-RT, cnc-RT, Jra-RT, Sp1-RT, and z-RT for readthrough candidates Abd-B, cnc, Jra, Sp1, and z, respectively. We chose readthrough candidates that are particularly unlikely due to dicistronic translation via an IRES: Two do not contain an ATG codon, and for two more the only ATG codon is not conserved, and all five are unlikely to function as independent ORFs given their short lengths (16, 47, 83, 11, and 30 amino acids, respectively). Moreover, between 266 and 2249 RNA-seq reads covering the readthrough regions of each of these genes in the specific development stages observed confirm that the stop codon is not bypassed via splicing or RNA editing.
We observed GFP expression in four of the five strains (Fig. 3B), demonstrating translation of eGFP, and thus confirming that readthrough occurs. Two of these (cnc-RT and Sp1-RT) were observed in embryos, one in larvae (Abd-B-RT), and one in both embryos and larvae (z-RT). We did not observe GFP expression in Jra-RT in either embryos or larvae, but this does not preclude readthrough of that gene in other stages. We confirmed that a wild-type strain used as a control does not show GFP expression (Supplemental Fig. S12).
Mass spectrometry evidence of protein translation
We also found evidence that several readthrough regions are translated by searching a mass spectrometry database for sequenced Drosophila peptides that match within a readthrough region. We used the Drosophila PeptideAtlas (Loevenich et al. 2009), which contains 75,753 distinct peptides between four and 55 amino acids long matching annotated coding regions, and an additional 971 peptides matching a whole-genome six-frame translation.
We found 14 distinct peptides that match our readthrough regions (Fig. 3C; Supplemental Data 1). We used BLAST to verify that these peptides do not match elsewhere within the D. melanogaster genome. Since most peptides in the database were sequenced by comparison to annotated coding regions, it is not surprising that there were no matches to most of our readthrough regions.
These sequenced peptides provide experimental evidence of translation within the readthrough regions of nine of our readthrough candidates, only two of which, syn and kel, had been previously experimentally observed. We note that most of the peptide matches are single hits, which are known to have a false-positive rate of 35% or more (Schrimpf et al. 2009), so some might not have been correctly identified. However, for those peptides that were correctly identified, our manual curation process provided strong evidence that downstream translation is not due to alternative splicing or dicistronic translation (Supplemental Text S3).
Single-species evidence of protein translation
Nucleotide k-mer composition, synonymous SNP bias, and periodicity of secondary structure pairing frequency match those of protein-coding regions
Seeking to complement our comparative genomics evidence with single-species evidence of readthrough, we obtained three additional lines of evidence that most of the predicted readthrough regions are, indeed, protein-coding.
First, we found that the nucleotide k-mer composition of readthrough regions matched that of protein-coding regions. We quantified protein-coding-like compositional properties using the Z curve score (Gao and Zhang 2004), which provides a single-species test for protein-coding regions using mono-, di-, and tri-nucleotide frequencies. Although most individual readthrough regions are too short to reliably call as protein-coding using the Z curve score alone, we found that it provided very strong group discrimination (Fig. 4A) between coding regions immediately upstream of stop codons (Fig. 4A, top panel) and non-coding regions immediately downstream from stop codons for non-readthrough transcripts (Fig. 4A, middle panel). For readthrough candidates, the distributions of scores upstream of the first stop was indistinguishable from that of coding regions for non-readthrough transcripts (Fig. 4A, top panel), and the distribution downstream from the second stop was indistinguishable from that of non-coding regions (Fig. 4A, middle panel), as expected. Strikingly, the distribution of scores for the readthrough portion of readthrough candidates matched that of coding regions (Fig. 4A, bottom panel). We conclude that single-species sequence composition strongly supports the comparative evidence that the candidate readthrough regions are predominantly protein-coding.
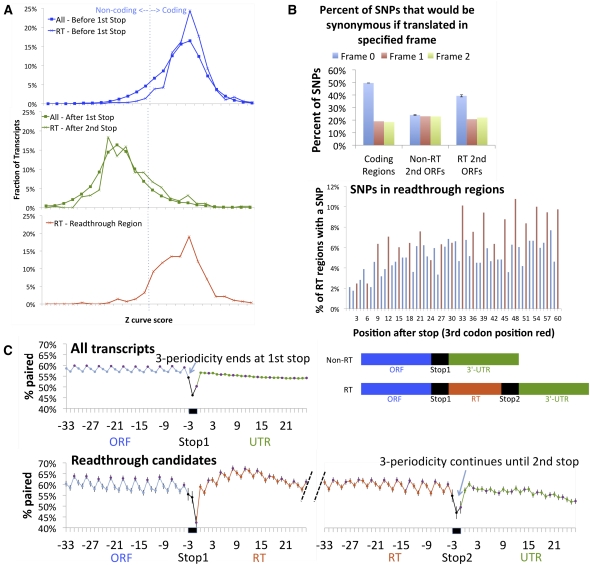
Figure 4.
Single-species evidence of readthrough region translation. (A) D. melanogaster sequence composition of readthrough regions as measured by the Z curve statistic (x-axis) suggests they are protein-coding (positive scores). (Top panel) Coding regions before the first stop for both readthrough candidates (crosses) and non-readthrough transcripts (squares) show positive Z curve scores typical of protein-coding regions. (Middle panel) Non-coding regions after the second stop for readthrough candidates (crosses) and after the first stop for typical transcripts (squares) show negative Z curve scores typical of non-coding regions. (Bottom panel) Readthrough regions show positives scores typical of protein-coding regions, providing single-species evidence that most readthrough regions are protein-coding. Evaluated regions in all panels were selected to match the length distribution of readthrough regions. (B) Single nucleotide polymorphisms (SNPs) show a strong bias to result in synonymous codon substitutions in readthrough regions (top right) and coding regions (top left), but no bias is seen in second ORFs downstream from non-readthrough stop codons (top middle), providing evidence that readthrough regions are under protein-coding selection within the D. melanogaster population. For each type of region we show the fraction of SNPs that would be synonymous if translated in each of three frames, with frame 0 matching the translated frame of the coding region of the gene. Error bars show the Standard Error of the Mean (SEM). As most third codon positions result in synonymous substitutions, the exclusion of non-synonymous substitutions is also visible as a periodicity in the fraction of readthrough candidates that have an SNP at each position of the second ORF (bottom panel), with third-codon-position SNPs (red) more prevalent than first or second-codon position SNPs (blue). This plot also shows an overall decrease in the number of SNPs near the readthrough stop codon, likely due to additional signals involved in regulating readthrough, such as RNA structures, encoded within the protein-coding signal. (C) Periodic base-pairing frequency in readthrough regions (red) matches that of known coding regions (blue) but is different from that of UTRs (green). Fraction of transcripts for which a given nucleotide is paired in predicted RNA secondary structures (y-axis) at each position relative to a stop codon (x-axis). Third codon positions (purple) are paired more frequently than first or second positions, and stop codons (positions −3, −2, and −1) show decreased pairing, as previously observed computationally in humans and experimentally in yeast (top panel). Transition from periodic to non-periodic pairing happens at the second stop codon for readthrough candidates (bottom panel). Signal is averaged over five codon positions (see Methods), with raw data shown in Supplemental Figure S2. Error bars show the Standard Error of the Mean (SEM).
Second, we found that polymorphism evidence strongly supports continued protein-coding selection in readthrough regions within the D. melanogaster lineage. Single nucleotide polymorphisms (SNPs) show a strong bias to be synonymous in readthrough regions and coding regions, but not in non-readthrough second ORFs (Fig. 4B). Across more than 6 million D. melanogaster SNPs in the Drosophila Population Genomics Project Release 1.0 (dpgp.org, data obtained from ensembl.org), 3490 lie within readthrough regions. Of these, 1372 (39%) result in synonymous codon substitutions, a significant excess compared with the 21% if the same regions were translated in alternate reading frames. By comparison, 50% of substitutions within protein-coding regions are synonymous (vs. 19% on average in alternate reading frames), perhaps because protein-coding constraint is moderately weaker in readthrough regions. In contrast, no frame bias is seen in the second ORFs of non-readthrough transcripts, confirming the stronger protein-coding selection of readthrough regions.
As a third line of evidence, we found that the mRNA secondary structures of the readthrough regions exhibit a periodicity characteristic of coding regions. Previous studies suggest that nucleotides in the third codon position are more likely to be paired in the least-energy mRNA secondary structure than nucleotides in the other two positions (Shabalina et al. 2006; Kertesz et al. 2010), resulting in a three-periodic pairing frequency signal averaged across protein-coding regions, which stops abruptly at the stop codon. This signal has been shown computationally in the human genome (Shabalina et al. 2006) and experimentally in the yeast genome (Kertesz et al. 2010). We reproduced the periodicity result for predicted structures within D. melanogaster coding regions, suggesting that it is likely a general feature of eukaryotic genomes. We found that while for non-readthrough genes the signal stops abruptly at the first stop codon, the three-periodicity continues beyond the annotated stop codon for readthrough candidates and abruptly ends after the second stop codon, consistent with the intervening region being protein-coding (Fig. 4C; Supplemental Fig. S2).
Protein domain evidence of protein translation
To obtain further evidence of protein-coding function in readthrough regions, we searched for homology with known protein domains. Because domain homology is more likely to be found in protein-coding regions than non-coding ones, this strategy was used in an earlier attempt to identify readthrough genes computationally (Sato et al. 2003) and provided further support for our candidates here. Using Search Pfam (Finn et al. 2010), we found 13 significant matches to Pfam protein domain families within the readthrough regions, compared with only five in non-readthrough controls with similar second ORF length (p = 0.045). For example, CG14669, itself annotated as an RAS GTPase in FlyBase, has a readthrough region that also shows a match to an RAS family GTPase domain in the curated Pfam-A database (e-value = 6.9 × 10−21), suggesting roles in vesicle formation, motility, and fusion.
Discerning readthrough mechanism
Several lines of evidence support that the observed protein-coding translation is the result of stop codon readthrough rather than alternative mechanisms.
Reading frame bias confirms readthrough mechanism
First, comparing the PhyloCSF score distributions downstream from the stop codon between the three possible reading frames provided evidence that the observed phenomenon is not due to splicing, dicistronic translation at an internal ribosomal entry site (IRES), ribosomal bypassing (“hopping”), or chance, and also provided an independent estimate of the number of readthrough genes in D. melanogaster.
We found that many more second ORFs have positive PhyloCSF score in the annotated reading frame (reading frame 0, n0 = 662) than in the other two reading frames (n1 = 219 and n2 = 225) (Fig. 5A), as would be expected if there were many readthrough genes. We next analyzed whether other explanations for a positive PhyloCSF score could explain the frame bias (Fig. 5B). Because splice variants, internal ribosome entry sites, overlaps with other genes, antisense genes, ribosomal hopping, and chance high-scoring regions can occur in any of the three frames and do not show a bias toward frame 0 (Supplemental Text S4), they would not explain the frame 0 excess. Moreover, programmed frameshifting or recent frameshifting indels can only lead to a protein-coding region in frame 1 or frame 2, thus the presence of these effects would imply even more readthrough genes to account for the observed frame 0 excess. Lastly, our manual curation determined that nonsense mutations, A-to-I editing, and selenocysteine insertion, which would explain a positive PhyloCSF score specifically in frame 0, account for at most 17 transcripts. Thus, the excess of high-scoring downstream regions in frame 0 implies more than 400 translational stop codon readthrough genes (n0 − n1|2 − 17 > 400). It is worth noting that this number is significantly more than the 283 readthrough candidates, suggesting that more than a hundred additional readthrough genes may exist, perhaps rejected by our stringent manual curation criteria.
Figure 5.
Evidence of readthrough mechanism. (A,B) Excess of high-scoring regions in-frame (frame 0) compared to out-of-frame (frame 1, frame 2) suggests readthrough as the likely mechanism and provides an estimate of readthrough count. (A) PhyloCSF score per codon (x-axis) of the regions starting 0, 1, or 2 bases after all D. melanogaster annotated stop codons (red, green, purple, respectively) and continuing until the next stop codon in that frame, excluding regions that overlap another annotated transcript. Frame 0 shows an excess of more than 400 predicted protein-coding regions compared with the other reading frames, suggesting abundant readthrough. In contrast, a similar plot for Caenorhabditis elegans shows no significant excess in frame 0 (Supplemental Fig. S11), suggesting that the abundance of readthrough in Drosophila is not universal. (B) Possible mechanisms associated with protein-coding function downstream from D. melanogaster stop codons (rows) and associated reading frame offsets where corresponding protein-coding function is expected (columns). Random fluctuations would lead to an even distribution among the three frames, as would unannotated alternative splice variants and unannotated IRESs (note that annotated splice variants and IRESs have already been excluded), while frameshift events and recent frameshifting indels would bias away from frame 0. A bias for in-frame protein-coding selection is expected only for stop codon readthrough, recent nonsense mutations, A-to-I editing, and selenocysteine, the latter three together accounting for at most 17 cases. This leaves readthrough as the only plausible explanation for an excess of ∼420 frame 0 regions with positive PhyloCSF scores. (C) Usage of stop codon context (stop codon and subsequent base) provides additional evidence of a readthrough mechanism. The 4-base contexts are sorted in order of decreasing frequency among the 14,928 non-readthrough stop codons (blue), with less frequent stop codons (top, e.g., TGA-C) experimentally associated with translational leakage in other species and most frequently associated with efficient termination (bottom, e.g., TAA-A). Context frequencies for readthrough candidates (red) are opposite of non-readthrough transcripts, suggesting a preference for leaky context, with one-third using TGA-C and almost none using TAA-A. (D) Increased stop codon conservation in readthrough candidates. Only ∼1/3 of D. melanogaster non-readthrough stop codons have aligned stops in all 12 species, and only ∼1/3 of those are perfectly conserved (i.e., have the same stop codon in all 12 species). In contrast, 83% of candidate readthrough stop codons have an aligned stop in all 12 species, and 97% of those are perfectly conserved. While all three stop codons are involved in readthrough of different genes, individual readthrough genes rarely show substitutions between different stop codons, suggesting that the three stop codons are not functionally equivalent. Moreover, the only eight substitutions observed are between TAA and TAG, with no substitutions involving TGA, even though it is the most frequent readthrough stop codon, suggesting that TAA and TAG are functionally similar.
Stop codon context supports translational leakage
Translation termination efficiency is known to be affected by several signals including the stop codon itself, the three nucleotide positions downstream, the final two amino acids in the nascent peptide, and the tRNA in the ribosomal P-site (Bertram et al. 2001). Of these signals, the strongest determinants consist of the 4-nt sequence that includes the stop codon and one downstream position, which we refer to as the stop codon “context.” In both yeast and mammals, the contexts most efficient for termination are the most common in the genome, while leaky contexts are the most rare, especially among highly expressed genes (Bonetti et al. 1995; McCaughan et al. 1995). Thus, the frequency of different contexts may be an indication of their efficiency at translation termination.
We found a striking inverse correlation between the usage of 4-base stop codon contexts among readthrough candidates and non-readthrough transcripts (Fig. 5C). For example, TGA-C, the least common context among non-readthrough transcripts (3.1%), is by far the most common among readthrough candidates (32.2%). In fact, genes containing a TGA-C stop context are nearly 10-fold more likely to be readthrough candidates than genes with other contexts (16.4% vs. 1.9%), suggesting that perhaps TGA-C is a leaky stop codon context enabling readthrough.
The relative frequencies of contexts suggest that the most leaky stop codons are TGA > TAG > TAA, while the most leaky downstream base is C>T>G>A. The frequency of TGA-C among readthrough candidates is not significantly different from the frequency of TGA (64%) times the frequency of C (51%), thus it appears the two may act independently. These results are consistent with TGA being the leakiest of stop codons in natural stop suppression in yeast (Firoozan et al. 1991), and stop codons followed by C showing preferential readthrough in eRF mutants in Drosophila (Chao et al. 2003).
The unusual stop codon contexts of the readthrough candidates provide further evidence that downstream translation requires identification of the stop codon, which occurs at the ribosome, and cannot be due to alternatives such as splicing (Supplemental Fig. S4).
Conservation pattern suggests stop codons encode functional amino acids
For non-readthrough transcripts, substitutions between the three stop codons are frequent, suggesting that they have little functional difference other than termination efficiency. However, the situation is strikingly different for readthrough candidates. While different readthrough genes use all three stop codons (182 use TGA, 73 TAG, and 28 TAA), any given readthrough candidate rarely showed substitutions between the three stop codons at the readthrough locus (Fig. 5D). Overall, 83% of readthrough stop codons show perfect conservation across the 12 Drosophila species, compared with 12% for non-readthrough genes. If we consider only transcripts that contain a stop codon at the first stop locus in all 12 species, the difference remains striking, with 97% of readthrough stops perfectly conserved, compared with 36% for non-readthrough genes. This suggests that the readthrough stop codons are not functionally equivalent.
A plausible functional difference between stop codons is that they may encode different amino acids if they are functionally translated. Although no cognate stop suppressor tRNA genes have been found in the Drosophila genome (Chan and Lowe 2009), experiments in Drosophila, mammals, and yeast have shown that specific near-cognate tRNAs can insert an amino acid when a stop codon is read through, with glutamine or tyrosine incorporated for UAA, glutamine, tyrosine, leucine, lysine, or possibly low levels of tryptophan for UAG, and arginine, cysteine, serine, or tryptophan for UGA (Bienz and Kubli 1981; Pure et al. 1985; Valle et al. 1987; Feng et al. 1990; Fearon et al. 1994; Chittum et al. 1998; Lao et al. 2009; Supplemental Text S5). Thus, the possible amino acids inserted for UGA are mostly different from the ones inserted for UAA or UAG, while UAA and UAG are more likely to result in the same amino acid incorporation.
The few substitutions that have occurred at readthrough loci all convert between UAA and UAG stop codons, suggesting that they may be typically translated into identical amino acids but that UGA encodes a different amino acid. In addition, the specific tRNA used may affect the rate of readthrough, providing a potential explanation for the small number of observed substitutions between UAA and UAG stop codons compared with what would be expected if they were entirely synonymous. We considered other explanations for the low rate of substitutions but found them lacking (Supplemental Text S6). Thus, we conclude that both the amino acid translation and the tRNA-dependent rate of readthrough likely contribute to the low rate of observed substitution between readthrough stop codons.
Possible regulatory mechanisms of readthrough rate
Several conserved RNA structures could enhance readthrough
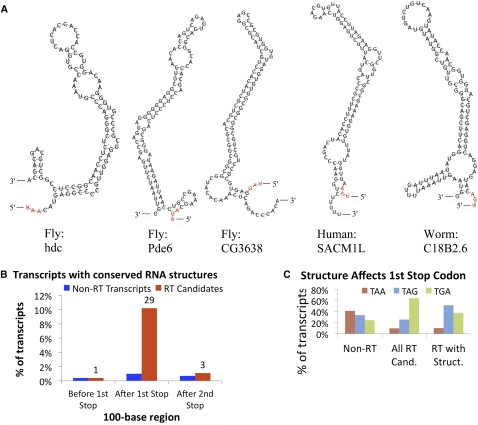
To determine if mRNA secondary structures participate in the readthrough mechanism, we used RNAz (Washietl et al. 2005), a widely used tool that tests an alignment for conserved and energetically stable predicted RNA structures. The 80 bases downstream from the Drosophila hdc stop codon are known to form a stem–loop that triggers readthrough, even when inserted after the stop codons of other genes, possibly through interference with the ribosome or release factors (Steneberg and Samakovlis 2001), and similarly located structures have been found to stimulate readthrough of viral genes (Firth et al. 2011). We therefore searched for evidence of RNA secondary structures specifically in the 100-base window downstream from the readthrough stop codon (including the stop codon itself).
RNAz reported conserved, stable RNA secondary structures for 29 (10%) of the readthrough candidates (Fig. 6A), compared with only 1% of non-readthrough stop codons. Furthermore, these structures are specifically found immediately downstream from the readthrough stop codons: Fewer than 1% of transcripts contain such structures in the 100-base window upstream of the first stop codon, or in the 100-base window downstream from the second stop codon, for either readthrough or non-readthrough genes (Fig. 6B).
Figure 6.
RNA structures associated with readthrough genes. (A) Fly, human, and worm examples of conserved, stable RNA structures predicted in the 100-nt regions downstream from (and including) candidate readthrough stop codons. The stop codon is highlighted in red. Twenty-nine structures were found in D. melanogaster, one in human, and one in C. elegans. The stem–loop in hdc was previously found to trigger readthrough. (B) Across 283 Drosophila readthrough candidates (red bars), 10% (n = 29) showed predicted structures in the 100-nt region downstream from the first stop codon compared with only 1% for non-readthrough transcripts (blue bars). The enrichment is exclusively found downstream from the first stop codon, with only one readthrough candidate showing a predicted structure in the 100 nt upstream of the first stop codon and three in the 100-nt downstream from the second stop codon, suggesting potential interactions with the ribosome during reading of the readthrough stop position. (C) Readthrough stop codon usage among readthrough candidates with and without predicted structures and non-readthrough genes. Although most readthrough candidates use TGA, readthrough candidates with structures show a preference for TAG, suggesting that a leaky stop codon context might not be necessary for readthrough in the presence of RNA structures.
Since the RNA structure in hdc is able to induce stop codon readthrough efficiently, independent of the specific stop codon context, one might expect that in the presence of other RNA structures, a leaky stop would be less strongly required. Indeed, among readthrough candidates with structures, TGA is a much less frequent stop codon (TGA: 38% vs. 67%, p = 0.002), and C is a somewhat less frequent next base (C: 45% vs. 52%, p = 0.30), in each case compared with readthrough candidates without structures (Fig. 6C).
Since RNA structures leading to translational readthrough can be condition-specific, the subset of readthrough candidates with structures could themselves be condition-specific, while readthrough events relying solely on sequence signals may be condition-invariant.
Long readthrough transcripts and predicted sequence motifs suggest extensive regulation
We observed that readthrough candidates have considerably longer primary transcripts on average than non-readthrough genes (19 kb vs. 5.8 kb) and also more exons (7.5 vs. 4.3) and splice variants (2.4 vs. 1.7) (Supplemental Fig. S5; Supplemental Text S7), suggesting that they may be subject to diverse forms of regulation. They are significantly larger in each of the four components that make up the primary transcript—coding sequence, introns, 5′ UTR, and 3′ UTR (even after subtracting the readthrough region)—and each may harbor regulatory elements involved in readthrough or other forms of gene regulation.
We found several indications of possible binding sites for cis-acting elements that may affect translation termination efficiency, possibly by interaction with the 18S rRNA (Namy et al. 2001; Williams et al. 2004). We found several bases near the stop codon with unusually high evolutionary conservation or unusual base frequencies, suggesting selection for specific sequences (Supplemental Text S9). We also found several enriched motifs in the six positions immediately downstream from candidate readthrough stop codons (Supplemental Text S10), including one related to the Skuzeski sequence, CAR-YYA, known to increase readthrough for viral mRNAs (Skuzeski et al. 1991). Finally, we found high enrichment of the 8-mer CAGCAGCA within a few hundred bases of the stop codon in all three reading frame offsets (Supplemental Text S11).
The long 3′ UTRs of the readthrough candidates could also be directly involved in the readthrough mechanism by physically separating the stop codon from the poly(A) tail. Indeed, termination efficiency can be affected by interaction between eRF3, a component of the termination complex, and the poly(A)-binding protein (Amrani et al. 2004; Kobayashi et al. 2004), and thus by separating the two, long 3′ UTRs might decrease termination efficiency.
Evolution of readthrough across animal species
Readthrough genes evolved as extensions, not truncations
We next studied the evolutionary history of readthrough loci to distinguish between two possibilities for how readthrough genes emerge.
Truncation scenario. An ancestral transcript originally terminated at the second stop locus, but a nonsense mutation introduced a new stop codon at the first stop locus. In this case, the readthrough mechanism could confer a selective advantage by partially rescuing the longer version of the protein.
Extension scenario. Alternatively, a readthrough transcript could have evolved as a recent extension of an ancestrally shorter gene by inclusion of a formerly untranslated region.
An analysis of alignments at the first and second stop loci suggests that most D. melanogaster readthrough genes evolved primarily by extension rather than truncation (Supplemental Text S12). In these cases, readthrough provides an efficient mechanism for evolving new domains, because the protein extension can evolve modularly without affecting the independent functionality of the shorter protein, whereas under the truncation scenario, there would be little opportunity to adapt the new shorter protein without potentially damaging the longer one.
In general, we would not expect the method used to find our 283 readthrough candidates to detect readthrough that has recently evolved by the extension scenario, because we evaluate our protein-coding signatures across all 12 flies. However, to gauge whether additional readthrough genes could have recently evolved in D. melanogaster that were not read through in the common ancestor of the 12 species, we also used PhyloCSF to score second ORFs using only the six closest relatives of D. melanogaster (up to and including Drosophila ananassae). Using our frame bias test to infer the number of recently evolved readthrough genes, we estimate that in addition to our 283 candidates, there are ∼100 to 200 genes that have evolved to be readthrough genes in the D. melanogaster lineage by the extension scenario since the divergence of the 12 flies (Supplemental Text S12).
Readthrough candidates in humans, nematodes, and other insects
We next used PhyloCSF to look for stop codon readthrough in four more groups of species for which multiple sequenced and aligned relatives allow us to apply our comparative methodology: mammals, worms, fungi, and insects.
Using a 29-way mammalian alignment (Lindblad-Toh et al. 2011), we found four non-selenocysteine readthrough candidates in the human genome: SACM1L, OPRK1, OPRL1, and BRI3BP. For one of these, SACM1L (Fig. 7A), RNAz predicted a conserved, stable stem–loop downstream from the readthrough stop codon (Fig. 6A). Although rabbit beta-globin is a known readthrough gene (Chittum et al. 1998), it was not found by our method since the second ORF does not show protein-coding constraint. Its 3′ UTR is generally not well conserved, even compared with its closest relatives among the aligned species (Supplemental Fig. S10), and the readthrough could be a species-specific event not conserved in other mammals.
Figure 7.
Examples of readthrough candidates in other species. (A) Alignment across 29 mammals for readthrough region in human gene SACM1L, one of four mammalian candidates. (B) Alignment across five worm species for the readthrough region in C. elegans gene C18B2.6, one of five nematode candidates. The stop codon context in all five is TGA-C and is perfectly conserved among Caenorhabditis species. (C) Alignment across 12 Drosophila and three other insect species, Anopheles gambiae (mosquito), Apis mellifera (honey bee), and Tribolium castaneum (red flour beetle), for the readthrough region of the D. melanogaster slo gene, one of 17 readthrough candidates conserved in mosquitoes, and one of four conserved across all 15 aligned insects. Although PhyloCSF cannot tell us whether the region is protein-coding in a particular subset of species, the large number of synonymous substitutions specifically in the other three insects, lack of non-synonymous substitutions and frameshifting indels, and perfectly conserved “leaky” TGA-C stop codon context suggest that readthrough also occurs in these other insects.
Using a six-way nematode alignment, we found five C. elegans readthrough candidates, namely, shk-1, F38E11.5, F38E11.6, K07C11.4, and C18B2.6 (Fig. 7B). RNAz predicted a conserved, stable RNA structure downstream from the C18B2.6 stop codon (Fig. 6A), but not for the other candidates. There is one known case of selenocysteine insertion in C. elegans (Buettner et al. 1999), and a search for SECIS elements and homology with selenocysteine genes in other species found no others (Taskov et al. 2005), thus we consider it unlikely that these five readthrough candidates are selenocysteine genes. Surprisingly, F38E11.5 and F38E11.6 are neighboring genes on opposite strands; we have no explanation for this coincidence, as they share no detectable homology.
However, outside these noteworthy individual candidates, neither mammalian nor worm species appear to have abundant readthrough. Comparing the PhyloCSF scores of second ORFs in three frames showed no excess in frame 0 in worm (Supplemental Fig. 11). In human, we found an excess of 86 second ORFs with a positive score in frame 0. Most of these did not have an aligned stop codon in most species and are probably due to recent nonsense mutations, but a handful of additional readthrough cases may exist.
Similarly in yeast, although several readthrough genes had been previously identified in S. cerevisiae (Namy et al. 2002, 2003), our method did not find any genes in either Saccharomyces or Candida with unambiguous signature of readthrough. We found protein-coding signatures in the second ORFs of two S. cerevisiae genes, BSC1 and BSC3, known to have readthrough rates higher than 20% (Namy et al. 2003). In BSC1 it appears that the stop codon arose from a nonsense mutation specific to S. cerevisiae and that readthrough provides partial rescue of the full-length protein, an example of readthrough evolution by the truncation scenario. In BSC3, the readthrough is seemingly conserved across all four aligned yeast species, although the protein-coding constraint could be partly explained by overlap with genes YLR462W and YLR464W on the opposite strand.
Because Anopheles gambiae (malaria mosquito), Apis mellifera (honey bee), and Tribolium castaneum (red flour beetle) lack alignments of multiple close relatives, we used their proximity to the 12 Drosophila species to find readthrough candidates in each by studying their alignments to our list of 283 readthrough candidates. We studied orthologous alignments of our readthrough regions and looked for a preponderance of synonymous substitutions in each of the three species, using a 15-way alignment including the 12 Drosophila species. We found that 17 of our candidates also show the signature of protein-coding selection in their readthrough regions in mosquitoes, of which seven are also found in beetles, and four of the latter also in bees (Fig. 7C; Supplemental Data 1).
The readthrough stop codons in human, nematodes, bees, and beetles were exclusively TGA, and 13 out of 16 have context TGA-C. In contrast, readthrough candidates in fruit fly and mosquito use all three stop codons. The TAA readthrough candidates are particularly notable because there are no others known in any other animal, and a search for conserved readthrough stop codons in prokaryotes using homology and dN/dS analysis did not find any TAA readthroughs either (Fujita et al. 2007), although TAA is used in the known yeast readthrough IMP3 (Namy et al. 2003).
Estimation of readthrough abundance across eukaryotic species
Beyond these individual candidates, we sought to estimate the abundance of readthrough across sequenced eukaryotic genomes, even for species lacking rich comparative evidence. We created a reading frame bias test similar to the one discussed above, but relying on the Z curve score instead of the PhyloCSF score. This test enabled us to recognize species with abundant readthrough even when individual readthrough transcripts were difficult to pinpoint due to lack of power. For each species, we estimated the excess of second ORFs with in-frame positive Z-scores, correcting for potential recent nonsense mutations and for nucleotide substitutions due to potential sequencing errors (see Methods).
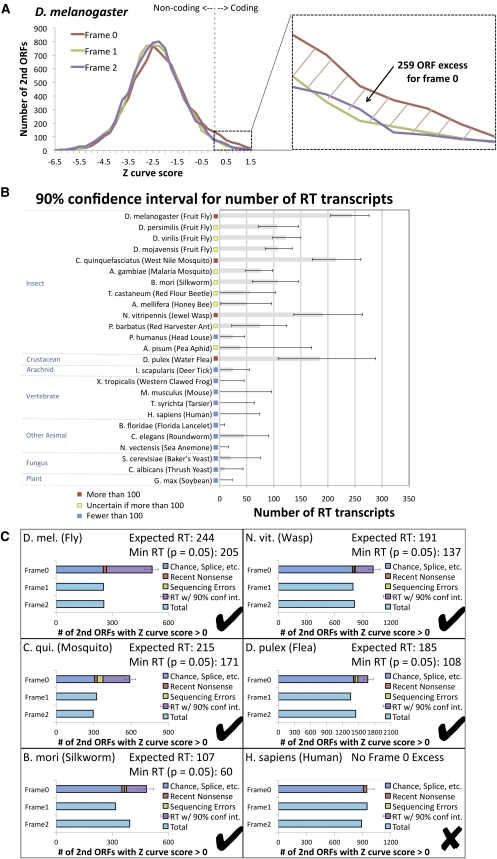
We first computed the distributions of Z curve scores for D. melanogaster second ORFs in three frames (Fig. 8A) and compared them with our previous comparative information (Fig. 5A) to confirm that the Z curve score has sufficient statistical power to distinguish the large excess of protein-coding regions in frame 0, even if it doesn't have sufficient power to detect individual readthrough candidates. Indeed, this test found an excess of 259 second ORFs with a positive Z curve score in frame 0, a lower bound consistent with our PhyloCSF-based estimate (Supplemental Text S13).
Figure 8.
Estimated abundance of readthrough in insects and other eukaryotic species using single-species evidence. Estimated number of readthrough transcripts in 25 species, calculated using single-species sequence-composition evidence quantified by Z curve scores for downstream ORFs in three frames to detect excess of positive scores in frame 0 associated with abundant readthrough (RT). (A) Distribution of Z curve scores in three frames providing a single-species estimate for D. melanogaster consistent with our PhyloCSF-based estimate (Fig. 5A). Even though the Z curve does not provide sufficient power to detect individual readthrough genes, the excess of 259 positive Z curve scores for frame 0 nonetheless provides a robust single-species estimate of the overall abundance of readthrough in D. melanogaster. Because the histogram excludes second ORFs shorter than 10 codons long and uses a conservative threshold for detecting coding regions, this number should be interpreted as a lower bound. (B) Estimated number of readthrough transcripts with 90% confidence intervals for 25 species. Estimated number of readthrough transcripts is dozens or more for each of the insects tested, and for three insects and one crustacean, even the low end of the confidence interval is more than 100 transcripts, whereas none of the other species tested has more than 100 readthrough transcripts even at the high end of the confidence interval, suggesting that this level of abundant readthrough is specific to insects and crustacea. (C) Contribution of several potential mechanisms to the number of positive-scoring frame 0 transcripts for humans and five species with abundant readthrough. Horizontal bars show the number of positive scores in each of the three frames, with the frame 0 bar divided into estimates of the number of transcripts resulting from each of four potential mechanisms: positive scores that could occur in any frame, such as chance or splicing, estimated using the counts for the other two frames (blue); recent nonsense mutations, estimated using comparative information from D. melanogaster (red); sequencing mismatches, estimated using a homology test and simulated sequencing errors (green); and readthrough, obtained by subtracting the others from the total (purple). The error bar shows the 90% confidence interval for the number of readthrough transcripts, measured from the start of the readthrough portion of the bar, with the expected number of readthrough transcripts and lower end of the confidence interval reported in the title.
Given the agreement with PhyloCSF at least in this bulk statistic, we applied this single-species test in 13 insect species and 12 other eukaryotic species including one plant and two fungi, to obtain 90% confidence intervals for the likely number of readthrough transcripts (Fig. 8B,C). Our results suggest more than ∼100 readthrough transcripts in Culex quinquefasciatus (West Nile vector mosquito) and Nasonia vitripennis (jewel wasp), and in one crustacean, Daphnia pulex (water flea) (P-value < 0.05 in each case). They also suggest at least ∼40 in Bombyx mori (silkworm) and A. gambiae. The 90% confidence intervals allow the possibility of a large number of readthrough transcripts in all insect species. In contrast, species outside insects and crustracea seem unlikely to have many readthrough transcripts, even the one arachnid tested, although there are several reasons our estimate could be low (Supplemental Text S13). The presence of abundant readthrough in the water flea suggests that abundant readthrough may have arisen prior to the root of the insect phylogeny, possibly associated with new regulatory mechanisms that tolerate readthrough transcripts and allow them to evade NMD.
In summary, while we have found individual examples of readthrough in species spanning much of the animal kingdom, abundant readthrough as seen in Drosophila may be confined to insects and crustacea, but not found elsewhere in the tree of life.
Discussion
We have found evolutionary signatures of translational stop codon readthrough in 283 D. melanogaster protein-coding genes and shown that other known mechanisms cannot plausibly explain most of these. Translation of the readthrough region has been previously experimentally confirmed for three of these genes with very long readthrough regions (hdc, kel, and syn), and we provide new experimental evidence of downstream translation for four additional genes and mass spectrometry evidence for seven more. Beyond the observation of protein translation, however, our approach using protein-coding evolutionary signatures confirms not only that hundreds of genes undergo readthrough, but also that the amino acid sequence downstream from the stop codon serves a conserved function that provides a selective advantage (both across related species and within the D. melanogaster population), even when the readthrough event leads to the inclusion of only a handful of additional amino acids. Our analysis also provides insights into the specific mechanism of readthrough, suggesting translational leakage; amino acid incorporation at the stop codon position; and RNA-based, length-based, and sequence-based regulation. Our results reveal that functional stop codon readthrough is considerably more prevalent among metazoa than previously recognized, affecting hundreds of genes in other insects and in at least one crustacean, and spanning the animal kingdom with candidates also found in nematodes and mammals, including in humans.
Readthrough could be a competitor to NMD for rescuing premature stop codons
While our evidence suggests that most readthrough genes evolved through extension, the readthrough mechanism itself could have arisen as a way to partially rescue nonsense mutations until a sense mutation at the same locus restores full function to the gene. The mechanism of readthrough could then serve as an evolutionary buffer for transient nonsense mutations that would eventually be replaced by sense codons. Genes that maintain readthrough over long periods of time would do so for other possibly regulatory roles, although initially they would have been non-readthrough genes whose stop codon was misidentified as premature, suggesting that the conditions that trigger readthrough would be ones that typically distinguish premature from normal stop codons. Evidence that an endogenous nonsense-rescue mechanism exists in Drosophila first came nearly 20 years ago, as suppressor tRNAs that can rescue nonsense mutations in Escherichia coli, yeast, and C. elegans have little effect in Drosophila, suggesting that an alternative mechanism may already be present (Washburn and O'Tousa 1992). Indeed, nonsense rescue in Drosophila has been observed for several naturally occurring and artificially introduced premature stop codons (Washburn and O'Tousa 1992; Samson et al. 1995; Chao et al. 2003; Yildiz et al. 2004).
The system for identifying which stop codons to read through might share some components with nonsense-mediated decay (NMD), another pathway that identifies premature stop codons, in order to degrade their transcripts. How NMD recognizes premature stop codons in Drosophila is not well understood, but there is evidence that NMD is more likely to be triggered if the 3′-UTR length is greater than ∼742 nt (Hansen et al. 2009). The average 3′-UTR length for readthrough transcripts is 1079 nt, compared with 381 for non-readthrough, so perhaps readthrough and NMD share this part of the recognition mechanism. If readthrough and NMD recognize premature stop codons in the same way, then the cell would have to choose one or the other; otherwise, NMD would eliminate the mRNA altogether. One possibility is that if the stop codon is read through on the first pass of translation, then the mRNA is marked so as to turn off NMD and facilitate readthrough on subsequent passes. NMD prevents formation of potentially toxic truncated proteins but does not restore production of the full-length protein, so a competing readthrough fate for non-toxic proteins with premature stops could be beneficial and selectively advantageous. However, both the common recognition signals with NMD and the unique signals that choose one pathway or the other remain to be elucidated.
Diversity in readthrough rate, context, and 3′-UTR length suggest complex regulation
While the evolutionary signature of protein-coding selection downstream from stop codons is a powerful method for finding readthrough genes, it is still unclear how the Drosophila cellular machinery determines which stop codons will be read through and which will not. A simple “leakage” probability that depends solely on the stop codon context is unlikely because non-readthrough genes with “leaky” stop codons would require a low readthrough rate, while this rate can be quite high for readthrough genes. In fact, readthrough protein isoform fractions as high as 20%, 50%, and 90% have been observed in Drosophila genes syn and kel, and nonsense alleles of elav, respectively (Samson et al. 1995; Klagges et al. 1996; Robinson and Cooley 1997), although these could be higher than the actual readthrough rate due to differential degradation of the two protein isoforms. Similarly, protein-coding selection in the third ORFs of the 16 double-readthrough candidates would require a high readthrough rate or coupled readthrough of the two stop codons. Lastly, some readthrough events are known to be tissue-specific (kel), stage-specific (elav), and temperature-specific (Chao et al. 2003), suggesting both precise and versatile regulation.
Our results suggest that many cis-acting signals influence the readthrough rate of individual genes, including stop codon context, 3′-UTR length, specific motifs, and RNA structures. In addition, trans-acting regulatory mechanisms can influence the global readthrough rate, including post-transcriptional modifications of tRNA anticodons (Bienz and Kubli 1981) and regulation of release factor activity and abundance (von der Haar and Tuite 2007). However, the currently known signals are insufficient to predict readthrough systematically, and the diversity of readthrough genes defies our expectations. For example, AlCR2, a double-readthrough candidate, has a frequent and presumably non-leaky stop codon context (TAA-G), a short 3′ UTR (227 bp), does not match our enriched 6-base motif, and lacks a conserved secondary structure after its stop codon, suggesting that non-negligible translation of the extended protein would require as yet unidentified regulatory signals.
Understanding of insect readthrough could have medical applications
The prevalence of conserved readthrough in Drosophila offers a rich opportunity to enhance our understanding of the underlying molecular mechanisms and regulation of the readthrough process more generally, which could lead to medical and biological engineering applications. For example, small molecules that induce stop codon readthrough have been used as a way to treat genetic diseases caused by nonsense mutations (Schmitz and Famulok 2007; Keeling and Bedwell 2010). A deeper understanding of readthrough regulation could enable targeting these drugs to trigger readthrough for specific genes carrying nonsense mutations, while allowing the NMD pathway to capture other aberrant transcripts, and allowing translation to terminate normally at genes lacking nonsense mutations. Biological engineering could also take advantage of readthrough, by designing fused domains to be translated at a specific ratio (e.g., as dictated by the stop codon context) or in specific conditions (e.g., as dictated by regulation of readthrough); in one such example, a transmembrane anchor downstream from a leaky stop codon was fused to a protein of interest to provide an external indicator of the protein level in the cell (Jostock 2010). Lastly, the recognition that numerous insect species including the malaria vector A. gambiae rely on abundant readthrough can offer new insights into their biology and regulation, as well as possible new avenues for anti-malarial targets.
Overall, the widespread use of readthrough across the animal kingdom suggests an additional level of regulatory complexity, requiring a deeper biological understanding but also providing exciting new opportunities to exploit its versatility.
Methods
Transcripts
We obtained the D. melanogaster genome and annotations from flybase.org (Tweedie et al. 2009), version 5.13, release FB2008-10. We used the Drosophila tree and divergences from Stark et al. (2007). We used nematode genome and annotations, from http://www.wormbase.org, release WS190, 16-May-2008. The 15-way dm3 insect alignments, the 29-way hg19 alignments, and the six-way ce6 nematode MULTIZ alignments (Blanchette et al. 2004) were obtained from the UCSC Genome Browser (Kent et al. 2002; Kuhn et al. 2009). Genomes and annotations for other species came from the UCSC Genome Browser or vectorbase.org except that the following species came from the specified websites: Bombyx mori, silkdb.org/silkdb; Candida albicans, http://www.candidagenome.org; Glycine max, http://www.phytozome.net; Tribolium castaneum, http://beetlebase.org; Tarsius syrichta and Mus musculus, http://www.ensembl.org; Drosophila persimilis, http://flybase.org; Daphnia pulex, http://genome.jgi-psf.org; Nematostella vectensis, http://genome.jgi-psf.org/Nemve1; Pogonomyrmex barbatus, http://hymenopteragenome.org; and Acyrthosiphon pisum, http://www.aphidbase.com/aphidbase. Publications reporting these genome sequences are listed in Supplemental Text S14.
Using PhyloCSF to find protein-coding second ORFs in D. melanogaster
For each protein-coding transcript, excluding mitochondrial DNA, trans-spliced transcripts, and transcripts whose final CDS does not end in a stop codon, we computed the PhyloCSF score for the second ORF excluding the final stop codon. For transcripts with no annotated 3′ UTR, or if the second ORF extended beyond the end of the annotated 3′ UTR, we defined the second ORF as continuing beyond the end of the annotated transcript without splicing. Among transcripts with identical second ORFs, we considered only one.
To obtain a P-value for a non-coding region of length L codons to have a PhyloCSF score above some threshold, we approximated the score distribution with a normal of mean and standard deviation C1 × L and C2 × LEX, where the coefficients C1 = 11.55, C2 = 10.81, and EX = 0.7184 were obtained empirically from second ORFs of all transcripts, excluding very high scores as likely coming from coding regions (Lin et al. 2011). For multiple hypothesis correction, we computed the Local FDR (Efron et al. 2001) using the localfdr R package, version 1.1-6, obtained from http://cran.r-project.org/src/contrib/Archive/locfdr/.
We excluded any transcript whose second ORF has a PhyloCSF score (in decibans) less than 16, indicating that it is less than 40 times as likely to occur in a protein-coding region as in a non-coding one. We included as protein-coding any of the remaining transcripts for which the P-value is <0.001; each of these second ORFs has a Local FDR <∼0.33. We individually examined each of the ∼100 remaining transcripts whose second ORFs have PhyloCSF score ≥ 16 and P-value ≥ 0.001, and considered it protein-coding or not based on various factors, including the presence of frameshifting insertions and deletions (indels).
For the D. melanogaster frame bias investigation, we computed the PhyloCSF scores of unique regions starting 0, 1, or 2 bases after the stop codon of an annotated protein-coding transcript and continuing until the next stop codon in that frame, excluding regions with no alignment and regions overlapping the coding portion of another annotated transcript, such as an alternative splice variant or second cistron.
RNA-seq
RNA-seq data were obtained from the October 2009 data freeze on modencode.org (Graveley et al. 2011). We used development timecourse data from submissions 2245-2259, 2262-2263, 2265-2267, and 2388-2397, consisting of 76-base Illumina reads from poly(A) RNA extracted from 30 development stages of D. melanogaster, mapped using Tophat, and processed using samtools (Li et al. 2009). When looking for reads overlapping a particular locus, we excluded reads for which the locus was within 7 nt of the end of the read, to avoid splice-mapping errors. When examining candidates for RNA editing, we checked for erroneous mapping to paralogs using BLASTN from blast.ncbi.nlm.nih.gov.
Splice prediction
We did not consider a transcript to be a readthrough candidate if any modENCODE RNA-seq reads or any EST or cDNA in the UCSC Genome Browser support a 3′-splice site early in the second ORF. We also used two splice prediction algorithms to look for additional splice sites. We used the web interface on fruitfly.org for splice site prediction using neural networks (Reese et al. 1997) and a maximum entropy method (Yeo and Burge 2004). If a candidate 3′-splice site has a neural network score above ∼0.7 or a maximum entropy score above ∼6, we considered the transcript to be a readthrough candidate or not based on a combination of factors including these scores and the PhyloCSF score of the region before the potential splice site.
SECIS elements
We searched for SECIS elements in the 3′ UTRs of D. melanogaster readthrough candidates with a TGA stop codon using SECISearch (Kryukov et al. 2003) with parameter values: Pattern = “Loose (canonical and non-canonical),” Core structure energy = −5, Overall structure energy = −11, and using all fine structural feature filters (Y, O, B, S). We also ran with Pattern = “Default (GTGA).” We used the recommended COVE score threshold of 15. If the 3′ UTR was not annotated, we searched the 1000 nt downstream from the stop codon. If the annotated 3′ UTR was <1000 nt long, we extended it without splicing to 1000 nt.
GFP constructs
BAC recombineering was performed as described in Venken et al. (2009) except that the recombineering enzymes on the pSIM6 plasmid were electroporated into the P[acman] library EPI300 bacteria rather than moving the BAC into a recombineering bacteria (gift from Donald Court, National Cancer Institute, Frederick, MD) (Datta et al. 2006). BACS from the P[acman] library (Venken et al. 2009) were chosen using the genome browser on the project home page (http://www.pacmanfly.org/) to include the gene of interest (GOI) and as much of the surrounding genomic DNA as possible (Supplemental Data 2). One BAC was tagged for each readthrough prediction tested, replacing the second stop codon with eGFP in-frame with a TGA stop (Fig. 3A), followed by a kanamycin-resistance marker (Poser et al. 2008). C-terminal laptags (a gift from Anthony Hyman, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) (Poser et al. 2008) were amplified with PCR and recombined into BACs from the CHORI-322 libraries using ∼70-bp primers: 50-bp homology arms specific to each gene of interest (GOI.readthrough.F and GOI.readthrough.R) (Supplemental Data 2) and 21–22 bp specific to the tag (tag sequence forward GATTATGATATTCCAACTACTG and tag sequence reverse TCAGAAGAACTCGTCAAGAAG). Tagged BACS were integrated into the Drosophila genome using the PhiC31 integrase system, screened for red eyes and balanced over CyO or TM6C, Sb if the integration site was on the second or third chromosome, respectively. BAC integration was checked with PCR as described in Venken et al. (2009). Embryonic expression patterns were determined with antibody staining using rabbit antiGFP from abcam ab290 at 1:200 dilution and goat anti-rabbit Alexa488 from Invitrogen at 0.75:5000 dilution. Larval expression patterns were determined from in vivo expression of the GFP. Larval tissues were dissected in saline and mounted in 50% glycerol/50% PBS. Images were collected on a Leica DM5000B microscope.
Z curve score
The Z curve score was computed as described in Gao and Zhang (2004) and trained using all annotated coding exons and similar sized intergenic regions as positive and negative examples. We offset the score so that the minimum average error threshold on the training examples was at 0. For the three-frames comparison test for abundant readthrough, we computed the linear discriminant using a prior probability for coding of 0.02, to account for the expectation that most second ORFs are non-coding. We excluded second ORFs of less than 10 codons because the Z curve score has high variance for such short regions, and also excluded second ORFs that overlapped the coding portion of another annotated transcript.
To estimate the number of recent nonsense mutations for the three-frames comparison test, we used an upper estimate for the number (n = 17) of such mutations in D. melanogaster found during manual curation of the readthrough candidates list, and then multiplied by the ratio of the number of annotated transcripts in the species being tested to the number in D. melanogaster.
To estimate the number of second ORFs with a positive score in frame 0 for which the stop codon was actually a sequencing mismatch error, we first applied a test that detects only a fraction of these errors and then estimated that fraction. For each second ORF with a positive score in frame 0, we used TBLASTX against D. melanogaster chromosomes to determine all orthologs of the 25-amino-acid sequence immediately 5′ of the stop. If all matching regions were immediately followed by a sense codon and if at least one satisfied a 3′ matching criterion, then we considered this to be a sequencing error. We obtained similar results using three different criteria: (1) an e-value cutoff of 0.001 and exact match of two amino acids 3′ of the stop codon (shown in Fig. 8); (2) an e-value <0.001 and three of six amino acids 3′ of the stop; and (3) an e-value <0.1 and four of nine matching amino acids downstream. To estimate the fraction of sequencing mismatches that would be detected by this test, we applied the test to 1000 simulated sequencing mismatches obtained by randomly changing a base of a coding region and selecting those changes for which the result was a stop codon and for which the downstream ORF had positive a Z curve score. The fraction of simulated sequencing mismatches caught by this test ranged from 0.053 for A. pisum to 0.489 for D. mojavensis. (We only applied the test to arthropods, since no other species had large frame 0 excess.) Among second ORFs with a positive score in frame 0, the number of sequencing errors detected by this test was no more than seven in any of the arthropods tested, except that in A. aegypti we detected 94 errors, enough to make any estimation of readthrough impossible and warranting future investigation.
To generate the 90% confidence intervals, we used a stochastic model in which we assumed that each second ORF whose first stop codon was readthrough, recent nonsense, or a sequencing mismatch would have a positive score in frame 0, and for each other second ORF the probability of having a positive score would be the same in each of the three frames. We estimated this probability using maximum likelihood. We modeled our test for a sequencing mismatch stop codon as a Bernoulli random variable with the probability of detection determined by our simulation data.
Potential limitations of this method are discussed in Supplemental Text S13.
Base-pairing
To calculate the frequency of mRNA base-pairing, we used RNAfold (Zuker and Stiegler 1981) from the Vienna RNA Package version 1.8.2 (Hofacker et al. 1994) with the default parameters to calculate the minimum free-energy secondary structure of the 403-base region of each transcript centered at the stop codon (extending without splicing if there was no annotated 3′ UTR or it was <200 nt long). Each distinct region was counted only once even if it is contained in multiple transcripts. To decrease the variance arising from the relatively small number of readthrough candidates, codons before the stop have been averaged with the previous four codons, and codons after the stop have been averaged with the next four codons.
Length, composition, and enrichment statistics
In computing length and composition statistics for readthrough candidates and non-readthrough transcripts, we eliminated duplicates (transcripts with identical second ORFs), leaving 15,211 transcripts. For computing UTR lengths, we included only transcripts that have annotated UTRs.
To adjust some statistic for one or more other statistics (e.g., computing average 5′-UTR length of transcripts with similar length and 3′-UTR length as readthrough candidates), we divided transcripts into bins and weighted the non-readthrough transcripts so as to make the total weight in each bin equal to the fraction of readthrough candidates in that bin. The bin size was 300 when adjusting for one statistic. When adjusting for two statistics, we used bins of size 750 for the first statistic and a bin of size 150 for the second statistic for each bin of the first statistic. For three statistics, we used bins of size 750, 150, and 15 and averaged the results over all permutations to make the result order-independent.
GO classifications were obtained from http://www.sb.cs.cmu.edu/stem/ (Ernst and Bar-Joseph 2006).
RNA structures
To look for conserved RNA structures, we used RNAz (Washietl et al. 2005), which combines a measure of secondary structure conservation with a measure of thermodynamic stability relative to other sequences of the same length and base composition. We used the -d option, which compares to a null model that also preserves dinucleotide frequencies. We used alignments as described above, but excluded the non-Drosophilidae from the dm3 alignment and for each region excluded species for which the alignment was not fully defined. Although we used the default threshold for SVM RNA-class probability of 0.5 for considering a structure to be significant, all of the significant structures found had probability >0.7, and the human, worm, and half of the fly structures had probability >0.9. Some of our preliminary investigation also used quickfold on the DINAMelt server (http://dinamelt.bioinfo.rpi.edu/quikfold.php) (Markham and Zuker 2005).
Homology with known protein domains
We searched for homology with known protein domain families using Search Pfam from http://pfam.sanger.ac.uk/search?tab=searchProteinBlock#tabview=tab1 (Finn et al. 2010). We excluded regions less than 10 codons long and regions that overlapped a known CDS, both for readthroughs and controls. We also excluded control regions with positive PhyloCSF, since these could be actual readthroughs excluded from our readthrough list by overly conservative curation. We considered a match to be significant if e-value < 0.05/(number of regions tested).
Acknowledgments
We thank Stacy Holtzman and Thom C. Kaufman for transgenic Drosophila production and Lionel Senderowicz and Sarah El Mouatassim Bih for protein expression validation. We thank William M. Gelbart for suggesting this line of investigation during the 12 flies project; the members of the modENCODE project for early release of data; members of dpgp.org and Ensembl for access to SNP data from the Drosophila Population Genomics Project; members of the International Ixodes scapularis Sequencing Committee for access to the deer tick genome sequence; Erich Brunner for help analyzing mass spectrometry data; Jade Vinson for use of his splice prediction code; and Matt Rasmussen, Stefan Washietl, Pouya Kheradpour, Loyal Goff, Jason Ernst, Rogerio Candeias, Chris Bristow, Pasha Baranov, Marco Mariotti, Ben Holmes, Alex Lancaster, Dace Ruklisa, Michal Rabani, Lukas Reiter, Sabine Schrimpf, Grigoriy Kryukov, Nick Patterson, Jessica Wu, Peter Everett, and Rintaro Saito for helpful discussions and suggestions. This work was supported by the National Institutes of Health (U54 HG00455-01), NSF CAREER 0644282, and a Sloan Foundation award.
Authors' contributions: I.J., M.L., C.C., and M.K. designed the study, performed the computational analysis, and wrote the manuscript. R.S., N.N., A.V., and K.W. performed the experimental verification by transgenic flies.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.119974.110.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Aphasizhev R 2007. RNA editing. Mol Biol 41: 227–239 [Google Scholar]
- Beier H, Grimm M 2001. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29: 4767–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, Baranov PV 2010. Recode-2: New design, new search tools, and many more genes. Nucleic Acids Res 38: D69–D74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DE, Merli CA, Cygan JA, Shelby R, Blackman RK 1995. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics 139: 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G, Innes S, Minella O, Richardson J, Stansfield I 2001. Endless possibilities: Translation termination and stop codon recognition. Microbiology 147: 255–269 [DOI] [PubMed] [Google Scholar]
- Bienz M, Kubli E 1981. Wild-type tRNA-Tyr-G reads the TMV RNA stop codon but Q base-modified tRNA-Tyr-Q does not. Nature 294: 188–190 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B, Fu L, Moon J, Bedwell DM 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251: 334–345 [DOI] [PubMed] [Google Scholar]
- Buettner C, Harney JW, Berry MJ 1999. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J Biol Chem 274: 21598–21602 [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM 2009. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Dierick H, Addy T, Bejsovec A 2003. Mutations in eukaryotic release factors 1 and 3 act as general nonsense suppressors in Drosophila. Genetics 165: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CE, Guigo R 2008. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE 3: e2968 doi: 10.1371/journal.pone.0002968.t002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum H, Lane W, Carlson B, Roller P, Lung F, Lee B, Hatfield D 1998. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37: 10866–10870 [DOI] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL 2006. A set of recombineering plasmids for Gram-negative bacteria. Gene 379: 109–115 [DOI] [PubMed] [Google Scholar]
- DeSimone SM, White K 1993. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol Cell Biol 13: 3641–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina VA, Brown JD 2006. Non-canonical decoding events at stop codons in eukaryotes. Mol Biol (Mosk) 40: 731–741 [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium; Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R, Storey J, Tusher V 2001. Empirical Bayes analysis of a microarray experiment. J Am Stat Assoc 96: 1151–1160 [Google Scholar]
- Ernst J, Bar-Joseph Z 2006. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7 doi: 10.1186/1471-2105-7-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon K, McClendon V, Bonetti B, Bedwell DM 1994. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast SteGp, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem 269: 17802–17808 [PubMed] [Google Scholar]
- Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG 1990. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag–pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci 87: 8860–8863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R, Mistry J, Tate J, Coggill P, Heger A, Pollington J, Gavin O, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211 doi: 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozan M, Grant C, Duarte J, Tuite M 1991. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast 7: 173–183 [DOI] [PubMed] [Google Scholar]
- Firth AE, Wills NM, Gesteland RF, Atkins JF 2011. Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 39: 6679–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Mihara H, Goto S, Esaki N, Kanehisa M 2007. Mining prokaryotic genomes for unknown amino acids: A stop-codon-based approach. BMC Bioinformatics 8: 225 doi: 10.1186/1471-2105-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang CT 2004. Comparison of various algorithms for recognizing short coding sequences of human genes. Bioinformatics 20: 673–681 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Lareau L, Blanchette M, Green R, Meng Q, Rehwinkel J, Gallusser F, Izaurralde E, Rio D, Dudoit S, et al. 2009. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet 5: e1000525 doi: 10.1371/journal.pgen.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa-Takamori M, Ossipov D, Novoselov S, Turanov A, Zhang Y, Gladyshev V, Krol A, Vorbruggen G, Jackle H 2009. A novel stem loop control element-dependent UGA read-through system without translational selenocysteine incorporation in Drosophila. FASEB J 23: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I, Fontana W, Stadler P, Bonhoeffer L, Tacker M, Schuster P 1994. Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Jostock T 2010. Cell surface display of polypeptide isoforms by stop codon readthrough. U.S. Patent WO/2010/022961. http://www.wipo.int/patentscope/search/en/WO2010022961
- Keeling KM, Bedwell DM 2010. Recoding therapies for genetic diseases. In Recoding: Expansion of decoding rules enriches gene expression (ed. Atkins JF, Gesteland RF), pp. 123–146 Springer, New York [Google Scholar]
- Kent W, Sugnet C, Furey T, Roskin K, Pringle T, Zahler A, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn J, Nutter R, Chang H, Segal E 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E 1996. Invertebrate synapsins: A single gene codes for several isoforms in Drosophila. J Neurosci 16: 3154–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Funakoshi Y, Hoshino SI, Katada T 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J Biol Chem 279: 45693–45700 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN 2003. Characterization of mammalian selenoproteomes. Science 300: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao NT, Maloney AP, Atkins JF, Kavanagh TA 2009. Versatile dual reporter gene systems for investigating stop codon readthrough in plants. PLoS ONE 4: e7354 doi: 10.1371/journal.pone.0007354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. 2007. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 17: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Deoras AN, Rasmussen MD, Kellis M 2008. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput Biol 4: e1000067 doi: 10.1371/journal.pcbi.1000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. Evolutionary constraint in the human genome based on 29 eutherian mammals. Nature 478: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loevenich SN, Brunner E, King NL, Deutsch EW, Stein SE, Consortium F, Aebersold R, Hafen E 2009. The Drosophila melanogaster PeptideAtlas facilitates the use of peptide data for improved fly proteomics and genome annotation. BMC Bioinformatics 10: 59 doi: 10.1186/1471-2105-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2005. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33: W577–W81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci 92: 5431–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Rousset JP 2010. Specification of standard amino acids by stop codons. In Recoding: Expansion of decoding rules enriches gene expression (ed. Atkins JF, Gesteland RF), pp. 79–100 Springer, New York [Google Scholar]
- Namy O, Hatin I, Rousset JP 2001. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2: 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Duchateau-Nguyen G, Rousset JP 2002. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol Microbiol 43: 641–652 [DOI] [PubMed] [Google Scholar]
- Namy O, Duchateau-Nguyen G, Hatin I, Denmat S, Termier M, Rousset J 2003. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res 31: 2289–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole ES, Major LL, Mannering SA, Tate WP 1998. Translational termination in Escherichia coli: Three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res 26: 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JRA, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. 2008. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pure GA, Robinson GW, Naumovski L, Friedberg EC 1985. Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. J Mol Biol 183: 31–42 [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D 1997. Improved splice site detection in Genie. J Comput Biol 4: 311–323 [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cooley L 1997. Examination of the function of two kelch proteins generated by stop codon suppression. Development 124: 1405–1417 [DOI] [PubMed] [Google Scholar]
- Samson ML, Lisbin MJ, White K 1995. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics 141: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels M, Schedl P, Cline T 1991. The complex set of late transcripts from the Drosophila sex determination gene Sex-lethal encodes multiple related polypeptides. Mol Cell Biol 11: 3584–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Umeki H, Saito R, Kanai A, Tomita M 2003. Computational analysis of stop codon readthrough in D. melanogaster. Bioinformatics 19: 1371–1380 [DOI] [PubMed] [Google Scholar]
- Schmitz A, Famulok M 2007. Ignore the nonsense. Nature 447: 42–43 [DOI] [PubMed] [Google Scholar]
- Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmström J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, et al. 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7: e48 doi: 10.1371/journal.pbio.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov NA 2006. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res 34: 2428–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF 1991. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218: 365–373 [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneberg P, Samakovlis C 2001. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep 2: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiharas H, Andrisani V, Salvaterrae PM 1990. Drosophila choline acetyltransferase uses a non-AUG initiation codon and full length RNA is inefficiently translated. J Biol Chem 265: 21714–21719 [PubMed] [Google Scholar]
- Takahashia K, Maruyamaa M, Tokuzawaa Y, Murakamia M, Odaa Y, Yoshikanea N, Makabeb KW, Ichisakaa T, Yamanakaa S 2005. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85: 360–371 [DOI] [PubMed] [Google Scholar]
- Taskov K, Chapple C, Kryukov GV, Castellano S, Lobanov AV, Korotkov KV, Guigó R, Gladyshev VN 2005. Nematode selenoproteome: The use of the selenocysteine insertion system to decode one codon in an animal genome? Nucleic Acids Res 33: 2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, Vagner S 2003. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95: 169–178 [DOI] [PubMed] [Google Scholar]
- True H, Lindquist S 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483 [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: Enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle RP, Morch MD, Haenni AL 1987. Novel amber suppressor tRNAs of mammalian origin. EMBO J 6: 3049–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Tuite MF 2007. Regulated translational bypass of stop codons in yeast. Trends Microbiol 15: 78–86 [DOI] [PubMed] [Google Scholar]
- Washburn T, O'Tousa JE 1992. Nonsense suppression of the major rhodopsin gene of Drosophila. Genetics 130: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF 2005. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci 102: 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams I, Richardson J, Starkey A, Stansfield I 2004. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res 32: 6605–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM 2010. Translational bypassing—peptidyl-tRNA repairing at non-overlapping sites. In Recoding: Expansion of decoding rules enriches gene expression (ed. Atkins JF, Gesteland RF), pp. 365–381 Springer, New York [Google Scholar]
- Yeo G, Burge C 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yildiz O, Kearney H, Kramer BC, Sekelsky JJ 2004. Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics 167: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Stiegler P 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]