Abstract
Melanopsin confers photosensitivity to a subset of retinal ganglion cells and is responsible for many non-image-forming tasks, like the detection of light for circadian entrainment. Recently, two melanopsin genes, Opn4m and Opn4x, were described in non-mammalian vertebrates. However, only one form, Opn4m, has been described in the mammals, although studies to date have been limited to the placentals and have not included the marsupials.
We report here the isolation and characterization of an Opn4 gene from an Australian marsupial, the fat-tailed dunnart (Sminthopsis crassicaudata), and present evidence which suggests that the Opn4x gene was lost before the placental/marsupial split. In situ hybridization shows that the expression of Opn4 in the dunnart eye is restricted to a subset of ganglion cells, a pattern previously reported for rodents and primates. These Opn4-positive cells are randomly distributed across the dunnart retina. We also undertook a comparative analysis with the South American marsupial, the grey short-tailed opossum (Monodelphis domestica), and two placental mammals, mouse and human. This approach reveals that the two marsupials show a higher sequence identity than that seen between rodents and primates, despite separating at approximately the same point in time, some 65–85 Myr ago.
Keywords: melanopsin, marsupial, retina, ganglion cell, phylogeny, circadian entrainment
1. Introduction
In placental mammals, both image and irradiance detection are mediated by the eye (Nelson & Zucker 1981) and it was assumed that non-image-forming tasks, like the detection of light for the entrainment of the circadian timing system, were performed by rod and cone photoreceptors. However, a series of recent studies indicated the presence of a third light-sensitive cell type located in the ganglion cell layer (GCL) of the inner retina (Freedman et al. 1999; Lucas et al. 1999). These photosensitive retinal ganglion cells (pRGCs) project to the circadian pacemaker, the suprachiasmatic nuclei (SCN) as well as several other photoresponsive centres in the brain (Berson et al. 2002; Hattar et al. 2002; Sekaran et al. 2003). It has since been demonstrated that the photosensitive pigment expressed in pRGCs is melanopsin (OPN4; Melyan et al. 2005; Panda et al. 2005; Qiu et al. 2005). Although two melanopsin genes, Opn4m and Opn4x, are found in non-mammalian vertebrates, only Opn4m has been described so far in mammals (Bellingham et al. 2006). Furthermore, unlike visual pigment opsins, vertebrate melanopsins show a high degree of sequence variability (Bellingham et al. 2006; Peirson & Foster 2006).
The presence of only Opn4m in mammals raises the question as to when the Opn4x gene was lost from their lineage. To date, the study of mammalian melanopsin has been limited to the placentals and has not included the marsupials. In addition, circadian entrainment and non-visual light detection in marsupials remain largely uncharacterized. Placental and marsupial mammals last shared a common ancestor approximately 130–150 Myr ago (Hope 1993). However, unlike their placental counterparts, the marsupials have retained a number of reptilian retinal characteristics such as double cones, oil droplets and cone photoreceptor dimensions (Ahnelt et al. 1995; Arrese et al. 1999; Ahnelt & Kolb 2000), and a number of Australian marsupials have trichromatic (rather than dichromatic) colour vision (Arrese et al. 2002, 2005, 2006). Such features are associated with a diurnal lifestyle and suggest that not all marsupials are strictly nocturnal, as commonly assumed (Morton 1978; Hope et al. 1997), and may have followed a different evolutionary path from that of placentals, which are thought to have taken refuge in nocturnality early in their evolution (Jacobs 1993; Bowmaker & Hunt 1999; Foster & Provencio 1999).
This study investigates the presence of Opn4 genes in an Australian marsupial, the fat-tailed dunnart (Sminthopsis crassicaudata). The dunnart, a small insectivorous polyprotodont, has been shown to exhibit intermittent periods of activity and rest during both day and night (Arrese et al. 1999).
We report here the isolation and characterization of an Opn4 gene from the fat-tailed dunnart. Furthermore, both molecular and bioinformatic approaches suggest that like their placental relatives, the marsupials have retained only the Opn4m gene, having lost the Opn4x form. The expression of Opn4m in the dunnart eye is restricted to a subset of retinal ganglion cells randomly distributed across the retina. Collectively, our data indicate a comparable role for Opn4 in the regulation of non-visual photoresponses in the marsupial retina, as has been reported for placental mammals.
2. Material and methods
(a) Isolation of dunnart Opn4 cDNA
Ex-breeder fat-tailed dunnarts were obtained from an established colony at the University of Adelaide, South Australia. All experimental procedures were approved by the University of Western Australia Ethics Committee and followed the guidelines of the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. Following euthanasia, eyes were removed and placed in RNAlater (Ambion) to minimize RNA degradation. RNA was extracted using TRIzol (Invitrogen) according to manufacturer's instructions and reverse transcribed using the RETROscript kit (Ambion).
A pair of degenerate primers, MelDegF3 (5′-GAGTTCTATGCCTTCTGTGG-3′) and MelDegR3 (5′-GGTGGGTRGAGCGGTAGCTG-3′), were designed to highly conserved regions of the melanopsin gene in human (NM_033282), mouse (NM_013887), rat (NM_138860) and cat (AY382594). These were used for PCR using whole-eye cDNA isolated from adult dunnarts and generated a 736 bp fragment which was cloned into pGEM-T easy (Promega) according to the manufacturer's instructions and sequenced. A further PCR was carried out using DR1 (5′-GGTGATCACAAAGTACCGGTCCA-3′), a dunnart-specific primer, and a primer designed to the start site of the Opn4 gene of Monodelphis domestica, MONF1 (5′-ATGAATCCTTCTCCCATGCTAA-3′). The 519 bp fragment obtained was again cloned into pGEM-T easy and sequenced. The 3′-end of the gene was isolated using a modified RACE procedure (Frohman et al. 1988) using a gene-specific primer (GSPDunF1 5′-CACGTCCTGACTCCCTATATGAA-3′) and adapter primer (5′-GACTCGAGTCGACATCGA-3′). The 877 bp product obtained was cloned into pGEM-T easy and sequenced. 5′-RACE was carried out using the RLM-Race kit (Ambion) according to the manufacturer's instructions with primers GSPDunR1 5′-GTAGGCATCGACTTCTGTGTTGTT-3′ and NGSPDunR1 5′-CTGAGCTGGACAGGAGAGATG-3′. This produced a 266 bp fragment that was subsequently cloned and sequenced.
The M. domestica Opn4 nucleotide sequence was derived using a Blast search of the opossum genome database (http://www.ensembl.org/Monodelphis_domestica/index.html) using the mouse Opn4 sequence (NM_013887) as the query sequence. This identified 10 exons, which were compiled to generate the predicted coding region (sequence available on request).
(b) Genomic structure
Oligonucleotide primers (sequences available on request) were designed flanking the putative exon–intron boundaries. The intronic sequences were then amplified using these primer pairs and dunnart genomic DNA as template. The products were analysed on agarose gels to determine the approximate sizes of the introns, then either subcloned into pGEM-T easy and sequenced, or directly sequenced to verify the exact boundaries.
(c) Phylogenetic analysis
A neighbour-joining (Saitou & Nei 1987) phylogenetic tree was constructed with all the available melanopsin amino acid sequences using the Mega3.1 program (Kumar et al. 2004). The amino acid sequences, excluding the variable C and N termini, were aligned using ClustalW (Higgins et al. 1996). The degree of support for internal branching was assessed by bootstrapping with 1000 replicates. Drosophila Rh1 (ninaE) was used as the out-group. Accession numbers: Homo sapiens, NM_033282; Mus musculus, NM_013887; Rattus norvegicus, NM_138860; Phodopus sungorus, AY726733; Bos taurus, XM_593123; Pan troglodytes, XM_001135445; Felis catus, AY382594; Canis familiaris, XM_848642; Monodelphis domestica, Halford & Bellingham (2007, unpublished data); Sminthopsis crassicaudata, DQ383281; Danio rerio, XM_695228; Rutilus rutilus, AY226847; Gallus gallus Opn4m, AY882944; Gallus gallus Opn4x, AY036061; Podarcis sicula, DQ013043; Xenopus laevis Opn4m, DQ384639; Xenopus laevis Opn4x, AF014797; Gadus morhua Opn4x1, AF385823; Gadus morhua Opn4x2, AY126448; Branchiostoma belcheri, AB205400; and Drosophila melanogaster Rh1, NM_079683.
(d) In situ hybridization
A 519 bp in situ probe was generated from dunnart whole-eye cDNA using the primers DunF 5′-ATGAATCCTTCTCCCATG-3′ and DunR 5′-GGTGATCACAAAGTACCG-3′ and cloned into pBS SK+ vector (Stratagene) and sequence verified. The plasmid was linearized using XhoI for the antisense probe and SacI (NEB) for the sense probe. Sense and antisense riboprobes were labelled using the DIG RNA labelling kit (Roche) and T7 and T3 RNA polymerases, respectively, according to the manufacturer's protocol.
Eyes were dissected from adult dunnarts and fixed in 4% paraformaldehyde (PFA) for 2 hours at 4°C, after which they were cryoprotected overnight in 10% sucrose at 4°C. Corneas and lenses were removed and the eye cups were embedded and frozen in 7.5% gelatin in 10% sucrose. Cryostat sections (15 μm) were mounted onto RNase-free polylysine-coated slides and stored at −80°C until required.
Sections were fixed for 10 min in 4% PFA, followed by several washes in PBS. The tissue sections were then permeabilized in 5 μg ml−1 proteinase K for 3 min, followed by a brief 5 min fixation in 4% PFA and additional PBS washes. The slides were then acetylated in acidified triethanolamine for 10 min, washed for 30 min in 1% Triton in PBS, and finally rinsed in PBS. Pre-hybridization was carried out at room temperature for 2 hours in hybridization buffer (50% formamide, 5× SSC, 5× Denhardt's solution, 5% dextran sulphate and 1 mg ml−1 tRNA). The pre-hybridization solution was then replaced with a 1 : 200 dilution of the probe in hybridization buffer and incubated in a humid chamber for 16 hours at 68°C. After hybridization, slides were placed in 0.2× SSC (preheated to 72°C), for 90 min, followed by two subsequent room temperature washes in 0.2× SSC and buffer 1 (150 nM NaCl, 0.1% Tween20 and 100 mM Tris (pH 7.5)). The sections were blocked for 60 min at room temperature with 10% normal goat serum (NGS) in buffer 1 and incubated overnight with an alkaline phosphatase-conjugated digoxigenin antibody (Roche) diluted 1 : 4000 in 1% NGS buffer. This incubation was followed by several 5 min washes in buffer 1 and a final 2 min wash in buffer 2 (100 mM NaCl, 50 mM MgCl2, 0.24 g l−1 levamisole and 100 mM Tris (pH 9.5)). Finally, slides were incubated for 2–5 hours in NBT/BCIP (Roche) at room temperature. Following colour development, the reaction was stopped using 0.1% Tween20 in PBS. This protocol was also used on flat-mount retinae except that permeabilization was carried out using 20 μg ml−1 proteinase K for 7 min. Both sections and whole retinae (GCL up) were mounted in water-soluble mounting media (Vector Labs) and stored at 4°C for subsequent image collection. The sections were examined using a Zeiss Axioplan2 microscope and images were acquired using a Spot digital camera (Diagnostic Instruments).
(e) Analysis of melanopsin-positive retinal ganglion cell distributions
Melanopsin-positive ganglion cells were counted using transmitted light microscopy at a magnification of 68×. Distribution plots of these cells in selected areas of the retinal wholemounts were made using a camera lucida drawing tube attachment. The plots were scanned and converted into digital (.jpeg) on-scale maps that were subsequently used to perform the distribution analysis.
For each cell, both X and Y coordinates were determined and used to calculate the distance to the nearest neighbour (NN). Cells located near the limit of the sampled area were not considered as a subject of NN calculation since the NN of these cells could potentially be located outside the boundaries of the area in question. For that reason, the cells present in this margin were only considered as NNs of cells included in the array.
Several methods were used to investigate the distribution characteristics of melanopsin-positive retinal ganglion cells in the dunnart retina. Cell distributions were analysed using the regularity index (RI; Wässle & Riemann 1978), calculated by dividing the mean NN measurement by its standard deviation. As such, a high RI is indicative of regular cell distribution. The dispersion index (DI; Cook 1996) was also calculated, which provides a measure of whether a cellular population is clustered or regularly distributed. The theoretical mean DI for random distributions is 1. Monte Carlo comparisons were also used to generate 100 random distributions for each map, with identical dimensions and cell densities. RI and DI values were calculated for each random distribution and compared against the observed data to determine the probability of obtaining such a distribution by chance.
Finally, a test for normality (Shapiro–Wilk's W test) was used on the melanopsin distributions. Shapiro–Wilk's test results provided a good concordance with R2 values obtained from fitting the observed data against a normal distribution.
3. Results and discussion
(a) Isolation of dunnart Opn4 cDNA
The full-length sequence of the fat-tailed dunnart, S. crassicaudata, Opn4 was generated from four overlapping fragments: a 736 bp product obtained using degenerate primers based on an alignment of mammalian (human, mouse, rat and cat) melanopsin gene sequences; a second fragment, 518 bp long, using a primer designed to the ATG region of Opn4 from the South American grey short-tailed opossum (M. domestica; Halford & Bellingham 2007, unpublished data); and 5′- and 3′-RACE products of 266 and 877 bp respectively. The compiled nucleotide sequence of the dunnart Opn4 gene of 2175 bp (GenBank accession number DQ383281) encodes a predicted protein of 487aa showing many of the characteristics of an opsin.
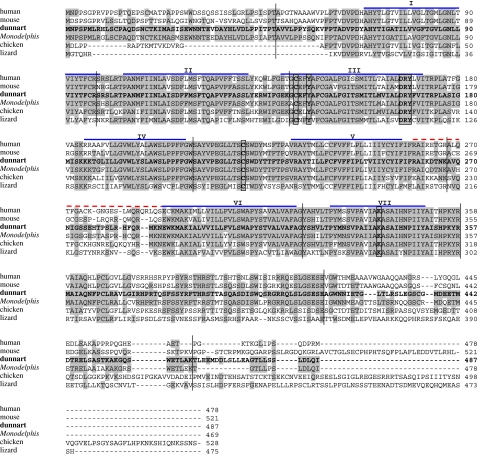
Opsins are members of the superfamily of G-protein-coupled receptors, which function through the activation of a guanine nucleotide-binding protein (G-protein) and an effector enzyme. Opsin proteins contain seven α-helical transmembrane regions connected by three intracellular and three extracellular loops. These seven α-helices form a bundle within the membrane, creating a hollow cavity on the extracellular side that serves as a binding site for the chromophore, retinal (Palczewski et al. 2000). The predicted amino acid sequence of dunnart Opn4 was examined using both the transmembrane hidden Markov model (TMHMM, http://www.cbs.dtu.dk/services/TMHMM; Sonnhammer et al. 1998) and the program HMMTOP (http://www.enzym.hu/hmmtop/). Both programs predict that this protein does contain seven transmembrane domains as would be expected in an opsin. These domains are shown in figure 1. A lysine residue at position 339 (K339) required to form a Schiff base linkage with the chromophore is at a position equivalent to 296 in bovine rod opsin. A counterion for this Schiff base is provided as in other melanopsin orthologues by the aromatic residue tyrosine at position 146 (Y146). Other characteristic features of opsins that are also present in the dunnart Opn4 sequence are a pair of cysteines at C143 and C221 that form a disulphide bridge between the first and the second extracellular loops to stabilize the tertiary structure and an aspartate, arginine and tyrosine (DRY) tripeptide at the cytoplasmic boundary of helix 3 that has been shown to be important for G-protein binding (figure 1; Bockaert & Pin 1999; Palczewski et al. 2000).
Figure 1.
Alignment of the deduced dunnart Opn4 amino acid sequence with those of human, mouse, opossum, chicken and lizard. Residues that are identical to the dunnart sequence are shaded. The seven probable transmembrane domains are marked with blue lines above the sequence and numbered using roman numerals. The third cytoplasmic loop is marked with a dotted red line above the sequence. The characteristic features of an opsin are shown boxed: lysine (K) to form a Schiff's base at position 339; tyrosine (Y), a possible counterion at position 146; aspartate, arginine and tyrosine (DRY) tripeptide for transducin binding at position 167–169; and cysteines (C) at positions 143 and 221 for disulphide bridge formation. The nucleotide sequence has been submitted to GenBank (accession number DQ383281). The positions of the intron insertion sites are also marked with vertical lines. Accession numbers: Sminthopsis crassicaudata, DQ383281; Homo sapiens, NM_033282; Mus musculus, NM_013887; Monodelphis domestica, Halford & Bellingham (2007, unpublished data); Gallus gallus, AY882944; and Podarcis sicula, DQ013043.
The dunnart Opn4 protein has a calculated molecular weight of 54.4 kDa and a predicted pI of 8.43. Prediction of putative post-translational modification sites was performed using the PredictProtein program (http://www.predictprotein.org/submit.html), which revealed the presence of two potential N-glycosylation sites (aa positions 18 and 30) and potentially 10 protein kinase C (aa positions 56, 100, 183, 231, 265, 280, 383, 387, 427 and 450) and seven casein kinase II (aa positions 25, 32, 131, 413, 464, 472 and 481) phosphorylation sites. An N-terminal site for glycosylation is also a feature characteristic of opsins (Kaushal et al. 1994).
Comparison of the sequences of the visual opsins from a broad range of vertebrates and non-vertebrates has helped to define regions of these proteins that are important in signal transduction. It was thought that a similar approach may help to elucidate the mechanism by which melanopsin transduces light information. However, the relative lack of sequence conservation across various species has led to speculation that there may be significant differences in function (Peirson & Foster 2006). The discovery that vertebrates have evolved two separate melanopsin genes has meant that comparisons between truly orthologous sequences can be undertaken. Alignment of the newly isolated dunnart sequence with those from human, mouse and opossum as well as two non-mammalian sequences, chicken and lizard, shows that all the characteristic features of an opsin described above are conserved (figure 1). Table 1 shows the percentage of identical residues shared between these species over the entire length of the protein. These results demonstrate that dunnart Opn4 is 57–60% identical to human, mouse and chicken but only 38% identical to lizard. This discrepancy arises from the fact that the lizard sequence represents the only Opn4x gene included in this comparison. This also emphasizes that the dunnart Opn4 sequence described here represents the Opn4m gene. This observation led us to design degenerate PCR primers to highly conserved regions of the known Opn4x genes (sequences available on request), but to date no products have been generated from dunnart retinal cDNA. Repeated Blast searches of the opossum genomic sequence (http://www.ensembl.org/Monodelphis_domestica/index.html) by us and others (Bellingham et al. 2006) have also failed to identify an Opn4x gene in the marsupial genome.
Table 1.
Percentage amino acid identity across the entire sequence of various vertebrate melanopsins.
| human | mouse | dunnart | opossum | chickena | lizard | |
|---|---|---|---|---|---|---|
| human | — | 75 | 60 | 62 | 53 | 38 |
| mouse | — | 58 | 59 | 51 | 40 | |
| dunnart | — | 87 | 57 | 38 | ||
| opossum | — | 59 | 38 | |||
| chicken | — | 42 |
Opn4m.
Melanopsin has been the subject of much discussion and interest owing to its remarkable variability at three different levels: first, the existence of two melanopsin genes in non-mammalian vertebrates as outlined above; second, the overall variability seen within the melanopsin family; and finally, the non-conserved amino acid sequence of the third intracellular loop.
In terms of their overall sequence, dunnart and opossum melanopsins are 87% identical (with a similarity of 92%) whereas human and mouse are 75% (table 1). If the hypervariable N and C termini are excluded and only the core regions are compared, then the identity between dunnart and opossum is increased to 92% and that of human and mouse to 86%. These results are unexpected given that dunnart/opossum and human/mouse both diverged approximately 65–85 Myr ago (Lee 1999).
The third intracellular loop is a region of the protein important for G-protein interaction and activation of the phototransduction cascade (Franke et al. 1992) and shows a remarkable conservation in other opsins. For example, in rod opsin, human, mouse and dunnart show 100% identity across this region. However, comparison of this region (marked with a red dotted line in figure 1) in dunnart and opossum melanopsins reveals a 77% identity, which falls to 65% in human and mouse. This variability raises the questions as to whether (i) melanopsin can accommodate unusually high number of variations in this region and still retain its function; (ii) melanopsin can signal light through different pathways (G-proteins) in different species; or (iii) the third intracellular loop of melanopsin is important in G-protein interaction at all.
The data presented in this study indicate that the similarity seen between dunnart and opossum Opn4 is slightly higher than that seen between human and mouse. This is intriguing given the facts that (as described above) these two groups diverged at approximately the same time and that the two marsupials have since evolved on two separate continents (Australian and American). This may suggest that a genetic drift has separated rodents and primates while unknown selective forces have acted to conserve several genetic features seen in the marsupials (Hope 1993).
(b) Genomic structure
To further characterize this marsupial gene, we have determined the position and nucleotide sequence of intron–exon boundaries. The genomic organization of the human melanopsin gene has been determined previously (Provencio et al. 2000), so we used this information to position the putative exon boundaries on the dunnart cDNA sequence. Oligonucleotide primers were designed flanking these putative boundaries. The intronic sequences were then amplified using these primer pairs and dunnart genomic DNA as template. Dunnart Opn4 consists of 10 exons and spans approximately 30 kb of genomic DNA. All of the intron–exon boundaries are consistent with the GT–AG rule (Mount 1982). All of the boundaries and intron and exon sizes are shown in table 2 and their positions are also marked in figure 1. We also determined the genomic structure of the opossum Opn4 gene by comparing the cDNA sequence of known melanopsins with the available genomic sequence. The opossum gene also consists of 10 exons, spanning approximately 30 kb of genomic DNA, and shows a remarkable similarity to dunnart. The human OPN4 consists of 10 exons, but spans only 11.8 kb (Provencio et al. 2000).
Table 2.
Exon–intron boundaries of dunnart Opn4.
| exon | exon size (bp) | 5′ intron | exon sequence | 3′ intron | intron size (bp) |
|---|---|---|---|---|---|
| 1 | 144a | 5′UTR------ | ATGAATCCTT ----- CACCCCCACG | gtaagccggctcatggaatc | ∼5000b |
| 2 | 146 | ctttgtctctctccccatag | GCAGTGGTAT ----- CCTTCTGCAG | gtacttggccctgggcaccc | ∼10 000b |
| 3 | 134 | caataacttgacctccgcag | GAGTCGCAGT ----- GGAGAGAAAG | gtaggagggaacggcagcca | 296 |
| 4 | 204 | cttcatacatttacccatag | GATGTGAATT ----- TTTGGATGGA | gttgagtgtgttttcctgag | 397 |
| 5 | 172 | ttctctggatacctcccaag | GTGCCTACGT ----- ACACCAACAA | gtgagcctccgggggcagcc | 701 |
| 6 | 162 | ttccatgtttgtttccctag | GGCCGTTCAG ----- CCTTTGCTGG | gtaattattgagaaggaaag | 890 |
| 7 | 108 | tctggtttctccccctccag | GTATTCCCAC ----- CCAAGTACAG | gtaagcgccccccagcactc | 1240 |
| 8 | 181 | ttgggtttcttcatccccag | AATGGCCATC ----- GAGTGAAGCC | gtaagtattcctgatgctcc | 3300 |
| 9 | 139 | ctcttcatggttccttgcag | GGCTGGAACA ----- TCTGGCGAAG | gtgactgggcgggatggtgg | 1304 |
| 10 | 75c | ctcttgttttttgcctttag | ACACTGGAAG ----- TCAAATATAA | ------3′UTR |
Length from translation start site.
Introns 1 and 2 have not been sequenced in their entirety and their approximate sizes are shown.
Length to stop codon.
(c) Phylogenetic analysis
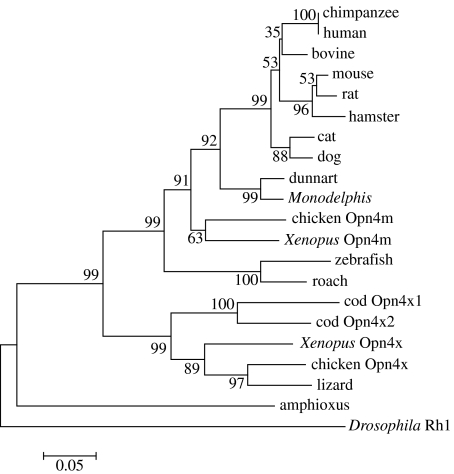
A phylogenetic analysis derived by the neighbour-joining method (Saitou & Nei 1987) places dunnart Opn4 on the expected marsupial branch with opossum and separated from the mammals on the placental branch (figure 2). The analysis also includes the known Opn4x genes which as expected clade together but are separate from all the Opn4m genes. These results provide further evidence that the dunnart Opn4 sequence is part of the Opn4m family.
Figure 2.
Phylogenetic tree of melanopsin proteins. The amino acid sequences were aligned using ClustalW (Higgins et al. 1996) and the tree was generated by the neighbour-joining method (Saitou & Nei 1987) using the Mega3.1 program (Kumar et al. 2004). The variable C and N termini were excluded. Drosophila Rh1 (ninaE) was used as the out-group. Branch confidence levels (per cent based on 1000 bootstrap replicates) are marked. Scale bar indicates substitutions per site.
(d) Expression and distribution
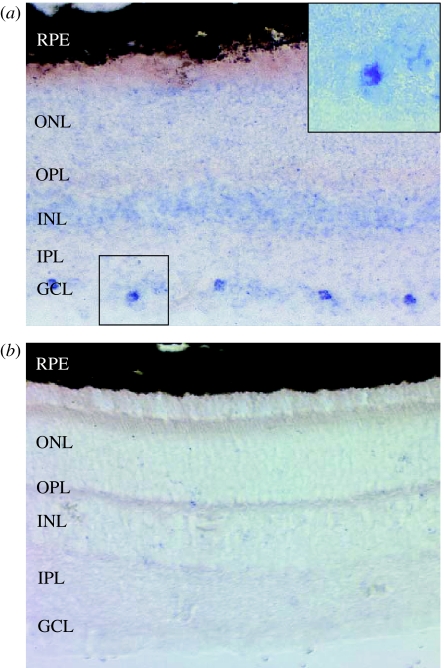
The localization of dunnart Opn4 was determined using in situ hybridization on adult dunnart retinal sections using sense and antisense riboprobes labelled with digoxigenin. The results in figure 3a show that melanopsin expression is restricted to a subset of cells in the GCL. This pattern of expression correlates with that previously reported for mouse and primate Opn4m genes (Provencio et al. 2000). No staining is seen with the sense probe (figure 3b). Occasionally, we have seen staining in the inner nuclear layer (INL) (data not shown), a finding previously described by Hattar et al. (2002) and attributed to displaced ganglion cells.
Figure 3.
Tissue in situ hybridization. Adult dunnart retinal sections probed with (a) Opn4 antisense riboprobe and (b) sense control riboprobe. Opn4 shows staining restricted to a subset of cells in the ganglion cell layer. No signal is seen with the sense probe. Inset in (a) is a magnification of the highlighted area. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; RPE, retinal pigmented epithelium.
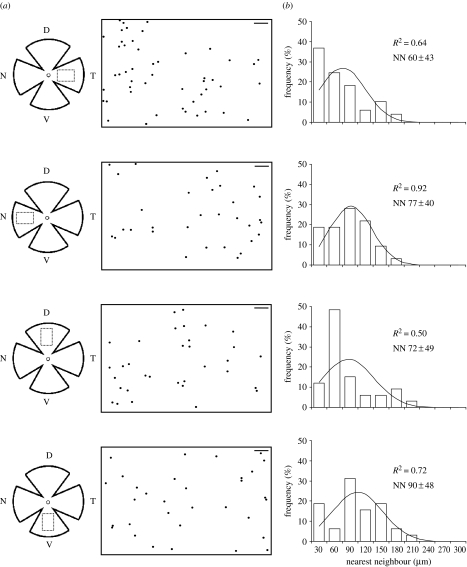
The same technique was used on flat-mounted retinae to determine the distribution of melanopsin-positive retinal ganglion cells within the dunnart retina. All four retinal regions analysed contained melanopsin-positive ganglion cells at an average density of approximately 46 cells mm−2, with a slight increase observed in the temporal region. The dunnart retina has an average area of 36 mm2 and approximately 81 400 RGCs (s.d.±3360; Arrese et al. 1999). Using the cell density of melanopsin-positive cells calculated in this study, we estimate the percentage of these to be between 1 and 2.7%, which is very similar to that determined for rodents by Hattar et al. (2002). This concordance suggests that the labelling technique described here has identified the majority of the cells expressing melanopsin. The distribution of melanopsin cells in table 3 is shown in figure 4. The RI values are in general low; the highest score is 0.72 and the average only 0.62. This implies that the standard distribution of the nearest-neighbour measurements is high and therefore indicates that cells are not regularly distributed. The dispersion indices obtained were close to 1, further supporting a random distribution. The RI and DI values obtained with our data are not significantly different from simulated random distributions. Shapiro–Wilk's W test for a normal distribution shows that the melanopsin-positive cells in the temporal and dorsal fields were non-normally distributed. The non-normal distribution observed in these regions may to be due to a skewed distribution, as nearest-neighbour data may more closely resemble a Poisson distribution. In summary, these data support a random distribution of melanopsin-positive retinal ganglion cells in the dunnart, broadly similar to the pattern observed in the mouse (Hattar et al. 2002), and comparable to the random distribution previously characterized in the cat retina (Semo et al. 2005). This is in contrast to the distribution of the non-photoreceptive population of ganglion cells in dunnart that show a non-random distribution with a mid-temporal region of high cell density (area centralis) embedded in a visual streak (Arrese et al. 1999). The mechanisms underlying the development of the pRGC system remain largely uncharacterized, but a random distribution of pRGCs may be expected of an irradiance detector, enabling environmental light levels to be sampled across the whole retina.
Table 3.
Summary of the analysis of nearest-neighbour distributions for melanopsin-labelled cellsa.
| region | number of cells | nearest neighbour, s.d. (μm) | cell density (cells mm−2) | RIa | DIa | Shapiro–Wilk's W statistic |
|---|---|---|---|---|---|---|
| temporal | 49 | 60.2±43.3 | 61.3 | 0.72 | 0.94 | W=0.872 |
| p=0.98 | p=0.27 | p<0.001 | ||||
| (0.56) | (0.97) | non-normal | ||||
| nasal | 32 | 77.2±39.8 | 40.0 | 0.52 | 0.98 | W=0.948 |
| p=0.30 | p=0.50 | p=0.124 | ||||
| (0.57) | (0.98) | normal | ||||
| dorsal | 33 | 71.9±49.1 | 41.3 | 0.68 | 0.92 | W=0.823 |
| p=0.85 | p=0.27 | p<0.001 | ||||
| (0.59) | (0.98) | non-normal | ||||
| ventral | 32 | 89.5±48.0 | 40.0 | 0.54 | 1.13 | W=0.952 |
| p=0.46 | p=0.99 | p=0.161 | ||||
| (0.56) | (0.96) | normal |
The p-values assigned from Monte Carlo tests as described in §2. Values in brackets show the mean value for 100 simulated random distributions (probability of getting equal to or less than the value observed with random distributed data). DI, dispersion index; RI, regularity index.
Figure 4.
Distribution of pRGCs in the dunnart retina. (a) Schematic diagrams of flat-mounted retinae. The circle illustrates the approximate position of the optic nerve and the dotted rectangle delimits the sampled areas. D, dorsal; T, temporal; V, ventral; N, nasal. The corresponding distribution plots are shown to the right (scale bar, 100 μm). (b) Histograms represent the nearest-neighbour distributions for melanopsin RGCs in each region. The fit of the data to a normal distribution is indicated in each plot by the R2 value.
(e) Conclusions
We have successfully isolated an Opn4 gene from the fat-tailed dunnart (S. crassicaudata) and have demonstrated that it belongs to the Opn4m family of melanopsins. We have been unable to isolate an Opn4x gene from the fat-tailed dunnart and can find no evidence of its presence in the marsupial genome. We also provide evidence that Opn4m is expressed in a subset of ganglion cells, a finding that correlates with previous studies in placental mammals. Finally, we have analysed the distribution of these cells across the dunnart retina, showing that they are random. This study provides further evidence that the Opn4x gene has been lost from the mammalian lineage. As to when and why this has occurred remains unanswered. Analysis of Opn4 in the monotreme lineage, which diverged prior to the placental/marsupial split (approx. 280 Myr ago), may shed some light on this issue.
Acknowledgments
All experimental procedures were approved by the University of Western Australia Ethics Committee and followed the guidelines of the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
This work was supported by an Australian Research Council Discovery Project Grant to J.S., R.G.F. and C.A., and by Fundação para a Ciência e Tecnologia (FCT) to S.S.P. This work was also in part funded by a Wellcome Trust programme grant to R.G.F. We thank Frederic Causeret (Imperial College) for his assistance with the in situ hybridization and Donna Mackay (UCL) for help and advice. We are grateful to Prof. Lyn Beazley (UWA) for her support and advice during the investigation.
References
- Ahnelt P.K, Hokoc J.N, Rohlich P. Photoreceptors in a primitive mammal, the South American opossum, Didelphis marsupialis aurita: characterization with anti-opsin immunolabeling. Vis. Neurosci. 1995;12:793–804. doi: 10.1017/s0952523800009366. [DOI] [PubMed] [Google Scholar]
- Ahnelt P.K, Kolb H. The mammalian photoreceptor mosaic-adaptive design. Prog. Retin. Eye Res. 2000;19:711–777. doi: 10.1016/s1350-9462(00)00012-4. doi:10.1016/S1350-9462(00)00012-4 [DOI] [PubMed] [Google Scholar]
- Arrese C, Dunlop S.A, Harman A.M, Braekevelt C.R, Ross W.M, Shand J, Beazley L.D. Retinal structure and visual acuity in a polyprotodont marsupial, the fat-tailed dunnart (Sminthopsis crassicaudata) Brain Behav. Evol. 1999;53:111–126. doi: 10.1159/000006588. doi:10.1159/000006588 [DOI] [PubMed] [Google Scholar]
- Arrese C.A, Hart N.S, Thomas N, Beazley L.D, Shand J. Trichromacy in Australian marsupials. Curr. Biol. 2002;12:657–660. doi: 10.1016/s0960-9822(02)00772-8. doi:10.1016/S0960-9822(02)00772-8 [DOI] [PubMed] [Google Scholar]
- Arrese C.A, Oddy A.Y, Runham P.B, Hart N.S, Shand J, Hunt D.M, Beazley L.D. Cone topography and spectral sensitivity in two potentially trichromatic marsupials, the quokka (Setonix brachyurus) and quenda (Isoodon obesulus) Proc. R. Soc. B. 2005;272:791–796. doi: 10.1098/rspb.2004.3009. doi:10.1098/rspb.2004.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese C.A, Beazley L.D, Neumeyer C. Behavioural evidence for marsupial trichromacy. Curr. Biol. 2006;16:R193–R194. doi: 10.1016/j.cub.2006.02.036. doi:10.1016/j.cub.2006.02.036 [DOI] [PubMed] [Google Scholar]
- Bellingham J, et al. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. doi:10.1371/journal.pbio.0040254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson D.M, Dunn F.A, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. doi:10.1126/science.1067262 [DOI] [PubMed] [Google Scholar]
- Bockaert J, Pin J.P. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. doi:10.1093/emboj/18.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker J.K, Hunt D.M. Molecular biology of photoreceptor spectral sensitivity. In: Archer S.N, Djamgoz M.B.A, Loew E.R, Partridge J.C, Vallerga S, editors. Adaptive mechanisms in the ecology of vision. Kluwer Academic Press; Dordrecht, The Netherlands: 1999. pp. 439–462. [Google Scholar]
- Cook J.E. Spatial properties of retinal mosaics: an empirical evaluation of some existing measures. Vis. Neurosci. 1996;13:15–30. doi: 10.1017/s0952523800007094. [DOI] [PubMed] [Google Scholar]
- Franke R.R, Sakmar T.P, Graham R.M, Khorana H.G. Structure and function in rhodopsin. Studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J. Biol. Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- Freedman M.S, Lucas R.J, Soni B, von Schantz M, Munoz M, David-Gray Z, Foster R. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:502–504. doi: 10.1126/science.284.5413.502. doi:10.1126/science.284.5413.502 [DOI] [PubMed] [Google Scholar]
- Foster R.G, Provencio I. The regulation of vertebrate biological clocks by light. In: Archer S, Djamgoz M, Loew E, editors. Adaptive mechanisms in the ecology of vision. Chapman & Hall; London, UK: 1999. pp. 223–243. [Google Scholar]
- Frohman M.A, Dush M.K, Martin G.R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. doi:10.1073/pnas.85.23.8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao H.W, Takao M, Berson D.M, Yau K.W. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. doi:10.1126/science.1069609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T. Using Clustal for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hope R.M. Selected features of marsupial genetics. Genetica. 1993;90:165–180. doi: 10.1007/BF01435038. doi:10.1007/BF01435038 [DOI] [PubMed] [Google Scholar]
- Hope P.J, Wittert G.A, Morley J.E. Feeding patterns of S. crassicaudata: role of gender, photoperiod and fat stores. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1997;272:78–83. doi: 10.1152/ajpregu.1997.272.1.R78. [DOI] [PubMed] [Google Scholar]
- Jacobs G.H. The distribution and nature of colour vision among the mammals. Biol. Rev. Camb. Philos. Soc. 1993;68:413–471. doi: 10.1111/j.1469-185x.1993.tb00738.x. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Ridge K.D, Khorana H.G. Structure and function in rhodopsin: the role of asparagine-linked glycosylation. Proc. Natl Acad. Sci. USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. doi:10.1073/pnas.91.9.4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lee M.S. Molecular clock calibrations and metazoan divergence dates. J. Mol. Evol. 1999;49:385–391. doi: 10.1007/pl00006562. doi:10.1007/PL00006562 [DOI] [PubMed] [Google Scholar]
- Lucas R.J, Freedman M.S, Mun¨oz M, Garcia-Ferna´ndez J.-M, Foster R.G. Regulation of the mammalian pineal by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:505–507. doi: 10.1126/science.284.5413.505. doi:10.1126/science.284.5413.505 [DOI] [PubMed] [Google Scholar]
- Melyan Z, Tarttelin E.E, Bellingham J, Lucas R.J, Hankins M.W. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. doi:10.1038/nature03344 [DOI] [PubMed] [Google Scholar]
- Morton S.R. An ecological study of Sminthopsis carasicaudata (Marsupialia: Dasyuridae) III. Reproduction and life history. Austr. Wildl. Res. 1978;5:183–211. doi:10.1071/WR9780183 [Google Scholar]
- Mount S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. doi:10.1093/nar/10.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.J, Zucker I. Absence of extraocular photoreception in diurnal and nocturnal rodents exposed to direct sunlight. Comp. Biochem. Physiol. Part A Physiol. 1981;69:145–148. doi:10.1016/0300-9629(81)90651-4 [Google Scholar]
- Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. doi:10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- Panda S, Nayak S.K, Campo B, Walker J.R, Hogenesch J.B, Jegla T. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. doi:10.1126/science.1105121 [DOI] [PubMed] [Google Scholar]
- Peirson S, Foster R.G. Melanopsin: another way of signaling light. Neuron. 2006;49:331–339. doi: 10.1016/j.neuron.2006.01.006. doi:10.1016/j.neuron.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Provencio I, Rodriguez I.R, Jiang G, Hayes W.P, Moreira E.F, Rollag M.D. A novel human opsin in the inner retina. J. Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Kumbalasiri T, Carlson S.M, Wong K.Y, Krishna V, Provencio I, Berson D.M. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. doi:10.1038/nature03345 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sekaran S, Foster R.G, Lucas R.J, Hankins M.W. Calcium imaging reveals a network of intrinsically light-sensitive inner-retinal neurons. Curr. Biol. 2003;13:1290–1298. doi: 10.1016/s0960-9822(03)00510-4. doi:10.1016/S0960-9822(03)00510-4 [DOI] [PubMed] [Google Scholar]
- Semo M, Munoz Llamosas M, Foster R.G, Jeffery G. Melanopsin (Opn4) positive cells in the cat retina are randomly distributed across the ganglion cell layer. Vis. Neurosci. 2005;22:111–116. doi: 10.1017/S0952523805001069. doi:10.1017/S0952523805001069 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Wässle H, Riemann H. The mosaic of nerve cells in the mammalian retina. Proc. R. Soc. B. 1978;200:441–461. doi: 10.1098/rspb.1978.0026. doi:10.1098/rspb.1978.0026 [DOI] [PubMed] [Google Scholar]