Abstract
Using an integrative genomics approach called Amplification Breakpoint Ranking and Assembly (ABRA) analysis, we nominated KRAS as a gene fusion with the ubiquitin-conjugating enzyme UBE2L3 in the DU145 cell line, originally derived from prostate cancer metastasis to the brain. Interestingly, analysis of tissues revealed that 2 of 62 metastatic prostate cancers harbored aberrations at the KRAS locus. In DU145 cells, UBE2L3-KRAS produces a fusion protein, specific knock-down of which, attenuates cell invasion and xenograft growth. Ectopic expression of the UBE2L3-KRAS fusion protein exhibits transforming activity in NIH 3T3 fibroblasts and RWPE prostate epithelial cells in vitro and in vivo. In NIH 3T3 cells, UBE2L3-KRAS attenuates MEK/ERK signaling, commonly engaged by oncogenic mutant KRAS, and instead signals via AKT and p38 MAPK pathways. This is the first report of a gene fusion involving Ras family suggesting that this aberration may drive metastatic progression in a rare subset of prostate cancers.
Keywords: KRAS, gene fusion, prostate cancer, genomic amplification, bioinformatics
Introduction
To understand the characteristic features of driving gene fusions in cancer, we previously carried out a large-scale integrative analysis of cancer genomic datasets matched with gene rearrangement data (1). As part of this analysis, we observed that in many instances, a small subset of tumors or cancer cell lines harboring an oncogenic gene fusion displays characteristic amplification at the site of genomic rearrangement (2–6) (Supplementary Fig. 1a–b). Such amplifications usually affect a portion of the fusion gene, and are generally considered secondary genetic lesions associated with disease progression, drug resistance, and poor prognosis (2, 4–8). In contrast, high level copy number changes that result in the marked over-expression of oncogenes usually encompass the target genes at the center of overlapping amplifications across a panel of tumor samples. Thus, a “partially” amplified cancer gene may suggest that this gene participates in a genomic fusion event important in cancer progression. This is the result of several independent genetic accidents including the formation of the gene fusion and subsequent amplification, suggesting possible selective pressure in cancer cells for this aberration. Toward this end, we developed an integrative genomic approach called amplification breakpoint ranking and assembly (ABRA) to discover causal gene fusions from cancer genomic datasets.
To uncover driving gene fusions contributing to prostate cancer progression, we applied ABRA analysis to genomic data from ten prostate cancer cell lines. Most interestingly, this led to the identification of a KRAS gene fusion in a rare subset of metastatic prostate cancer. RAS proteins play a critical role in cellular physiology, development, and tumorigenesis (9, 10). Mutations in RAS have been identified in a wide spectrum of cancers (9), but rarely in prostate cancer (11). To date, oncogenic alterations in the Ras pathway have been exclusively restricted to activating point mutations, including the most commonly studied Gly-to-Val substitution at codon 12 and substitutions at codons 13 and 61 of the different RAS isoforms (9, 12, 13). Gene fusions involving RAS genes have thus far not been described as a class of cancer-related mutations. This is the first description of a mutant chimeric version of KRAS and thus may represent a new class of cancer-related alterations.
Materials and Methods
Amplification Breakpoint Ranking and Assembly. Cell lines used for aCGH analysis were obtained from either ATCC or collaborators and authenticated by providers (detailed in the supplementary methods)
The microarray CGH data from prostate cancer cell lines were segmented by the circular binary segmentation (CBS) algorithm (14), and the genomic position of each amplification breakpoint was mapped with the genomic regions of all human genes. The 3′ amplified genes were rated by their rConSig Score, which identified KRAS as the top candidate. Matching the amplification level of 3′ KRAS with 5′ amplified genes from DU145 cells nominated UBE2L3, SOX5, and C14orf166 as 5′ partner candidates. The array CGH data used in this study has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus with the accession number GSE26447.
Reverse Transcription PCR, Nuclease protection assay, and Fluorescence In Situ Hybridization
RT-PCR with the fusion primers UBE2L3-S2 and KRAS-R2 (Supplementary Table 1) confirmed the UBE2L3-KRAS fusion in DU145 cells. Fusion qPCR was performed on a panel of prostate cancer cell lines using primers UBE2L3-q1 and KRAS-q2 (StepOne Real Time PCR system, Applied Biosystems). Ribonuclease protection assays were performed utilizing a 230 bp fragment spanning the UBE2L3-KRAS fusion junction. Interphase FISH was done on cell lines, paraffin-embedded tissue sections, and tissue microarrays using bacterial artificial chromosome probes.
Western Blotting and Multiple Reactions Monitoring Mass Spectrometry
Lysates from DU145, PrEC, RWPE, 22RV1, VCaP, and PC3 cells, either untreated or treated with 500nM bortezomib for 12 hours, were probed with anti-RAS monoclonal (Millipore) and anti-KRAS rabbit polyclonal antibodies (Proteintech Group Inc). Cell lysates from DU145 and LnCaP cells treated with bortezomib were separated by SDS-PAGE and subject to trypsin digestion.. Transitions of tryptic digested peptides were compared to those of labeled internal standard peptides (spanning the fusion junction) by Multiple Reactions Monitoring Mass Spectrometry to identify the fusion peptides, see supplementary methods for more details..
In Vitro Overexpression and Stable Knockdown of UBE2L3-KRAS Fusion
Expression plasmids for UBE2L3-KRAS were generated with the pDEST40 (with or without 5′ FLAG) and pLenti-6 vectors (without 5′ FLAG). The expression plasmids were introduced into HEK (5′ FLAG-UBE2L3-KRAS pDEST40 vector), NIH/3T3 (UBE2L3-KRAS pDEST40 vector), and RWPE cells (UBE2L3-KRAS pLenti-6 vector) using standard protocols, detailed in the supplementary Materials and Methods. The prostate cancer cell line DU145 was infected with lentiviruses with scrambled shRNA or UBE2L3-KRAS shRNA, and stable cell lines were generated by selection with puromycin (Invitrogen).
Cell Proliferation, Invasion and Pathway Analysis, Xenograft Mouse Model
Cell counting analysis and basement membrane matrix invasion assays were performed as described previously (15, 16). Protein lysates from NIH/3T3 stable cell lines expressing UBE2L3-KRAS, V600E mutant BRAF, G12V mutant KRAS, and vector controls were probed with phospho and total MEK1/2, p38 MAPK, Akt, and ERK antibodies (Cell Signaling Technologies). The stable NIH/3T3 and RWPE cells expressing UBE2L3-KRAS, and pooled or single clone population of DU145 cells with the stable knockdown of UBE2L3-KRAS were implanted subcutaneously into nude mice.
Additional Details
Additional details can be found in Supplemental Information.
Results
Based on the fusion breakpoint principle previously described (1), amplifications associated with gene fusions usually involve the 5′ region of 5′ partners, and 3′ region of 3′ partners. Further, the amplification levels of 5′ and 3′ fusion genes will be identical due to their co-amplification as a single fusion gene. This observation provided the rationale to assemble putative gene fusions from amplification breakpoints by matching the amplification levels of candidate 5′ and 3′ partners. We therefore developed ABRA analysis, which leverages the in vivo amplification and breakpoint analysis in cancer cells to assemble novel gene fusions and predict their tumorigenicity. Concept signature analysis was developed in a previous study (17) and provides a Consig score, which is helpful in ranking biologically relevant candidates based on prior knowledge and has been incorporated into ABRA analysis.
The detailed methodology of ABRA analysis is depicted in Supplementary Fig. 1c and discussed in Supplementary Methods. We initially focused this analysis on cancer cell lines, as breakpoint analyses are more reliable in uniform cellular populations as opposed to tumors, which comprise multiple cell types, many of which are not malignant.
Next, we tested the ABRA approach using a published single nucleotide polymorphism microarray (aSNP) dataset (2) generated from 36 leukemia cell lines including the K-562 chronic myeloid leukemia cell line known to harbor the amplified BCR-ABL1 fusion (18). We inferred the relative DNA copy number data and identified all 5′ and 3′ amplified genes from the 36 cell lines (≥2 copies). In this dataset, ABL1 was the top ranking gene with a 3′ copy number increase (Supplementary Fig. 1d, left panel, Supplementary Table 2). The amplification levels of all 5′ amplified genes in K-562 cells were then matched with ABL1 to nominate potential 5′ partners. In total, six 5′ amplified genes were found in K-562 and five matched the level of ABL1 3′ amplification. After curation of the amplification breakpoints, BCR and NUP214 were nominated as ABL1 fusion partner candidates (Supplementary Fig. 1d, right panel, see methods and Supplementary Fig. 2 for the criteria of candidate selection). This demonstrated the feasibility of this method in nominating driver gene fusions from genomic datasets.
To nominate novel gene fusions contributing to the progression of advanced prostate cancer, we applied this method to an array comparative genomic hybridization (aCGH) dataset of 10 prostate cancer cell lines. Interestingly, the top candidate nominated in the ETS gene fusion-negative prostate cancer cell line, DU145, was the KRAS locus, which exhibited a clear breakpoint accompanied by a 3′ amplification of KRAS (Figure 1a, left panel, Supplementary Table 3). This result was particularly intriguing due to the well known role of KRAS as an oncogene (9) and the observation that activating point mutations of KRAS are found in a number of tumor types, but rarely seen in prostate cancer (11). Interestingly, the activation of downstream signaling intermediaries of the RAS-MAPK pathway has been observed in prostate cancer by a number of studies (19–21).
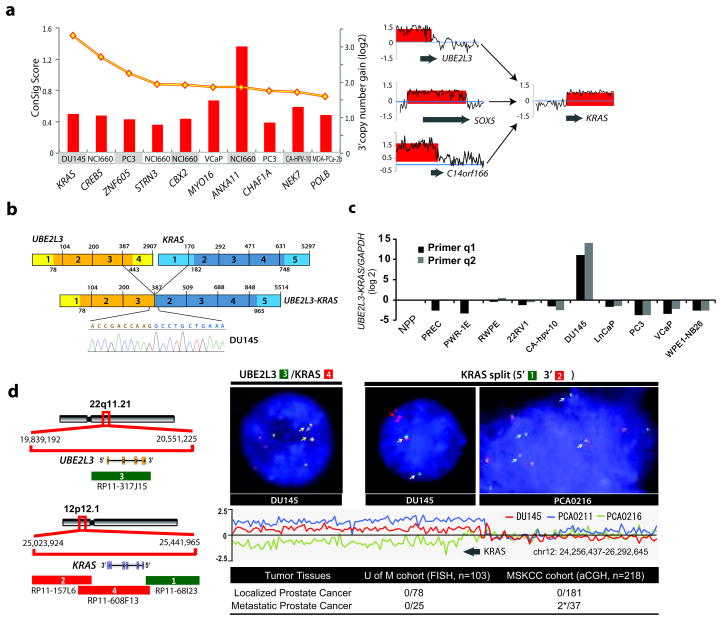
Figure 1. Identification and characterization of a novel KRAS rearrangement in metastatic prostate cancer.
(a) Left panel, amplification breakpoint analysis and ConSig scoring of 3′ amplified genes from a panel of advanced prostate cancer cell lines nominated KRAS as a fusion gene candidate with 3′ amplification in the DU145 prostate cancer cell line. The ConSig scores are depicted by the yellow line and the level of 3′ amplification for each 3′ fusion gene candidate is depicted by red columns. Right panel, matching the amplification level of 5′ amplified genes in DU145 cells nominates SOX5, C14orf166, and UBE2L3 as 5′ fusion partner candidates for KRAS. The relative quantification of DNA copy number data from the genomic regions 1Mb apart from the candidate fusion genes is shown. The x-axis indicates the physical position of the genomic aberrations. The fusion partners are indicated by grey arrows. (b) Schematic of sequencing results from Reverse Transcription PCR revealing fusion of UBE2L3 with KRAS in DU145. Structures for the UBE2L3 and KRAS genes have their basis in the Genbank reference sequences. The numbers above the exons (indicated by boxes) indicate the last base of each exon. Open reading frames are shown in darker shades. The exons of UBE2L3-KRAS fusion are numbered from the original reference sequences. Line graphs show the position and DNA sequencing of the fusion junction. (c) A panel of prostate cancer cell lines was analyzed for UBE2L3-KRAS mRNA expression by SYBR assay with the fusion primers. * NPP, normal prostate pool. (d) Left panel, the genomic organizations of UBE2L3 and KRAS loci are shown in the schematic, with red and green bars indicating the location of BAC clones. Genes are shown with the direction of transcription indicated by the arrows and exons indicated by bars. Right panel, FISH assay (upper) and copy number data analysis (lower) confirms the fusion of UBE2L3 to KRAS in DU145 cells and recurrent rearrangements at the KRAS locus. The left FISH figure shows three copies of fusion signals as indicated by yellow arrows, using co-localizing probes for the fusion. The right FISH figure shows triplicate KRAS 3′ signals in DU145, and 3′ deletion of KRAS in a metastatic prostate tumor, PCA0216, using probes that tightly encompass the KRAS locus. Relative quantification of copy number array CGH data at the KRAS locus in DU145, and metastatic prostate tumors PCA0211 and PCA0216 are shown in the middle panel. The lower panel displays a summary of KRAS rearrangements revealed by FISH and copy number analysis of a series of prostate cancer tissues from the University of Michigan (UM) and Memorial Sloan-Kettering Cancer Center (MSKCC). Table indicates the number of positive cases divided by the total number of cases evaluated. *Positive cases: PCA0211 (spine metastasis), 3′ amplification; PCA0216 (bladder metastasis), 3′ deletion.
To assemble amplification breakpoints in the KRAS gene with more confidence, we carried out replicate array CGH hybridizations for DU145 cells. Matching the amplification level of KRAS with the 5′ amplified genes from DU145 cells, we identified 10 potential 5′ partner candidates that were suggested by either of the two array CGH hybridizations. After curation, C14orf166, SOX5, and UBE2L3 remained as the top 5′ partner candidates for KRAS (Figure 1a, right panel, Supplementary Table 4), based on the criteria detailed in Supplementary Fig. 2.
To experimentally validate the predicted fusions of C14orf166-KRAS, SOX5-KRAS, and UBE2L3-KRAS, we designed primer pairs from the first exons of candidate 5′ partners and the last exon of KRAS, as well as the exons next to the breakpoints (Supplementary Table 1). Reverse transcription polymerase chain reaction (RT-PCR) analysis of DU145 cells identified a specific fusion band for UBE2L3-KRAS, but not for the others. Subsequent sequencing of the RT-PCR product confirmed fusion of UBE2L3 exon 3 to KRAS exon 2, as schematically depicted in Figure 1b. To assess the abundance of the UBE2L3-KRAS fusion transcripts, we analyzed a panel of prostate cell lines by SYBR green quantitative PCR (QPCR) (Figure 1c) and RNase protection assay (Supplementary Fig. 3). UBE2L3-KRAS was highly expressed in DU145 cells, but not in the other prostate cancer cell lines tested; this result was confirmed by subsequent paired-end transcriptome sequencing of DU145 cells (Supplementary Fig. 4). Moreover, mutation analysis of the fusion sequences from DU145 did not reveal canonical point mutations in the fusion allele of KRAS (Supplementary Fig. 5).
To examine the chromosomal rearrangements involving UBE2L3 and KRAS loci in DU145 cells, we carried out fluorescence in situ hybridization (FISH) analysis. By both KRAS split probe and UBE2L3-KRAS fusion probe FISH analysis, DU145 cells clearly showed a rearrangement at the KRAS genomic loci and fusion with UBE2L3 (Figure 1d). In addition, we observed low level amplification (3 copies) of the UBE2L3-KRAS fusion consistent with its nomination by the ABRA approach. To extend our studies to prostate tumors, we carried out a combination of KRAS split probe FISH (n= 103 total cases) and array CGH breakpoint (n=218 total cases) analysis of 259 clinically localized prostate cancers, and 62 metastatic prostate cancers from the University of Michigan and Memorial Sloan Kettering Cancer Center. Interestingly, while clinically localized cases did not show aberrations at the KRAS locus, we identified 2 of 62 metastatic prostate cancers that harbored a rearrangement at the KRAS locus (Figure 1d). One of the index cases, PCA0216, which was a soft tissue metastasis, was validated by both array CGH and FISH; the other index case, PCA0211, was a bone metastasis and was only validated by arrayCGH (FISH analysis was attempted, but due to decalcification, adequate hybridization was not possible) (Figure 1d, Supplementary Fig. 6a). Further investigation of the available gene and exon expression data for case PCA0211 suggested that this case did not over-express ETS family genes, but instead exhibited high expression of KRAS exons 2–6 (except exon 1) similar to the DU145 cell line (Supplementary Fig. 6b–c).
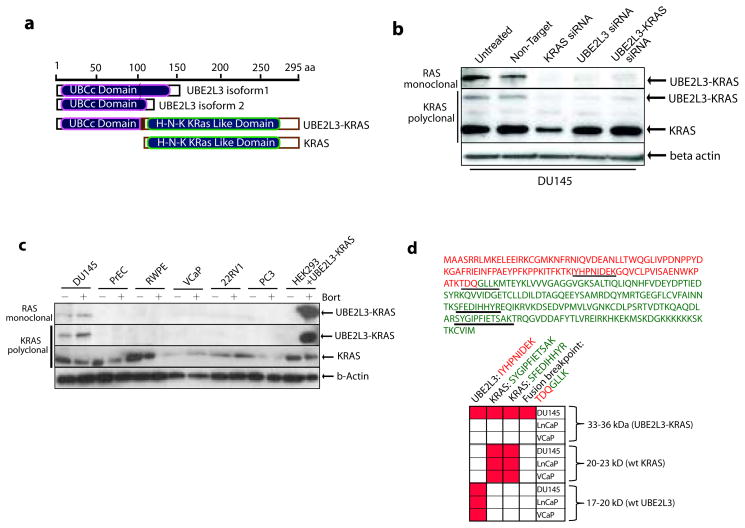
We next examined expression of the UBE2L3-KRAS protein. The predicted 296 amino acid fusion protein trims 17 amino acids from the C-terminus of UBE2L3 (Figure 2a). The full length KRAS protein is preserved, with a 4 amino acid insertion between UBE2L3 and KRAS. Using both a monoclonal antibody raised against the Ras family and a polyclonal antibody raised against KRAS specifically, we detected a 33 kDa fusion protein in addition to the 21 kDa band corresponding to wild-type KRAS in DU145 cells (Figure 2b). Specificity of the band attributed to the UBE2L3-KRAS protein was shown by knocking down expression using RNA interference against KRAS, UBE2L3, and the chimeric junction of UBE2L3-KRAS (Supplementary Fig. 7a). The UBE2L3-KRAS protein was found specifically in DU145 cells and not in a panel of other prostate cell lines (Figure 2c). Specific expression of the protein was independently confirmed by mass spectrometric assessment of DU145 cells using a multiple reaction monitoring (MRM) assay (which does not require antibody based detection) (Figure 2d). Over-expression of UBE2L3-KRAS in HEK293 cells, however, did not result in detectable fusion protein. Interestingly, in the presence of the proteosomal inhibitor, bortezomib, expression of the fusion protein was clearly apparent, suggesting decreased stability of the fusion protein in the over-expression system (Figure 2c).
Figure 2. Characterization of the UBE2L3-KRAS fusion protein.
(a) Schematic representations of UBE2L3, KRAS, and the predicted UBE2L3-KRAS fusion protein. (b) Expression of the UBE2L3-KRAS fusion protein in DU145 cells. Immunoblot analysis of DU145 cells using an anti-RAS mouse monoclonal antibody and an anti-KRAS rabbit polyclonal antibody detects a 33kDa fusion protein specific to DU145 cells. siRNA duplexes employed are indicated. β-actin was used to demonstrate equal loading. (c) Survey of the UBE2L3-KRAS fusion protein in a panel of prostate cancer cell lines and stabilization of protein expression with a proteosome inhibitor, bortezomib. Cell lines are indicated and treated in the presence or absence of 500nM bortezomib for 24 hours. HEK293 cells were transfected with an expression construct encoding UBE2L3-KRAS. Immunoblot analysis was carried out using KRAS polyclonal and RAS monoclonal antibodies. (d) Mass spectrometric assay for the detection of the UBE2L3-KRAS protein in DU145 cells. An MRM-MS assay was developed to detect the UBE2L3-KRAS fusion protein. Upper panel, sequence of the UBE2L3-KRAS fusion protein with amino acids colored in red from UBE2L3 and colored in green from KRAS. Tryptic peptides used for MRM-MS analysis are underlined. Matrix represents positive measurement (highlighted in red) of peptides from corresponding gel fractions of DU145, LNCaP, and VCaP whole cell lysates.
One area that needs to be explored and may explain the protein stability issues is the function of the UBE2L3 ubiquitin-conjugating (E2) enzyme portion of the fusion (22). We therefore attempted to detect ubiquitination of UBE2L3-KRAS protein. We identified a Rat anti-Ras monoclonal antibody that precipitated the 33kd UBE2L3-KRAS protein as well as additional bands in the 40–55kd region, which were specific to HEK 293T cells expressing the fusion (Supplementary Fig. 8a). These shifted bands are recognized by both anti-Ras and anti-HA tagged ubiquitin antibodies, and their molecular weights match the prediction for ubiquitinated fusion proteins. We further validated these ubiquitinated UBE2L3-KRAS proteins in DU145 cells (Supplementary Fig. 8b). These data suggest that the UBE2L3-KRAS protein is ubiquitinated, which may contribute to its decreased stability.
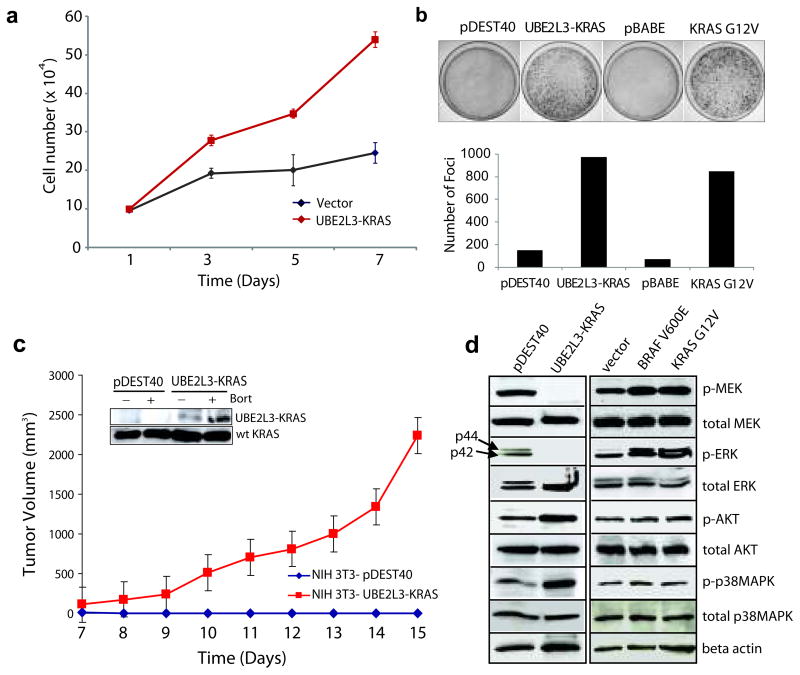
To determine the function of the UBE2L3-KRAS fusion, we over-expressed the fusion protein in NIH 3T3 cells (Supplementary Fig. 7b), a system classically used to study RAS biology (12, 23). Of note, enforced expression of UBE2L3-KRAS induced loss of fibroblast morphology, and increased cell proliferation and focus formation (Supplementary Fig. 9a, Figure 3a-b). Cell cycle analysis revealed an increase in the S phase fraction of cells (Supplementary Fig. 9b). To determine the effects of UBE2L3-KRAS expression on tumor growth in vivo, we implanted nude mice with stable NIH 3T3 vector control cells or NIH 3T3 UBE2L3-KRAS fusion expressing cells. We observed robust tumor formation by the UBE2L3-KRAS expressing cells but not the vector transfected cells (Figure 3c, Supplementary Fig. 10). To interrogate the potential RAS-related signaling pathways engaged by UBE2L3-KRAS in NIH 3T3 cells, we carried out a series of immunoblot analyses on key signaling intermediaries. As reported in the literature for NIH 3T3 cells, KRAS is a stronger inducer of the MEK/ERK cascade; whereas HRAS is a stronger activator of the PI3K/AKT pathway (24). Interestingly, UBE2L3-KRAS over-expression resulted in attenuated endogenous MEK and ERK phosphorylation (Figure 3d) in NIH 3T3 cells; instead, the signaling was directed to AKT and p38 MAP Kinase cascades, both of which have been implicated in prostate cancer (19, 21).
Figure 3. Transforming activities of the UBE2L3-KRAS fusion in NIH 3T3 cells.
(a) Overexpression of UBE2L3-KRAS in NIH 3T3 cells increases cellular proliferation. pDEST40 represents an empty vector. (b) Overexpression of UBE2L3-KRAS in NIH 3T3 cells induces focus formation. Oncogenic KRAS G12V was used as a positive control with respective empty vectors as negative controls (pDEST40 and pBABE). Photographs of representative plates are shown in the upper panel and quantification of focus formation is shown by the bar graph in the lower panel. (c) UBE2L3-KRAS transfected NIH 3T3 cells form tumors in nude mice. Stable polyclonal populations of NIH 3T3 cells expressing either the vector or UBE2L3-KRAS fusion gene were injected subcutaneously into nude mice. Tumor growth was monitored from day 7 to day 15 as indicated. The insert shows the presence of the fusion protein in the stably transfected NIH 3T3 cells, which is further stabilized upon bortezomib treatment. (d) Investigation of the downstream signaling pathways engaged by the UBE2L3-KRAS fusion in NIH 3T3 cells. Lysates prepared from stably transfected NIH 3T3 polyclonal populations and vector controls were subject to immunoblot analysis for phospho- and total MEK, ERK, AKT, and p38 MAPK. Oncogenic BRAFV600E and KRASG12V were included as controls. β-actin was used as a loading control.
As activation of the MEK-ERK pathway is dependent on membrane attachment of Ras proteins, we investigated Ras sub-cellular localization using immunofluorescence assays. Interestingly, Ras proteins, which are normally distributed in the cytoplasm, were found to be highly enriched in the late endosome after ectopic expression of UBE2L3-KRAS fusion in NIH3T3 cells (Supplementary Fig. 11). We speculate that this relocation of Ras proteins may decrease their association with the cellular membrane, and possibly enhance growth-factor receptor signaling in the endosome.
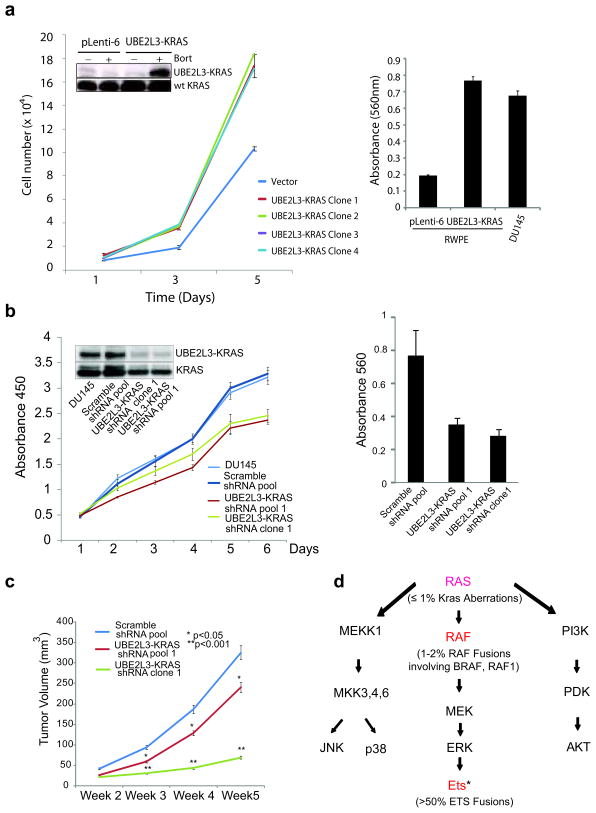
To investigate the role of the UBE2L3-KRAS fusion in a prostate background, we over-expressed the fusion in RWPE prostate epithelial cells (Supplementary Fig. 7c). The expression of the fusion protein was enhanced by incubation with bortezomib (Figure 4a, insert). Over-expression of the UBE2L3-KRAS fusion in RWPE cells led to increased cell invasion, proliferation, and a transient increase of tumor growth in nude mice (Figure 4a, Supplementary Fig. 12). It is notable that in the RWPE model, signaling pathway analysis did not reveal inhibition of the MEK/ERK pathway or activation of AKT/p38 MAPK, (data not shown). Although the MEK inhibitor U0126 inhibits the invasion of RWPE cells over-expressing either wild type or mutant KRAS, treatment of RWPE cells over-expressing the fusion continued to exhibit invasive properties in the presence of U0126 (Supplementary Fig. 13), suggesting that downstream effectors other than MEK/ERK may be engaged by the fusion in the prostate context.
Figure 4. The oncogenicity of UBE2L3-KRAS fusion in the prostate context.
(a) Expression of the UBE2L3-KRAS fusion in RWPE benign prostate epithelial cells leads to increased cellular proliferation and invasion. Left, the results of cell proliferation assays using stable RWPE cell clones infected with either the pLenti-6 vector or UBE2L3-KRAS. The inset shows the 33kd fusion protein detected only in the fusion transfected cells treated with bortezomib to enhance protein stability (data from a representative clone is shown (Clone 2)). Right, modified Boyden chamber-matrigel assays using the pLenti-6 vector and the fusion expressing cells (Clone 2). Invading cells were stained with crystal violet and quantitated. DU145 prostate cancer cells were used as a positive control. (b) Knockdown of the UBE2L3-KRAS fusion reduces cell proliferation and invasion in DU145 cells. Left, cell growth relative to the control shRNA was monitored using WST-1 assay for 6 days. Insert shows the immunoblot analysis for the 33kd fusion protein detected using Ras monoclonal antibody. Right, results of matrigel invasion assay for DU145 pool and clone with UBE2L3-KRAS knock-down. Scrambled shRNA duplexes are used as control. (c) Knock-down of the UBE2L3-KRAS fusion attenuates prostate tumor growth in mouse xenograft models. The figure shows a plot of mean tumor volume trajectories over time for mice inoculated with DU145 pool (red) or single clone (green) after UBE2L3-KRAS stable knock-down. Error bars represent the standard error of the mean at each time point. (d) A summary of RAS-RAF signaling pathways in relation to recurrent gene fusions characterized in prostate cancer. Genes that participate in fusion events are indicated in red. In parenthesis are the percentage of prostate cancers harboring aberrations in the ETS family, RAF family, and KRAS gene locus. * ETS family members involved in gene fusions include ERG, ETV1, 4, and 5. Figure adapted and modified from: Gioeli, Kraus, Weber et al, Current Clinical Oncology: Prostate Cancer.
To further confirm the function of endogenous UBE2L3-KRAS in DU145 cells, we performed stable knock-down of UBE2L3-KRAS fusion and generated chicken embryo chorioallantoic membrane and mouse xenograft models. This resulted in decreased cell invasion and proliferation in vitro, as well as the inhibition of tumor formation in the in vivo models (Figure 4b-c, Supplementary Figure 14).
Discussion
The addiction of cancer cells to causal gene fusions often results in amplification, in vivo, which may be exploited to reveal unidentified recurrent gene rearrangements. Based on this observation, we developed an integrative genomic-based approach called ABRA to explore driving gene fusions contributing to the progression of prostate cancer. This led to the nomination of an oncogenic UBE2L3-KRAS fusion in DU145 prostate cancer cells, which was delineated by amplification breakpoints at KRAS and UBE2L3 loci, validating the ABRA analysis approach. Although the ConSig score was not necessary to nominate KRAS as an oncogenic partner, it remains a useful approach for the identification of potential novel oncogenes involved in gene rearrangements in large genomic datasets.
UBE2L3-KRAS encodes a protein encompassing most of the UBE2L3 protein and full length KRAS, which is ubiquitinated and exhibits a high turn-over rate. Importantly, recurrent genomic rearrangements at the KRAS locus were found in 2 of 62 metastatic prostate tumors in addition to the DU145 metastatic prostate cancer cell line. While a number of oncogenic activating point mutations of KRAS have been identified, this is the first description of a mutant chimeric version of KRAS that is oncogenic and thus may represent a new class of cancer-related alteration. Consistent with this finding, we recently described gene fusions of BRAF and RAF1 in 1–2 % of prostate tumors, further implicating the RAS-RAF-MAPK signaling pathway in subsets of prostate cancer (25). A summary of gene fusions involving this pathway that have been discovered in prostate cancer is shown in Figure 4d. The rarity of KRAS rearrangement will limit its clinical relevance to a small subset of metastatic prostate cancer patients.
While both KRAS G12V and UBE2L3-KRAS exhibit an oncogenic phenotype in vitro and in vivo, UBE2L3-KRAS over-expression leads to attenuation, rather than activation, of the MEK-ERK pathway in NIH 3T3 cells. Instead, it appears that the KRAS fusion enriches Ras proteins in the endosome, and switches signaling to the AKT and p38 MAPK cascades. This observation may have important implications in understanding the biology of this most studied proto-oncogene. These signaling switches were not observed in RWPE cells expressing the fusion, which bypasses the ERK pathway (unlike the KRAS G12V mutant), indicating that other downstream effectors are being engaged in the prostate background. KRAS rearrangements, although rare in the prostate cancer setting, could modulate metastasis using signaling pathways that have thus far not been associated with KRAS. Future studies will be needed to elucidate the details of how chimeric KRAS engages endogenous RAS-related signaling pathways in the context of prostate cancer
Supplementary Material
Statement of significance.
This is the first description of an oncogenic gene fusion of KRAS, one of the most studied proto-oncogenes. KRAS rearrangement may represent the driving mutation in a rare subset of metastatic prostate cancers, emphasizing the importance of RAS-RAF-MAPK signaling in this disease.
Acknowledgments
This work is supported in part by the Early Detection Research Network UO1 CA111275, Prostate SPORE P50CA69568, National Center for Integrative Bioinformatics (U54 DA21519-01A1), National Institutes of Health (R01CA132874), the National Functional Genomics Center (W81XWH-09-2-0014), R01 CA125612-01, and R01 CA132874. A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award and a Burroughs Welcome Foundation Award in Clinical Translational Research. A.M.C. is an American Cancer Society Research Professor.
We thank R. Ponnala, X. Jiang, H. Li, C. Brenner, A. Menon, L. Wang, C. Maher, C. Kumar-Sinha, F. Demichelis, A. K. Tewari, J. W. Locasale, and D. Anastasiou for their assistance, H. Choi and N. Shah for assistance on array CGH data analysis, B. Han, K. Suleman, Y. Wei, R. Mehra for pathology techniques, T. Barrette for hardware support, J. Siddiqui, R. Varambally, and R. Kuefer for providing tissue samples and information, Dr. C. L. Sawyers for facilitating collaboration with MSKCC, J. Yu and S. Tomlins for helpful discussions, Dr. B. Weinberg for providing the pBABE puro plasmid, W. Hahn for the pBABE-puro-BRAF-V600E and pBABE puro K-RAS V12 plasmids.
Abbreviations
- FISH
fluorescence in situ hybridization
- ABRA
amplification breakpoint ranking and assembly
- ConSig
concept signature analysis
- CBS
circular binary segmentation
- aSNP
single polymorphism microarray
- aCGH
comparative genomic hybridization
- QPCR
quantitative PCR
- RT-PCR
Reverse transcription polymerase chain reaction
- MRM
multiple reaction monitoring
Footnotes
Potential conflict of interests: none.
References
- 1.Wang XS, Prensner JR, Chen G, et al. An integrative approach to reveal driver gene fusions from paired-end sequencing data in cancer. Nat Biotechnol. 2009;27:1005–11. doi: 10.1038/nbt.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 3.Graux C, Cools J, Melotte C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 4.Barr FG, Nauta LE, Davis RJ, Schafer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira BI, Alonso J, Carrillo J, et al. Array CGH and gene-expression profiling reveals distinct genomic instability patterns associated with DNA repair and cell-cycle checkpoint pathways in Ewing’s sarcoma. Oncogene. 2008;27:2084–90. doi: 10.1038/sj.onc.1210845. [DOI] [PubMed] [Google Scholar]
- 6.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stergianou K, Fox C, Russell NH. Fusion of NUP214 to ABL1 on amplified episomes in T-ALL--implications for treatment. Leukemia. 2005;19:1680–1. doi: 10.1038/sj.leu.2403877. [DOI] [PubMed] [Google Scholar]
- 8.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cancer targets in the Ras pathway. Cold Spring Harb Symp Quant Biol. 2005;70:461–7. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 11.Moul JW, Friedrichs PA, Lance RS, Theune SM, Chang EH. Infrequent RAS oncogene mutations in human prostate cancer. Prostate. 1992;20:327–38. doi: 10.1002/pros.2990200407. [DOI] [PubMed] [Google Scholar]
- 12.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–5. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 13.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 14.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 15.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X-S, Cao Q, Prensner JR, et al. Integrative Analyses Reveal the Functional and Genetic Associations of Gene Fusions in Cancer. 2009. submitted. [Google Scholar]
- 18.Wu SQ, Voelkerding KV, Sabatini L, Chen XR, Huang J, Meisner LF. Extensive amplification of bcr/abl fusion genes clustered on three marker chromosomes in human leukemic cell line K-562. Leukemia. 1995;9:858–62. [PubMed] [Google Scholar]
- 19.Graff JR, Konicek BW, McNulty AM, et al. Increased AKT activity contributes to prostate cancer progression by dramatically accelerating prostate tumor growth and diminishing p27Kip1 expression. J Biol Chem. 2000;275:24500–5. doi: 10.1074/jbc.M003145200. [DOI] [PubMed] [Google Scholar]
- 20.Daniel Gioeli SK, Weber Michael J, Weber Michael J. Signal Transduction by the Ras MAP Kinase Pathway in Prostate Cancer Progression. In: Nevalainen RGPaMT., editor. Current Clinical Oncology: Prostate Cancer: Signaling Networks, Genetics, and New Treatment Strategies. Totowa: Humana Press; 2008. pp. 223–56. [Google Scholar]
- 21.Xu L, Chen S, Bergan RC. MAPKAPK2 and HSP27 are downstream effectors of p38 MAP kinase-mediated matrix metalloproteinase type 2 activation and cell invasion in human prostate cancer. Oncogene. 2006;25:2987–98. doi: 10.1038/sj.onc.1209337. [DOI] [PubMed] [Google Scholar]
- 22.Moynihan TP, Cole CG, Dunham I, O’Neil L, Markham AF, Robinson PA. Fine-mapping, genomic organization, and transcript analysis of the human ubiquitin-conjugating enzyme gene UBE2L3. Genomics. 1998;51:124–7. doi: 10.1006/geno.1998.5257. [DOI] [PubMed] [Google Scholar]
- 23.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:3637–40. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Zhu T, Guan KL. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J Biol Chem. 2004;279:37398–406. doi: 10.1074/jbc.M405730200. [DOI] [PubMed] [Google Scholar]
- 25.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.