Summary
Single stranded DNA binding proteins play many roles in nucleic acid metabolism, but their importance during transcription remains unclear. Quantitative proteomic analysis of RNA polymerase II (RNApII) pre-initiation complexes (PICs) identified Sub1 and the Replication Protein A complex (RPA), both of which bind single-stranded DNA (ssDNA). Sub1, homolog of mammalian coactivator PC4, exhibits strong genetic interactions with factors necessary for promoter melting. Sub1 localizes near the transcription bubble in vitro and binds to promoters in vivo dependent upon PIC assembly. In contrast, RPA localizes to transcribed regions of active genes, strongly correlated with transcribing RNApII but independently of replication. RFA1 interacts genetically with transcription elongation factor genes. Interestingly, RPA levels increase at active promoters in cells carrying a Sub1 deletion or ssDNA binding mutant, suggesting competition for a common binding site. We propose that Sub1 and RPA interact with the non-template strand of RNApII complexes during initiation and elongation, respectively.
Introduction
RNA Polymerase II (RNApII) mediated transcription requires multiple accessory proteins for initiation, elongation, and termination. Basal transcription factors position RNApII on promoters to form the Pre-Initiation Complex (PIC) but also function at later steps such as promoter melting and initiation site choice (Sikorski and Buratowski, 2009; Thomas and Chiang, 2006). Characterization of these factors remains an important goal. Using a quantitative proteomic screen to identify promoter-bound PIC components, we identified Sub1 and Replication Protein A (RPA) as two ssDNA binding proteins that associate with RNApII complexes.
Yeast Sub1 was originally identified genetically as a suppressor of specific TFIIB mutations (Knaus et al., 1996) and biochemically as a stimulator of in vitro basal transcription (Henry et al., 1996). Sub1 is homologous to the mammalian coactivator PC4, which is reported to physically interact with activators and components of the RNApII basal transcription machinery (Ge and Roeder, 1994; Malik et al., 1998). Sub1 has also been implicated in transcription elongation, mRNA 3′ end processing, and DNA repair (Conesa and Acker, 2010), but recent chromatin immunoprecipitation (ChIP) experiments place Sub1 primarily at RNApII and RNApIII promoters (Rosonina et al., 2009; Tavenet et al., 2009). Proposed roles for Sub1 include stimulating PIC recruitment, breaking contacts between basal factors during promoter escape, and modulating RNApII CTD phosphorylation, (Calvo and Manley, 2005; Garcia et al., 2010; Knaus et al., 1996), yet the molecular functions of Sub1 in transcription remain unclear.
RPA is an abundant, highly conserved heterotrimer (consisting of the proteins Rfa1, Rfa2, and Rfa3 in yeast) that binds ssDNA. Originally identified as a replication factor for Simian Virus 40, RPA has been implicated in multiple DNA repair pathways, homologous recombination, and telomere maintenance (Richard et al., 2009). While there is no functional evidence of a general role for RPA in transcription, Rfa1 was reported to associate with RNApII in high-throughput protein interaction studies (Krogan et al., 2006) and an RFA2 mutant exhibits a strong genetic interaction with deletion of the gene for RNApII subunit Rpb4 (Collins et al., 2007).
Here we show that Sub1 is a component of the PIC that localizes close to where promoter DNA unwinds for transcription. The SUB1 gene becomes essential in combination with TFIIE and TFIIH mutants that are thought to affect promoter melting. RPA, on the other hand, is primarily found downstream of the promoter in transcribed regions of genes. Genome-wide ChIP analysis shows that RPA localization strongly correlates with RNApII throughout the genome. RFA1 mutants exhibit phenotypes and genetic interactions that suggest an effect on gene expression. Interestingly, deletion of Sub1 leads to binding of Rfa1 at many promoters, suggesting these two factors compete for binding to ssDNA within RNApII transcription complexes, probably the single-stranded non-template strand. Sub1 may be preferentially recruited to initiation complexes via its interaction with TFIIB and other basal factors, while the more abundant RPA may interact with elongation complexes.
Results
Sub1 and RPA associate with the RNApII PIC in vitro
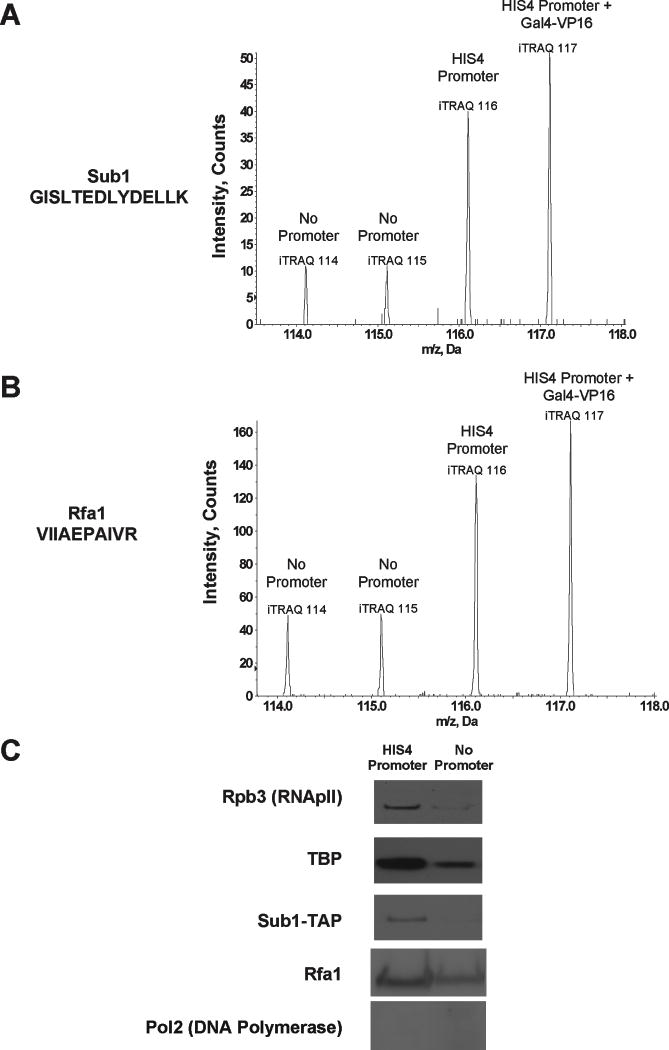
Transcription complexes can be purified by incubating nuclear extracts with promoter DNA immobilized on beads, followed by identification with mass spectrometry (Mittler et al., 2009; Ranish et al., 2003). However, non-specific DNA binding proteins complicate analysis, particularly when specific proteins are present at low levels relative to abundant contaminants. Quantitative tandem mass spectrometry (MS/MS) can circumvent these limitations by comparison of proteins bound to specific versus control DNA templates. Figure S1A illustrates the strategy used for enrichment of proteins associated with immobilized DNA templates (Ranish et al., 1999). Briefly, a biotinylated DNA fragment containing the basal HIS4 promoter and a single upstream Gal4 binding site was linked to streptavidin-coated magnetic beads. The template was incubated with S. cerevisiae nuclear extract, washed, and then the promoter and associated proteins eluted by restriction enzyme cutting just upstream of the Gal4 site. A second reaction was identical except for addition of recombinant Gal4-vp16 activator protein. A negative control used a DNA fragment lacking the HIS4 basal promoter. Proteins from each reaction were isolated and digested with trypsin, followed by labeling of free amines on the peptides with one of several distinct iTRAQ stable isotope labels (Ross et al., 2004). Peptides from all reactions were then mixed and subject to RP-RP MS/MS (Zhou et al., 2010). Peak intensities of the low mass iTRAQ reporter ions provide a measure of relative abundance of the peptide within each reaction (Figure S1A and S1B).
The relative abundances of 1867 unique peptides from 558 proteins associated with the promoterless and promoter-containing templates were compared. Proteins exhibiting Log2 ratios in channels 117:114 (comparing the Promoter plus Gal4-vp16 sample to the No Promoter control) greater than 3 standard deviations relative to the mean of the Log2 ratios in channels 115:114 (comparing two independent No Promoter controls) were flagged as possible HIS4 promoter-associated factors (Figure S1A, Table S1 and Supplementary Methods). Based on this classification, the majority of proteins showed no enrichment with HIS4 promoter DNA (Table S1 and Figures S1B and S1C). As expected, subunits of the basal initiation factors, Mediator, and several chromatin remodeling complexes were enriched on DNA templates that contained the HIS4 promoter. The full proteomic analysis will appear elsewhere.
Several peptides from the single-stranded DNA binding proteins Sub1 (Figure 1A), Rfa1 (Figure 1B), and Rfa3 (Figure S1D) were also detected at higher abundance on the HIS4 promoter relative to promoterless templates, a result verified by immunoblotting (Figure 1C). To verify that the association of RPA was specific relative to other replication factors, we repeated the quantitative proteomic analyses after enrichment of cysteine-containing peptides from the immobilized template eluents (full analysis to be presented elsewhere). RPA was the only replication associated complex that was found to be reproducibly enriched with the HIS4 promoter (Table S2). There was no evidence of DNA polymerase binding enrichment (Figure 1C), indicating RPA’s association with the HIS4 promoter is not dependent upon other components of the replisome. Therefore, Sub1, and possibly RPA, associate in vitro with proteins or DNA within the RNApII PIC.
Figure 1. Sub1 and RPA are enriched on promoter sequences in vitro.
Representative spectra from low mass iTRAQ reporter region of a (A) Sub1 peptide and (B) Rfa1 peptide. Enrichment is measured by taking the ratios of the areas under each of the peaks. Supplemental Figure S1C shows spectra for Rfa3 and a non-enriched peptide. (C) The preferential interaction between Sub1 and the HIS4 Promoter template was confirmed by immobilized template binding of nuclear extracts containing a TAP-tagged Sub1 on the HIS4 and No Promoter templates. Bound proteins were analyzed by immunoblotting using antibodies against Rpb3 (a subunit of RNApII), TBP, Rfa1, Pol2 (a subunit of DNA polymerase), and the TAP tag (for Sub1).
Sub1 associates with active RNApII-dependent promoters in vivo
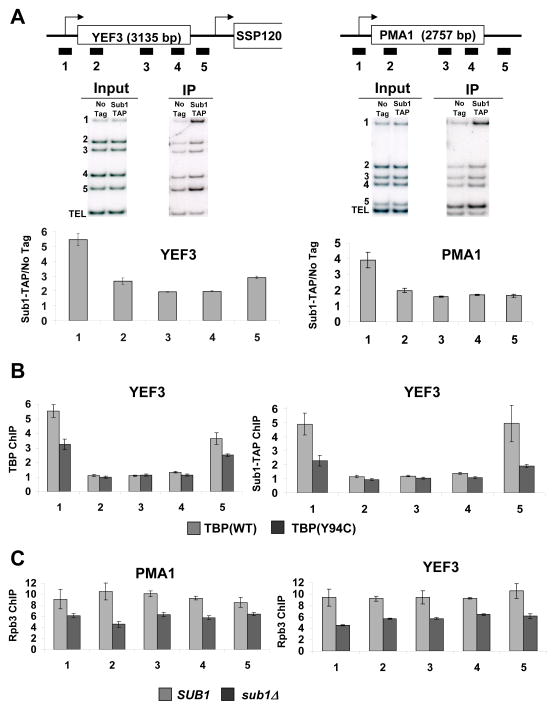
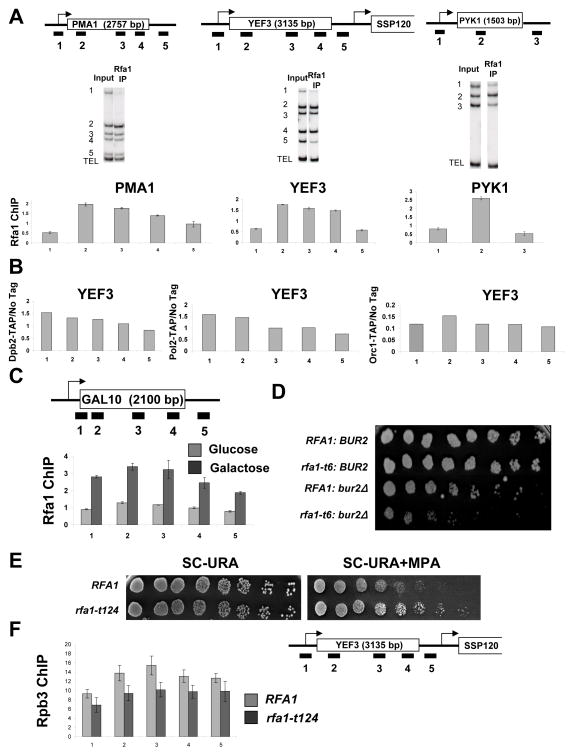
To assess Sub1 localization in vivo, four representative mRNA genes (PMA1, YEF3, PYK1, and ADH1) were assayed by chromatin immunoprecipitation (ChIP) in cells expressing TAP-tagged Sub1 (Figure 2A and Figure S2A). In agreement with several previous studies (Nedea et al., 2003; Rosonina et al., 2009), a strong Sub1 signal is seen at promoters. Strong crosslinking throughout the transcribed region was not seen (as reported in Calvo and Manley, 2005; Tavenet et al., 2009), but a low signal above background was apparent. A small peak of Sub1 was also observed at 3′ ends of many genes tested, probably due to adjacent downstream promoters or possibly gene looping (Ansari and Hampsey, 2005). In agreement with reports of Sub1 localizing to snoRNA genes (Tavenet et al., 2009; Yang et al., 2005), Sub1 also crosslinks strongly to SNR13 (Figure S2B). Sub1 binding was ~8-fold higher at the GAL10 promoter region under inducing relative to repressed conditions (Figure S2C), demonstrating that Sub1 occupancy at promoters is dependent on transcription.
Figure 2. Sub1 is found at promoters.
(A) Schematic representations of the YEF3 and PMA1 genes used in ChIP experiments are shown at top. The box shows the open reading frame and arrows indicate transcription start site. Bars below the genes represent the positions of the ChIP PCR products (see Table S4 for primer sequences). ChIP was performed using a TAP-tagged Sub1 strain and representative gels are shown in middle panels. DNA coprecipitated with IgG-agarose (to precipitate TAP-tagged Sub1) was analyzed by multiplex PCR using primers shown in the schematic. The TEL PCR product is an internal background control from a nontranscribed region. The input control is used to normalize for PCR amplification efficiency of each primer pair. Quantitations of experiments are shown at bottom. The values shown represent the averages and standard errors (bars) from three independent experiments. (B) ChIP analysis at the YEF3 gene was performed for TBP (left) and Sub1-TAP (right) in wild-type TBP and TBP(Y94C) strains. Quantitation of triplicate experiments is as in part A. (C) ChIP of the Rpb3 subunit of RNApII at the PMA1 and YEF3 genes was performed in strains with SUB1 or sub1Δ backgrounds. Quantitation of triplicate experiments is as in part A.
Because TATA-Binding Protein (TBP) is essential for the formation of RNApII PICs, Sub1 occupancy was analyzed in cells carrying TBP(Y94C), a temperature-sensitive TBP mutant with defective DNA binding (Matsui et al., 1995; Morehouse et al., 1999). We found that Sub1 crosslinking was reduced proportionally with binding of TBP (Figure 2B). Given previous indications that Sub1 and mammalian PC4 stimulate basal transcription in vitro (Ge and Roeder, 1994; Henry et al., 1996), the effect of deleting SUB1 on RNApII was tested. In agreement with recently published work (Rosonina et al., 2009), a significant drop in RNApII levels throughout the gene was observed at the constitutively active genes PMA1 and YEF3 (Figure 2C). Together, these results indicate that Sub1 is a functional component of the PIC.
A proposed role for Sub1 in the open complex
Addition of recombinant Sub1 to immobilized template reactions lacking Sub1 showed little effect on basal factor levels (data not shown), suggesting Sub1 functions after PIC assembly. Interestingly, SUB1 overexpression suppresses mutant phenotypes of specific TFIIB mutants that affect start site selection (Knaus et al., 1996; Wu et al., 1999). These TFIIB mutants form PICs in vitro but are blocked at a post-assembly step (Cho and Buratowski, 1999; Ranish et al., 1999). A recent TFIIB-RNApII crystal structure predicts that residues changed in these TFIIB mutants interact with the template DNA strand within the “open” PIC (Kostrewa et al., 2009). We hypothesize that Sub1 is recruited to the “closed” PIC and then stabilizes the unwound “open” PIC, perhaps by interacting with the melted non-template DNA strand. High levels of Sub1 might compensate for TFIIB mutants that are less efficient at capturing the template strand, explaining the observed suppression.
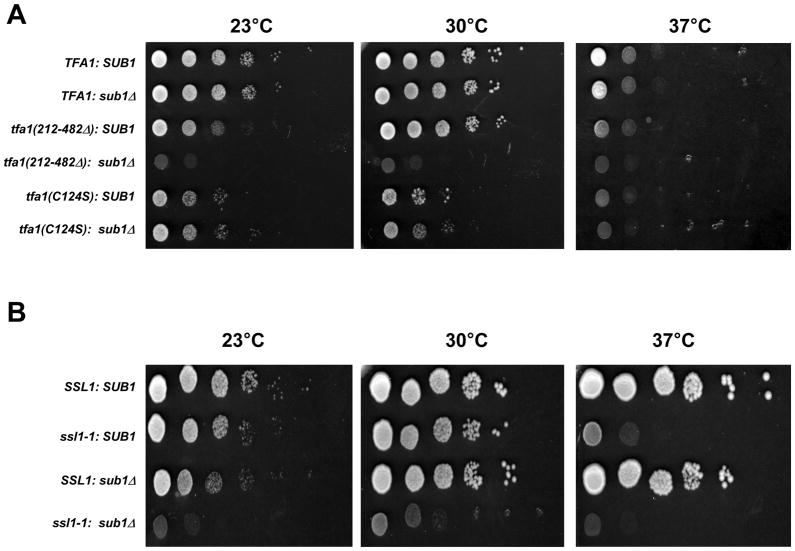
This model predicts genetic interactions between sub1Δ and mutant alleles of TFIIE and TFIIH, two initiation factors that play a role in promoter melting. Deleting a C-terminal region of the TFIIE large subunit (Tfa1) causes a cold sensitive growth defect (Kuldell and Buratowski, 1997) and this acidic region is thought to stimulate TFIIH binding and function (Watanabe et al., 2003). In contrast, mutations in cysteines of the Tfa1 N-terminal zinc finger confer sensitivity to high temperatures (Kuldell and Buratowski, 1997). Deletion of SUB1 in cells expressing the C-terminally truncated Tfa1 resulted in a severe synthetic growth defect (Figure 3A). A much weaker synthetic phenotype was seen with mutations in the zinc finger region. This allele specificity suggests Sub1 cooperates with the C-terminal region of Tfa1 for some essential function. In addition, we looked for a genetic interaction between Sub1 and Ssl1, a core subunit of TFIIH. Combining sub1Δ with the temperature sensitive mutant ssl1-1 (Yoon et al., 1992) resulted in a very strong synthetic growth defect (Figure 3B).
Figure 3. Sub1 interacts genetically with TFIIE and TFIIH.
(A) Growth of TFA1, SUB1(YSB290); TFA1, sub1Δ (YSB2530); tfa1(212–482Δ), SUB1 (YSB318); tfa1(212–482Δ), sub1Δ (YSB2532); tfa1(C124S), SUB1 (YSB335); and tfa1(C124S), sub1Δ (YSB2534) strains was assayed by 10-fold serial dilutions and incubation at 23, 30, or 37°C for three days. (B) Growth of SSL1, SUB1 (YF28); ssl1-1, SUB1 (YF29); SSL1, sub1Δ (YSB2578); and ssl1-1, sub1Δ (YSB2579) strains was assayed as in part A.
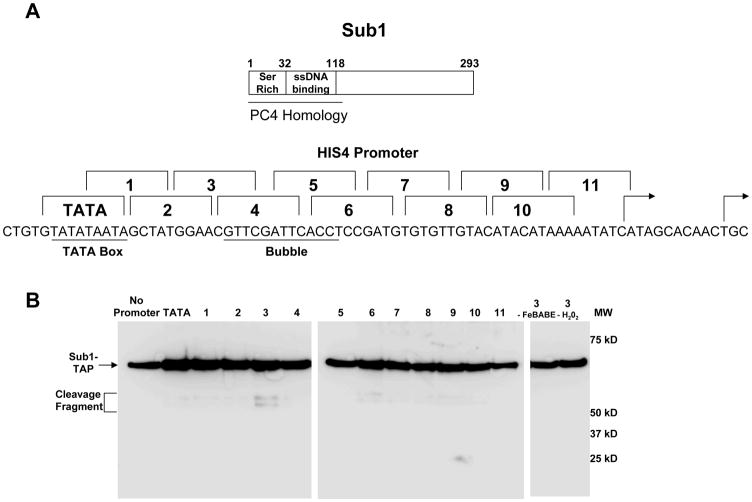
FeBABE cleavage probes tethered to specific sites within the HIS4 promoter DNA can be used to map surfaces of PIC proteins that lie in proximity to DNA (Miller and Hahn, 2006). We analyzed Sub1 using this approach. Twelve probes were created that were identical except that each carried a cluster of FeBABE moieties at different locations on the nontemplate strand, spanning a region from the TATA box to the transcription start site (Figure 4A). Nuclear extract containing TAP-tagged Sub1 was used to form initiation complexes on the bead-bound templates, and the purified PICs were treated with hydrogen peroxide to induce hydroxyl radical formation. Immunoblotting for the C-terminal tag revealed two strong cleavage sites for Sub1 when the FeBABE was downstream of the TATA box in a region overlapping the upstream boundary of the predicted transcription bubble (Figure 4B, probe 3). Some enhanced cleavage is also seen at the downstream edge at probe 6. Based on the size of the Sub1 fragments, the cleavage sites map to the region of the ssDNA binding domain. The fact that there are two probably reflects the fact that Sub1/PC4 is a dimer and its ssDNA binding surfaces will be asymmetrical with respect to the Fe-BABE. A low level of background cleavage was observed with most of the probes, but only a single weaker band is seen. We suspect this is due to non-specific binding of Sub1 to DNA, outside of the context of the PIC. FeBABE in the bubble region also cleaves TFIIE subunits Tfa1 and Tfa2, as well as TFIIB and the core of the RNApII clamp domain (Miller and Hahn, 2006). These factors are important for forming and stabilizing the transcription bubble, so Sub1 is appropriately positioned for a possible role in open-complex formation.
Figure 4. Sub1 localizes near the transcription bubble.
(A) The top panel shows a representation of Sub1 protein domain structure. Below are the positions of the FeBABE labels in each probe (numbered brackets) shown above the HIS4 promoter sequence. The TATA element and the predicted transcription bubble region are indicated. (B) Cleavage fragments of TAP-tagged Sub1 were resolved by gel electrophoresis, blotted, and visualized by probing for the TAP protein A domain at the Sub1 C-terminus. All 12 probes were conjugated to FeBABE and used in PIC formation/hydroxyl radical probing as described in Miller and Hahn, 2006. Negative controls include probe 3 reactions lacking either FeBABE or H2O2.
RPA localizes to transcribed regions of genes
RPA is an abundant complex, so it would be easy to dismiss its presence on the immobilized templates as contamination. However, its enrichment with promoter sequences (Figure 1B–C) led us to consider whether RPA might actually associate with RNApII complexes. RPA occupancy along several representative mRNA genes was assayed by ChIP using an antibody against the Rfa1 subunit (generously provided by Steven Brill, Rutgers Univ.). The Rfa1 localization pattern was strikingly different from, and complementary to, that of Sub1. Rfa1 was most strongly associated with the transcribed regions and generally excluded from promoters and intergenic regions (Figure 5A). As RPA is a component of the replisome, it was important to exclude the possibly that this crosslinking pattern was a result of ongoing replication. We were not able to detect crosslinking of other replication factors, including components of DNA polymerase (II) epsilon and the origin recognition complex, to highly transcribed genes (Figure 5B and S3B). Importantly, the same Rfa1 crosslinking pattern was seen in cells arrested at G1 with alpha-factor (Figure S4B), indicating that Rfa1 occupancy at transcribed loci is not dependent upon ongoing replication. To determine whether Rfa1 localization to downstream regions is dependent on transcription, we compared Rfa1 occupancy along the GAL10 gene when the gene was repressed or highly transcribed. Rfa1 crosslinking was seen under inducing but not repressed conditions (Figure 5C).
Figure 5. RPA associates with active genes.
(A) ChIP was performed using an antibody against the Rfa1 subunit of RPA. Cop-precipitated DNA was analyzed across the PMA1, YEF3, and PYK1 genes as described in Figure 2. The values shown represent the averages and standard errors (bars) from three independent experiments. (B) ChIP was performed using strains expressing TAP-tagged Dpb2 or Pol2, (components of DNA Polymerase (II) ε), or Orc1 (component of the replication origin recognition complex). DNA coprecipitated with the TAP-tagged proteins was analyzed by PCR for regions across the YEF3 gene. (C) Cells were grown in YP-Glucose or YP-Galactose to an OD ~0.5, and Rfa1 ChIP was performed as above except testing for regions across the GAL10 gene. Quantitation of triplicate experiments is as in part A. (D) Strains with the indicated genotypes were spotted in 3-fold dilutions on Synthetic Complete (SC) + 5-Fluoroorotic Acid to remove an RFA1-URA3 covering plasmid. Plates were incubated for 6 days at 30°C. (E) The indicated isogenic strains were spotted in 3-fold dilutions on SC plates lacking uracil (3 days growth shown) or SC-URA plates containing MPA (4 days). (F) ChIP of the Rpb3 subunit of RNApII at the YEF3 gene was performed in strains with wild-type Rfa1 (RFA1) or expressing the rfa1-t124 allele grown in galactose. Quantitation of triplicate experiments is as in part A.
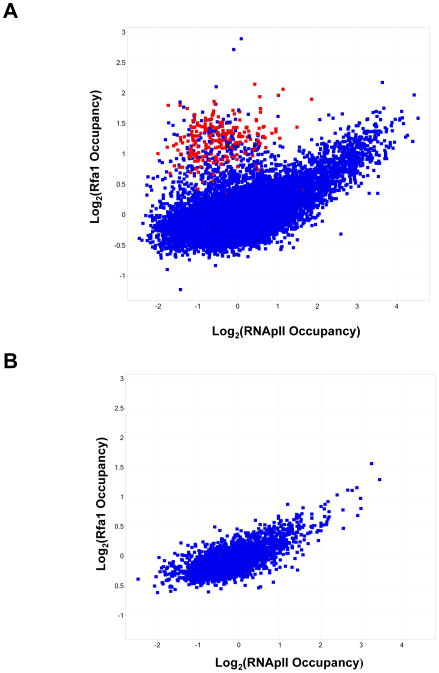
To assess the generality this observation, DNA crosslinked to and immunoprecipitated with Rfa1 was used to probe full genome tiled microarrays. Rfa1 associates with most active mRNAs and snoRNA genes. The occupancy of Rfa1 on individual probes was plotted against that of RNApII subunit Rpb3 (Figure 6). A clear positive correlation was seen, except for a cluster of probes that showed high levels of Rfa1 but not Rpb3. Interestingly, the majority of these were tRNA probes (colored red in Figure 6A), suggesting that Rfa1 also associates with RNA Polymerase III complexes. Notably, Sub1 has also recently been shown to function at RNApIII-transcribed genes (Rosonina et al., 2009; Tavenet et al., 2009), suggesting more general roles for these ssDNA-binding factors in transcription. To evaluate the correlation of RPA specifically with RNApII transcribed genes, only the subset of probes annotated as mid-coding sequences (i.e. removing both promoter and 3′ regions as well as other classes) were plotted (Figure 6B). Again, a clear positive correlation was observed, with a Pearson correlation coefficient of 0.731.
Figure 6. RPA associates with transcribed regions of the genome.
(A) Comparison of Rfa1 and Rpb3 (RNApII) occupancy at coding sequences throughout the genome. ChIP was performed as in Figure 5, except that DNA was labeled and used to probe tiled genome oligonucleotide arrays. Scatter plot represents all probes plotted for the log2 ratio between the immunoprecipitated DNA and a total DNA control. tRNA probes are marked in red. (B) Comparison of Rfa1 and Rpb3 (RNApII) occupancy at probes classified as “mRNA mid-coding” sequences.
Since Rfa1 associates with transcribed region of genes, we tested whether it affects elongation in vivo. The rfa1-t124 and rfa1-t6 alleles, which carry point mutations in the ssDNA binding domain (Umezu et al., 1998), synthetically enhance the slow growth phenotypes of strains lacking the elongation factors Spt4 or Bur2 (Figure 5D and S3C). RFA1 mutant strains were tested on media containing mycophenolic acid (MPA), a drug that reduces NTP pools and thereby slows transcription elongation (Desmoucelles et al., 2002). The rfa1-t124 allele and other ssDNA binding mutants conferred resistance to this drug (Figure 5E and data not shown), while mutants in other regions of the protein did not. MPA resistance has also been observed upon deletion of the histone methyltransferase Set2 or the Rpd3S histone deacetylase complex (Carrozza et al., 2005; Keogh et al., 2005), and RPA mutants genetically interact with components of these elongation factors as well as the RNApII subunit Rpb4 (Collins et al., 2007). RNApII occupancy along the YEF3 gene was tested in an rfa1-t124 strain, showing a small decrease in RNApII levels in transcribed regions (Figure 5F). While much more work will be required for a full mechanistic understanding, out data strongly suggest that the RPA complex can interact with and affect RNApII elongation complexes in vivo. This interaction could be with ssDNA generated by the transcription bubble or possibly R-loops (Huertas and Aguilera, 2003).
Rfa1 binds near mRNA transcription start sites in the absence of Sub1 ssDNA binding function
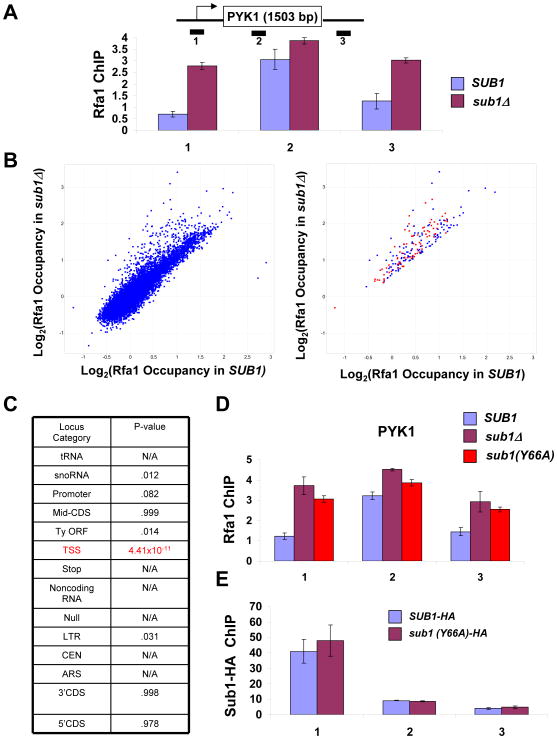
Since Rfa1 and Sub1 have complementary patterns of crosslinking across active genes, yet both bind ssDNA, we asked whether Sub1 and Rfa1 might compete for binding to RNApII complexes. Rfa1 occupancy across active genes was tested by ChIP in a sub1Δ strain. At PYK1, there was a dramatic increase in Rfa1 levels at the promoter region (Figure 7A). Other genes followed the same trend (Figure S4A). At 3′ ends of genes where Sub1 is normally found, there was also an increase of Rfa1 in the sub1Δ strain. This Rfa1 re-localization was independent of replication (Figure S4B).
Figure 7. RPA crosslinks to transcription start sites in the absence of Sub1 activity.
(A) ChIP analysis at the PYK1 gene was performed using antibody against Rfa1 in SUB1 and sub1Δ strains. The values shown represent the averages and standard errors (bars) from three independent experiments. (B) Left panel: Scatter plot of all probes on the genomic arrays, comparing Rfa1 occupancy in SUB1 and sub1Δ cells. Right panel: Probes that are > 1.5 fold increased in sub1Δ cells are shown. Loci annotated as “transcription start sites” (TSS) are marked in red. (C) Coincidence test for increase in Rfa1 occupancy in sub1Δ cells for each locus category. P-value indicates probability a particular locus category is represented in the increased category by chance. N/A refers to categories that had no loci increased in sub1Δ cells. (D) ChIP analysis for Rfa1 at the PYK1 gene was performed using chromatin samples from sub1Δ, or sub1Δ cells carrying plasmid-borne SUB1 or SUB1(Y66A) alleles. Quantitation of triplicate experiments is as in part A. (E) ChIP for Sub1-HA was performed in a strain bearing a tagged version of wild type Sub1 or the Y66A mutant, and PCR analysis of the precipitated DNA was carried out on the PYK1 gene. Quantitation of triplicate experiments is as in part A.
To test this phenomenon at the genome-wide scale, Rfa1 crosslinking was assayed in a sub1Δ strain using genomic arrays for ChIP-chip analysis. A plot of Rfa1 occupancy on individual probes in sub1Δ versus SUB1 cells revealed that most were unchanged (Figure 7B, left panel). However, those that showed increased binding (>1.5 fold) in cells lacking Sub1 were disproportionally annotated as RNApII transcription start sites (TSS) (red spots in Figure 7B, right panel). A large percentage of other probes in this group were annotated as promoters, snoRNAs, or LTRs, all of which would be very close to TSSs. Essentially all of the TSSs that showed increased Rfa1 in the absence of Sub1 are actively transcribed genes, although not all genes with high RNApII occupancy showed increased Rfa1 binding (Figure S4C). Coincidence tests (Tavazoie et al., 1999) were performed to calculate the probability of observing the number of increased probes within a particular category by chance. The increase in the TSS category had a p-value of 4×10−11, while all other categories had values indicating their changes were statistically insignificant (Figure 7C). These results suggest Rfa1 can opportunistically associate with Sub1 binding sites near TSSs when Sub1 is absent. Interestingly, deletion of SUB1 in rfa1-t124 cells resulted in a synthetic growth defect, suggesting some cooperative function (Figure S4D). However, it is very unlikely that RPA can perform all the functions of Sub1, as it presumably cannot interact with the basal factors during initiation (see Discussion).
A single point mutation in a key tyrosine residue abrogates ssDNA binding activity of the mammalian Sub1 homologue, PC4 (Werten et al., 1998). The corresponding mutation was made in Sub1 (Y66A; tyrosine 66 to alanine) and Rfa1 occupancy was assayed by ChIP. Rfa1 was now seen at the promoter, very much like in the sub1Δ strain (Figure 7D). Similar to sub1Δ (Koyama et al., 2008), the Y66A mutation caused increased levels of IMD2 transcripts (Figure S4F), indicating loss of Sub1 function. Moreover, combining sub1(Y66A) with the ssl1-1 allele resulted in a significant growth defect (Figure S4E), although not as strong as with complete deletion of SUB1. Importantly, Sub1 occupancy at the promoter was not significantly affected by the Y66A mutation (Figure 7E), suggesting that Sub1 is recruited to promoters by interaction with other PIC components rather than by ssDNA. These results show that ssDNA binding is necessary for the function but not recruitment of Sub1 in vivo, and that RPA interacts with the PIC only when the Sub1 interaction with ssDNA is lost.
Discussion
Single-stranded DNA binding proteins play key roles in most DNA metabolic processes (Kodadek, 1998). Proteins that bind ssDNA are found at replication bubbles, DNA damage sites, and recombination intermediates, where they protect ssDNA from reannealing and degradation as well as help recruit DNA synthesis and repair enzymes (Fanning et al., 2006; Richard et al., 2009). Transcription by RNA polymerases generates a ssDNA region known as the transcription bubble. Crystal structures show that the template strand enters the active site to base pair with the nascent RNA, but the nontemplate strand could not be modeled, presumably because of flexibility (Kostrewa et al., 2009; Liu et al., 2010). Recent FRET studies of reconstituted RNApII elongation complexes suggest that nontemplate strand bases in the transcription bubble are exposed outside the polymerase (Andrecka et al., 2009), providing a potential binding site for ssDNA-binding proteins.
It has been suggested that TFIIE may interact with the nontemplate strand within the PIC. TFIIE has ssDNA binding activity in vitro, interacts with and activates TFIIH functions (Kuldell and Buratowski, 1997; Watanabe et al., 2003), and is dispensable for in vitro transcription reactions using pre-melted templates (Goodrich and Tjian, 1994; Parvin et al., 1992; Tantin and Carey, 1994). However, this attractive hypothesis still lacks direct evidence and, if true, would not preclude the presence of additional ssDNA binding proteins. Our proteomic analysis of RNApII PICs revealed the presence of two such factors, Sub1 and the RPA complex. Sub1 is already implicated in transcription, but the RPA complex is typically thought of as a DNA replication and repair factor.
The SUB1 gene was originally isolated as a high-copy suppressor of specific mutations in the TFIIB linker region (Knaus et al., 1996) and Sub1/PC4 boosts basal and activated transcription in vitro (Ge and Roeder, 1994; Henry et al., 1996; Kretzschmar et al., 1994). Other studies have suggested roles for Sub1 or its mammalian counterpart PC4 in transcription elongation, termination, reinitiation, and DNA repair (Conesa and Acker, 2010). Our ChIP experiments suggest that Sub1 primarily localizes to 5′ ends of active genes dependent upon functional TBP, although low levels of crosslinking can be observed in some downstream-transcribed regions and 3′ ends (Figure 2). Notably, although Sub1/PC4 stimulates Gal4-VP16 dependent transcription in cell free systems (Ge and Roeder, 1994; Henry et al., 1996; Kretzschmar et al., 1994), our immobilized template experiments show that Sub1 recruitment does not depend on an activator for its recruitment to the PIC. Fe-BABE cleavage experiments (Figure 4) indicate that Sub1 is located near the 5′ edge of the HIS4 transcription bubble region, which is also close to the B-linker helix (Kostrewa et al., 2009; Liu et al., 2010). This finding is consistent with experiments suggesting direct interactions between the two proteins (He et al., 2003; Knaus et al., 1996). Cells lacking Sub1 have decreased RNApII levels throughout transcribed regions of genes (Figure 2D).
Based on all of these findings, we propose the following model (Figure S5). Sub1 is first recruited to the PIC by protein interactions, probably with TFIIB. Upon promoter melting, Sub1 can interact with the non-template strand or perhaps both strands at the upstream junction between single- and double-stranded DNA. This would stabilize the open complex, thereby making it more likely that transcription can initiate and proceed to promoter clearance. Supporting this model, the mammalian homolog of Sub1 stimulates promoter escape in vitro (Fukuda et al., 2004). This mechanism also nicely explains the observed genetic interactions. The specific TFIIB mutants suppressed by Sub1 overexpression assemble into PICs normally, but are defective for in vitro transcription (Cho and Buratowski, 1999; Ranish et al., 1999). They are also cold sensitive for viability (He et al., 2003; Knaus et al., 1996; Wu et al., 1999), consistent with a defect in promoter melting. The residues mutated in these TFIIB alleles are in the B-reader helix and linker region, which is proposed to be involved in capture of the template strand within the RNApII active site (Kostrewa et al., 2009). Saturating Sub1 may increase the duration of transient promoter melting so that initiation can still occur despite reduced TFIIB-template strand interaction. Sub1 is essential for viability in cells containing these TFIIB mutants (He et al., 2003; Knaus et al., 1996; Wu et al., 1999), further suggesting cooperation between these two factors in promoter melting. Similarly, the negative genetic interactions between sub1Δ and mutations in the basal factors TFIIE and TFIIH (Figure 3) are predicted if these three factors all contribute to open complex formation and stabilization. This model of Sub1 function is also consistent with its role in RNApIII transcription, which presumably requires similar capture of the template strand by Brf1, the TFIIB-related subunit of TFIIIB.
PC4 and Sub1 both contain a ssDNA binding domain, but the importance of this module has been unclear. Some in vitro experiments suggest that the PC4 ssDNA binding domain may be dispensable for coactivator function and actually inhibit basal transcription in the absence of TFIIH (Malik et al., 1998; Werten et al., 1998). However, in transfection assays, PC4 lacking the ssDNA binding domain lost coactivator function for TAT-dependent or p53-dependent transcription (Batta and Kundu, 2007; Holloway et al., 2000). Our data also indicate ssDNA binding is important for Sub1 function. One possible resolution is a model where Sub1/PC4 aids in promoter melting and early stages of promoter clearance, but then must be removed from the PIC before the polymerase moves into productive elongation. This may be dependent on TFIIH phosphorylation activity, as TFIIH can phosphorylate Sub1/PC4 to inhibit its interactions with the initiation machinery in vitro (Ge et al., 1994; Henry et al., 1996; Jonker et al., 2006; Kretzschmar et al., 1994).
We were surprised to identify members of the RPA complex in our mass spectrometry screen of proteins enriched in PICs. Rfa1 is an abundant protein that, together with its partners Rfa2 and Rfa3, comprises the major eukaryotic ssDNA binding complex that functions in replication, recombination, and DNA repair. Arguing for a bona fide interaction between RPA and transcription complexes in vivo, genome-wide ChIP analysis shows that Rfa1 crosslinks to the downstream transcribed regions of active RNApII genes as well as genes transcribed by RNApIII (Figure 6). Recent studies reported other replication proteins associated with protein coding genes (Azvolinsky et al., 2009; Shor et al., 2009; Snyder et al., 2009), and some have argued this is a result of replisome pausing at sites of heavy transcription. However, we find RPA associating with transcription units even in G1 arrested cells (Figure S4B), indicating that the interaction with transcribed loci is replication independent.
Given the apparent exclusion of Rfa1 from RNApII promoters in vivo, an obvious question is why enrichment was observed with in vitro assembled PICs. The genome-wide ChIP analysis did show lower levels of Rfa1 binding at some promoter regions. However, it is perhaps more likely that our in vitro PIC preparation contains both closed and open complexes, with Rfa1 interacting with ssDNA within the latter. Although we depleted ATP from our nuclear extracts to enrich for closed complexes, we cannot rule out some residual activation of the TFIIH helicases or some ATP-independent promoter melting. In this regard, it is worth noting that neither RNApI or RNApIII requires a helicase analogous to TFIIH for transcription. In vitro studies of an RNApII/RNA/DNA ternary complex predict the exposed region on the nontemplate strand to be roughly 5 base pairs (Andrecka et al., 2009). RPA can bind single-stranded bubbles in dsDNA as small as four nucleotides (Matsunaga et al., 1996). Extending our model (Figure S5), Sub1 may be positioned within the PIC for capture of ssDNA upon open complex formation, but RPA could quickly bind the non-template DNA strand if an open PIC forms without Sub1 or if Sub1 dissociates. This idea of Sub1 and RPA competing for ssDNA within transcription complexes is supported by the observation that Rfa1 crosslinking is increased at many TSSs in cells lacking Sub1 protein or ssDNA binding activity (Figure 7). We do not believe RPA can substitute for Sub1 function, particularly since ssDNA binding is likely just one function of Sub1 and RPA has never been reported to directly interact with the basal factors.
Does RPA directly function in transcription? There have been occasional reports of gene-specific roles for RPA in regulating transcription through promoter sequences (Tang et al., 1996), and a recent ChIP study placed RPA and multiple nucleotide excision repair enzymes at active mammalian promoters (Le May et al., 2010). Other than RPA, DNA repair factors were not enriched with PICs in our mass spectrometry screen (Table S2). RPA crosslinking primarily to downstream regions hints instead at a role in elongation. More relevant to our model, RPA was shown to recruit a cytidine deaminase to DNA transcribed by T7 RNA polymerase in an in vitro model for immunoglobulin somatic hypermutation and class switch recombination (Chaudhuri et al., 2004). This paper speculated that an RPA-deaminase complex might interact with RNApII transcription bubbles during expression of immunoglobulins. Furthermore, mammalian RPA showed crosslinking to coding regions of two mRNA genes, leading to the suggestion that it can associate with stalled RNApII complexes as part of a co-transcriptional scanning mechanism for DNA damage (Jiang and Sancar, 2006).
Our ChIP and genetic results strongly suggest the possibility of a more general role for RPA in transcription. We observed synthetic genetic interactions between ssDNA binding domain mutants of Rfa1 and two positive elongation factors: Spt4 and Bur2 (Figures 5D and S3C) and Rfa1 physically interacts with the Bur1/Bur2 kinase complex (Clausing et al., 2010). These same RFA1 mutant alleles confer partial resistance to mycophenolic acid (Figure 5E), a phenotype also seen with mutations in factors that are normally inhibitory for RNApII elongation (Carrozza et al., 2005; Keogh et al., 2005). Interestingly, RFA1 and RFA2 hypomorphic alleles show negative genetic interactions with mutants in many of these regulators of elongation, including those in the HIR complex and the Set2/Rpd3S pathway (Collins et al., 2007). These negative elongation factor mutants also typically exhibit internal initiations at cryptic promoters within transcribed sequences, presumably due to improper chromatin reassembly after transcription, but we did not see this phenotype in rfa1 mutants (data not shown). Interestingly, recent structural studies suggest the Spt4/5 elongation factor is positioned near the nontemplate strand of the transcription bubble during elongation (Klein et al., 2011; Martinez-Rucobo et al., 2011). It will be of interest to see whether the genetic interactions we see between this complex and RPA might reflect either cooperative or competitive binding to the non-template strand.
Another interesting possibility is that RPA association with transcription elongation suppresses transcription-coupled recombination. Transcription can promote recombination, probably by R-loop formation between the transcript and template strand behind the elongation complex, i.e. distinct from the transcription bubble (Huertas and Aguilera, 2003). The unpaired non-template strand is presumably the recombinogenic agent, and RPA binding might suppress strand invasion (Figure S5). Finally, plant and mammalian genomes contain multiple paralogs of the RPA subunits (Sakaguchi et al., 2009) and it will be important to determine if higher eukaryotes contain multiple RPA-like complexes with specific functions in transcription, repair, or replication.
EXPERIMENTAL PROCEDURES
Yeast strains and extracts
S. cerevisiae strains used in this study are listed in Table S3. Yeast nuclear extracts were prepared as described (Johnson et al., 2009; Ranish et al., 1999). Spotting analyses for resistance to MPA (50 μg/ml) were performed as previously described (Kim and Buratowski, 2009).
Recombinant Proteins
Recombinant Gal4-VP16 (Cho et al., 1997) and TBP (Buratowski and Zhou, 1993) were expressed and purified as previously described.
Immobilized Promoter Templates
Immobilized promoter templates were prepared as described (Kim et al., 2007; Ranish et al., 1999). Briefly, a 600-bp fragment containing upstream vector sequence, one Gal4 binding site, 71 base pairs of the yeast HIS4 promoter centered around the TATA box, and 100 nucleotides downstream of the transcription start site was PCR amplified from pSH515 with primers p965 and pNot (Ranish 1999, 2003). The “promoterless” template was created by PCR amplifying from the same template a similar fragment that ends 14 bp downstream of the Gal4 site. Binding assays were performed as described (Ranish 1999, 2003). See Supplemental Information for details of PIC purification and mass spectrometry analysis.
FeBABE Catalyzed Hydroxyl Radical Cleavage Assay
Reactions were preformed essentially as described (Miller and Hahn, 2006). A detailed description appears in the Supplemental Information.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as previously described (Keogh and Buratowski, 2004; Kim and Buratowski, 2009). All yeast strains were grown to OD600 ~0.5 in YPD or YP-Galactose as indicated. For G1-arrest experiments, bar1Δ strains were grown to OD600 ~0.25, then incubated with alpha factor at a final concentration of 600 nM for 2 hours before crosslinking. Primers for ADH1, PYK1, PMA1, YEF3, and GAL10 (Houseley et al., 2008; Kim and Buratowski, 2009) and snR13 (Kim et al., 2006) are listed in Table S4. Twenty-five PCR cycles were performed for anti-Rpb3, anti-TBP, anti-Rfa1 (from Steven Brill) and IgG ChIPs.
DNA Amplification and Microarray Hybridization
ChIP was performed as above with Rfa1 antibody for BY4741 and the isogenic strain deleted for Sub1. Input and IP samples were amplified and hybridized to Agilent whole-genome arrays as previously described (Kim et al., 2010). Briefly, DNA was amplified by Round A-Round B PCR amplification, and ChIP material and input DNA were Klenow-labeled using Cy3 and Cy5-dCTP, respectively. Microarray hybridization and scanning were carried out according to manufacturer’s instructions. For downstream analysis, genomic loci were annotated with respect to coding regions and other genomic elements (tRNAs, etc.) as previously described (Kim et al., 2010; Liu et al., 2005). Genomewide Rpb3 data were obtained from Kim et al., 2010. Coincidence testing was performed with SpotFire Decision Site software using algorithms described in (Tavazoie et al., 1999).
Supplementary Material
Highlight.
A quantitative proteomics screen finds Sub1 and RPA in transcription complexes
Sub1 localizes near the transcription bubble for open complex formation
RPA associates with transcribed regions of the genome
Mutations in Sub1 ssDNA binding result in relocalization of RPA to promoters
Acknowledgments
We thank Olga Calvo, Steve Hahn, Richard Kolodner, Michael Hampsey, and Claire Moore for strains and plasmids, Steven Brill for the Rfa1 antibody, Manor Askenazi and Jignesh Parikh for help with mass spectrometry and microarray data analysis, and Minkyu Kim, Sebastian Marquardt, and Nihal Terzi for technical assistance. We also thank Steve Hahn, Michael Meisterernst, Olga Calvo, Alicia Martinez, Doug Koshland, and members of the Buratowski and Marto labs for helpful discussions. This work was supported by NIH grant GM46498 to SB, a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and NIGMS grant GM079205 to OJR, DFCI/Blaise Proteomics Center support to JM, and a NDSEG pre-doctoral fellowship to TS.
Footnotes
Accession numbers. The microarray data for the RPA ChIP experiments can be downloaded from the GEO database, series accession number GSE32416.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, Cramer P, Michaelis J. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic acids research. 2009;37:5803–5809. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes & development. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Molecular cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Kundu TK. Activation of p53 function by human transcriptional coactivator PC4: role of protein-protein interaction, DNA bending, and posttranslational modifications. Molecular and cellular biology. 2007;27:7603–7614. doi: 10.1128/MCB.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. The EMBO journal. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. The Journal of biological chemistry. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & development. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausing E, Mayer A, Chanarat S, Muller B, Germann SM, Cramer P, Lisby M, Strasser K. The transcription elongation factor Bur1-Bur2 interacts with replication protein A and maintains genome stability during replication stress. The Journal of biological chemistry. 2010;285:41665–41674. doi: 10.1074/jbc.M110.193292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Conesa C, Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA biology. 2010;7:4. doi: 10.4161/rna.7.3.11491. [DOI] [PubMed] [Google Scholar]
- Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. The Journal of biological chemistry. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic acids research. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Nakadai T, Shimada M, Tsukui T, Matsumoto M, Nogi Y, Meisterernst M, Hisatake K. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Molecular and cellular biology. 2004;24:6525–6535. doi: 10.1128/MCB.24.14.6525-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A, Rosonina E, Manley JL, Calvo O. Sub1 globally regulates RNA polymerase II C-terminal domain phosphorylation. Molecular and cellular biology. 2010;30:5180–5193. doi: 10.1128/MCB.00819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- Ge H, Zhao Y, Chait BT, Roeder RG. Phosphorylation negatively regulates the function of coactivator PC4. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12691–12695. doi: 10.1073/pnas.91.26.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes & development. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry NL, Bushnell DA, Kornberg RD. A yeast transcriptional stimulatory protein similar to human PC4. The Journal of biological chemistry. 1996;271:21842–21847. doi: 10.1074/jbc.271.36.21842. [DOI] [PubMed] [Google Scholar]
- Holloway AF, Occhiodoro F, Mittler G, Meisterernst M, Shannon MF. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. The Journal of biological chemistry. 2000;275:21668–21677. doi: 10.1074/jbc.M909058199. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Molecular cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Molecular and cellular biology. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Molecular cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker HR, Wechselberger RW, Pinkse M, Kaptein R, Folkers GE. Gradual phosphorylation regulates PC4 coactivator function. The FEBS journal. 2006;273:1430–1444. doi: 10.1111/j.1742-4658.2006.05165.x. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Buratowski S. Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods in molecular biology (Clifton, NJ. 2004;257:1–16. doi: 10.1385/1-59259-750-5:001. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Molecular cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando OJ. RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome biology. 2010;11:R75. doi: 10.1186/gb-2010-11-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. The EMBO journal. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- Kodadek T. Mechanistic parallels between DNA replication, recombination and transcription. Trends in biochemical sciences. 1998;23:79–83. doi: 10.1016/s0968-0004(97)01165-1. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- Koyama H, Sumiya E, Nagata M, Ito T, Sekimizu K. Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells. 2008;13:1113–1126. doi: 10.1111/j.1365-2443.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuldell NH, Buratowski S. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Molecular and cellular biology. 1997;17:5288–5298. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Molecular cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS biology. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science (New York, NY. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Guermah M, Roeder RG. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. The EMBO journal. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui P, DePaulo J, Buratowski S. An interaction between the Tfb1 and Ssl1 subunits of yeast TFIIH correlates with DNA repair activity. Nucleic acids research. 1995;23:767–772. doi: 10.1093/nar/23.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Park CH, Bessho T, Mu D, Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. The Journal of biological chemistry. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nature structural & molecular biology. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome research. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse H, Buratowski RM, Silver PA, Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. The Journal of biological chemistry. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Timmers HT, Sharp PA. Promoter specificity of basal transcription factors. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. The study of macromolecular complexes by quantitative proteomics. Nature genetics. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes & development. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Critical reviews in biochemistry and molecular biology. 2009;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Willis IM, Manley JL. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Molecular and cellular biology. 2009;29:2308–2321. doi: 10.1128/MCB.01841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Ishibashi T, Uchiyama Y, Iwabata K. The multi-replication protein A (RPA) system--a new perspective. The FEBS journal. 2009;276:943–963. doi: 10.1111/j.1742-4658.2008.06841.x. [DOI] [PubMed] [Google Scholar]
- Shor E, Warren CL, Tietjen J, Hou Z, Muller U, Alborelli I, Gohard FH, Yemm AI, Borisov L, Broach JR, et al. The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication. PLoS genetics. 2009;5:e1000755. doi: 10.1371/journal.pgen.1000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Current opinion in cell biology. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Huang XY, Zhang JJ. The minichromosome maintenance proteins 2–7 (MCM2-7) are necessary for RNA polymerase II (Pol II)-mediated transcription. The Journal of biological chemistry. 2009;284:13466–13472. doi: 10.1074/jbc.M809471200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CM, Tomkinson AE, Lane WS, Wold MS, Seto E. Replication protein A is a component of a complex that binds the human metallothionein IIA gene transcription start site. The Journal of biological chemistry. 1996;271:21637–21644. doi: 10.1074/jbc.271.35.21637. [DOI] [PubMed] [Google Scholar]
- Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. The Journal of biological chemistry. 1994;269:17397–17400. [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nature genetics. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Tavenet A, Suleau A, Dubreuil G, Ferrari R, Ducrot C, Michaut M, Aude JC, Dieci G, Lefebvre O, Conesa C, et al. Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14265–14270. doi: 10.1073/pnas.0900162106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Critical reviews in biochemistry and molecular biology. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hayashi K, Tanaka A, Furumoto T, Hanaoka F, Ohkuma Y. The carboxy terminus of the small subunit of TFIIE regulates the transition from transcription initiation to elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:2914–2926. doi: 10.1128/MCB.23.8.2914-2926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werten S, Stelzer G, Goppelt A, Langen FM, Gros P, Timmers HT, Van der Vliet PC, Meisterernst M. Interaction of PC4 with melted DNA inhibits transcription. The EMBO journal. 1998;17:5103–5111. doi: 10.1093/emboj/17.17.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Pinto I, Chen BS, Hampsey M. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Molecular and cellular biology. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Miller SP, Pabich EK, Donahue TF. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes & development. 1992;6:2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]
- Zhou F, Cardoza JD, Ficarro SB, Adelmant GO, Lazaro JB, Marto JA. Online nanoflow RP-RP-MS reveals dynamics of multicomponent Ku complex in response to DNA damage. J Proteome Res. 2010;9:6242–6255. doi: 10.1021/pr1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.