Summary
Transcriptional pausing by RNA polymerase (RNAP) plays an essential role in gene regulation. Pausing is modified by various elongation factors, including prokaryotic NusA, but the mechanisms underlying pausing and NusA function remain unclear. Alternative models for pausing invoke blockade events that precede translocation (on-pathway), enzyme backtracking (off-pathway), or isomerization to a non-backtracked, elemental pause state (off-pathway). We employed an optical-trapping assay to probe the motions of individual RNAP molecules transcribing a DNA template carrying tandem repeats encoding the his pause, subjecting these enzymes to controlled forces. NusA significantly decreased the pause-free elongation rate of RNAP while increasing the probability of entry into short- and long-lifetime pauses, in a manner equivalent to exerting a ~19 pN force opposing transcription. The effects of force and NusA on pause probabilities and lifetimes support a reaction scheme where non-backtracked, elemental pauses branch off the elongation pathway from the pre-translocated state of RNAP.
Introduction
Transcription elongation by RNA polymerase (RNAP) is a highly regulated cellular process that plays an essential role in gene expression. During transcription, RNAP moves discontinuously, pausing in response to signals encoded intrinsically in the DNA template or generated by the nascent RNA transcript (Artsimovitch and Landick, 2000; Uptain et al., 1997). Transcriptional pausing is critical to gene regulation in several ways. Pausing mediates overall RNA synthesis rates and plays an important role in synchronizing transcription and translation (Landick et al., 1985; Richardson and Greenblatt, 1996). Pausing also facilitates proper co-transcriptional folding of RNA and serves as a precursor to termination or arrest (Kassavetis and Chamberlin, 1981; Komissarova and Kashlev, 1997a; Pan and Sosnick, 2006). Finally, paused elongation complexes (ECs) provide a target for other regulation by transcription factors that can modulate pause duration and subsequent pause susceptibility (Roberts et al., 2008; Uptain et al., 1997).
In the eubacteria and archaea, NusA is a universally conserved transcription factor that is essential for cell viability (Ingham et al., 1999). In E. coli, NusA forms a 55 kDa monomer comprised of five domains: the N-terminal RNAP-binding domain, three RNA-binding domains (S1, KH1 and KH2), and a C-terminal autoinhibitory domain (Mah et al., 2000; Worbs et al., 2001). Although first identified over three decades ago, NusA remains incompletely understood, in part because of its pleiotropic effects on cellular and viral processes (Friedman and Baron, 1974). For example, NusA not only increases the efficiency of intrinsic termination, but also affects Rho-dependent termination and mediates antitermination by λN, λQ and ribosomal RNA (Greenblatt et al., 1981; Schmidt and Chamberlin, 1987; Vogel and Jensen, 1997). NusA also moderates the elongation rate of RNAP, which has been suggested as a mechanism for modulating termination efficiency in vivo (Artsimovitch and Landick, 2000; Bar-Nahum et al., 2005; Roberts et al., 2008), and enhances certain pauses more than others (Artsimovitch and Landick, 2000; Burns et al., 1998; Theissen et al., 1990). However, the kinetic details and molecular mechanisms underlying these effects are poorly understood.
An understanding of the mechanism by which NusA regulates transcriptional elongation (including both pausing and active elongation) will inevitably depend upon our understanding of the mechanism of elongation itself. Although there is no general consensus on the details or the relative order of some of the transitions, elongation is generally thought to occur in a cycle of reaction steps, consisting minimally of elongation complex (EC) translocation from the pre-translocated to the post-translocated register, substrate NTP binding, nucleotidyl transfer, and pyrophosphate release.
The mechanisms responsible for of pausing are also under debate. One view is that all pauses arise from backtracking (the backtracking-only model; Mejia et al., 2008). Another is that at least some pauses reflect on-pathway blocks to translocation that affect all ECs, but do not necessarily backtrack (Bai et al, 2004; Bai et al., 2007). Our prior results favor a third view, where pauses arise initially from the formation of a short-lived, elemental pause state that branches from the elongation pathway without backtracking, and that can subsequently be stabilized into longer-lived pauses that can (in some cases) backtrack (Ederth et al., 2006; Herbert et al., 2006; Toulokhonov et al., 2007). Three classes of stabilized pauses have been characterized in ensemble-based biochemical experiments (Artsimovitch and Landick, 2000; Kireeva and Kashlev, 2009). One class of such pauses, observed to date only in prokaryotes, involves the formation of a hairpin structure in the nascent RNA (Landick, 2006). The hairpin must be energetically stable, properly spaced from the enzyme active site to interact with RNA exit channel of RNAP, and not disrupt the RNA:DNA hybrid (Toulokhonov et al., 2001; Toulokhonov and Landick, 2003). Hairpin-stabilized pauses have been found, for example, in the leader regions of biosynthetic operons in bacteria, where they synchronize RNAP and the ribosome during transcriptional attenuation (Henkin and Yanofsky, 2002). One example is the his pause, found near the beginning of the histidine biosynthetic operon in E. coli. NusA has been shown to enhance such hairpin-stabilized pauses (Chan and Landick, 1989; Farnham and Platt, 1981), and the magnitude of this effect depends on the structure of the hairpin (Toulokhonov et al., 2001). A second established mechanism of stabilized pauses, backtracking, is found in both prokaryotes and eukaryotes, and involves RNAP moving backwards along the nucleic acid scaffold, disrupting the alignment of the 3′-end of the nascent RNA with the active site of the enzyme. RNAP backtracking can be promoted, for example, when enzyme in its pre-translocated state encounters an energetically weak RNA:DNA hybrid (Komissarova and Kashlev, 1997a, b; Palangat and Landick, 2001). A third class of pauses occurs without stabilization by either hairpins or backtracking, and appears to result from an active-site rearrangement (Kireeva and Kashlev, 2009).
Because they can directly probe the transient, unsynchronized behavior of individual reaction cycles, single-molecule experiments have provided additional insights into transcriptional pausing that are otherwise inaccessible to ensemble experiments (Abbondanzieri et al., 2005a; Adelman et al., 2002; Herbert et al., 2008; Larson et al., 2011). Two types of pause have been characterized by single-molecule approaches, distinguished operationally on the basis of their lifetimes. One type consists of comparatively rare, long-lived pauses (lasting >20 s-30 min). Long pauses can be enhanced by the misincorporation of non-complementary NTPs, and at least some portion of these events are known to result from enzyme backtracking (Shaevitz et al., 2003). The other type consists of comparatively frequent, sequence-dependent, short-lived pauses (~3 s avg. duration), constituting ~95% of all pausing events observed in single-molecule experiments (Dalal et al., 2006; Herbert et al., 2006; Neuman et al., 2003). The mechanism responsible for these so-called ‘ubiquitous’ pauses remains unclear. Evidence suggests that the ubiquitous pauses correspond to elemental pauses; i.e., they reflect an off-pathway state that forms without backtracking, but can subsequently lead to pauses of a longer lifetime via stabilizing mechanisms, such as backtracking or hairpin-based interactions (Toulokhonov et al., 2007). Although NusA is known to decrease the overall elongation rate, it is not established whether NusA achieves this reduction chiefly by lowering the nucleotide incorporation rate of the active elongation complex, by changing the rates into, or out of, any paused states (elemental pauses or long-lived pauses), or through some combination of these effects. Probing such questions using ensemble transcription assays is difficult. First, determinations of pause-free elongation rates in gel-based assays are problematic because all known transcription templates encode pauses. Second, the kinetic characteristics of the pause sites are challenging to characterize, since the occupancy of a given pause site depends in a complicated manner on the pause efficiency, the pause lifetime, and the fraction of complexes that actually reach the pause site. Finally, entry into the paused state is in kinetic competition with elongation at the pause site, which affects both the pause efficiency and the pause lifetime (Herbert et al., 2006). The only practical way to measure independently the rates for active elongation, pause entry, and pause escape is to observe the kinetics of individual molecules.
To investigate the mechanism of pausing and determine how NusA modulates active elongation, we employed an optical trapping assay in which the motion of single molecules of RNAP can be monitored. High spatiotemporal resolution allows us to detect pauses as short as 1 s and spaced by as little as ~2 base pairs (bp). The kinetic rates for pause entry, average pause-free elongation rates, and pause lifetimes can all be extracted from the individual records. The application of controlled forces on elongating complexes can be used to tilt the energy landscape, modulating the reaction rates involving translocation and thereby providing quantitative information about how NusA affects the kinetics of elongation and sequence-dependent pausing. Using these methods to study RNAP transcribing a template engineered to carry repeated motifs, which allows single-molecule records to be aligned with the underlying sequence with near base-pair precision (Herbert et al., 2006), allowed us to measure the effect of NusA on translocation by RNAP and to gain insight into the fundamental mechanism of transcriptional pausing.
Results
Pause-free elongation rate can be modulated by both NusA and applied force
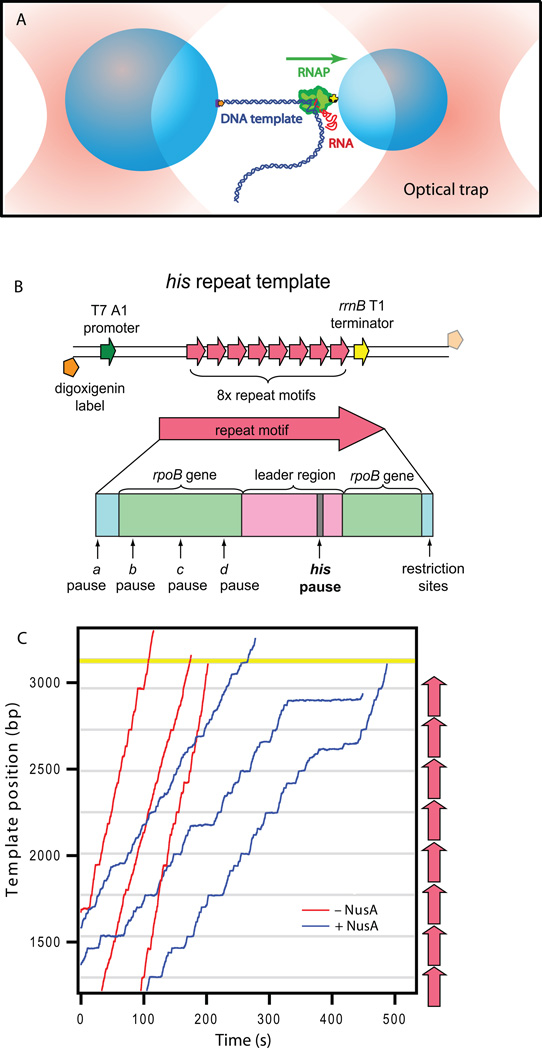
Transcription by single RNAP molecules was recorded using a dumbbell optical trapping assay, in which a transcriptionally stalled elongation complex is tethered between two polystyrene beads, each held in a separate optical trap (Figure 1A) (Shaevitz et al., 2003). The polymerase molecule is attached via a biotin linkage to the surface of one bead, and either the upstream or the downstream end of the template DNA is attached to the opposite bead, allowing application to the elongating enzyme of an assisting or hindering force, respectively. Immediately prior to recording, stalled complexes were restarted by introducing ribonucleoside triphosphates at physiological concentrations (1mM each NTP). A DNA template engineered to contain eight direct repeats of a sequence motif carrying the well-characterized his pause and several adjacent “ubiquitous” pauses (Figure 1B) (Herbert et al., 2006) was used to explore the effects of NusA. Aligned, single-molecule records illustrate the motion of RNAP along the repetitive template (Figure 1C), and clearly reveal pausing at the locations of the his and other sequence-dependent pause sites, as well as a significant reduction in overall elongation rates in the presence of saturating levels of NusA (0.5 µM), mainly as a consequence of increased pausing.
Figure 1. Single-molecule Transcription Assay.
(A) Experimental geometry for the DNA-pulling dumbbell assay (not to scale). Two polystyrene beads (light blue) are held in twin optical traps (pink) above the surface of a coverglass. RNAP (green) is attached to the smaller of the two beads via a biotin-avidin linkage (yellow, black), and the DNA template (dark blue) is attached to the larger bead via a digoxigenin-antidigoxigenin linkage (purple, orange). The RNA transcript (red) emerges co-transcriptionally. Transcription proceeds in the direction shown (green arrow) such that applied tension assists translocation. By attaching the digoxigenin label to the 3′-downstream end of the DNA instead, the direction of the applied load may be reversed in assays.
(B) Engineered transcription templates carrying the his pause element, as previously described (Herbert et al., 2006). Transcription templates consist of 8 repeat motifs (red) located ~1100 bp beyond a T7 A1 promoter (dark green), from which transcription was initiated, and ~80 bp in front of the rrnB T1 terminator (yellow). Templates were labeled with digoxigenin on the transcriptionally upstream end (for assisting loads; solid orange) or the transcriptionally downstream end (for hindering loads; transparent orange). Single, 239-bp repeat motifs (red arrow) consist of leader sequence (pink) with the his pause sequence (gray), along with rpoB gene-derived sequences (light green) showing positions of the b–d pause sites, and flanking DNA corresponding to restriction sites used for cloning (blue) containing the a pause site.
(C) Six representative records of RNAP elongation on the his repetitive template in the presence of 0.5 µM NusA (blue) (N = 34) or absence of NusA (red) (N = 45) under 7.5 pN assisting load at 1 mM NTPs. The gray lines indicate the positions of his pause sequences, and the yellow bar indicates the rrnB T1 terminator sequence. Locations of the 8 repeat motifs are indicated (red arrows, right side).
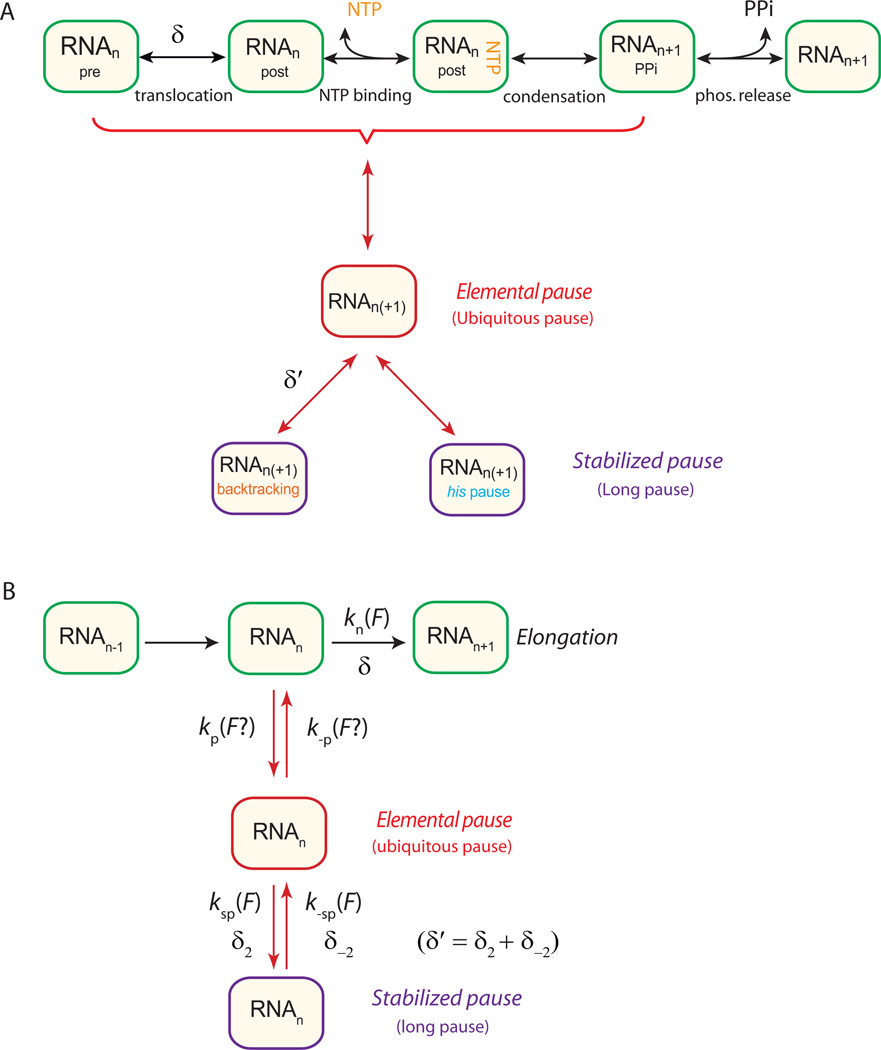
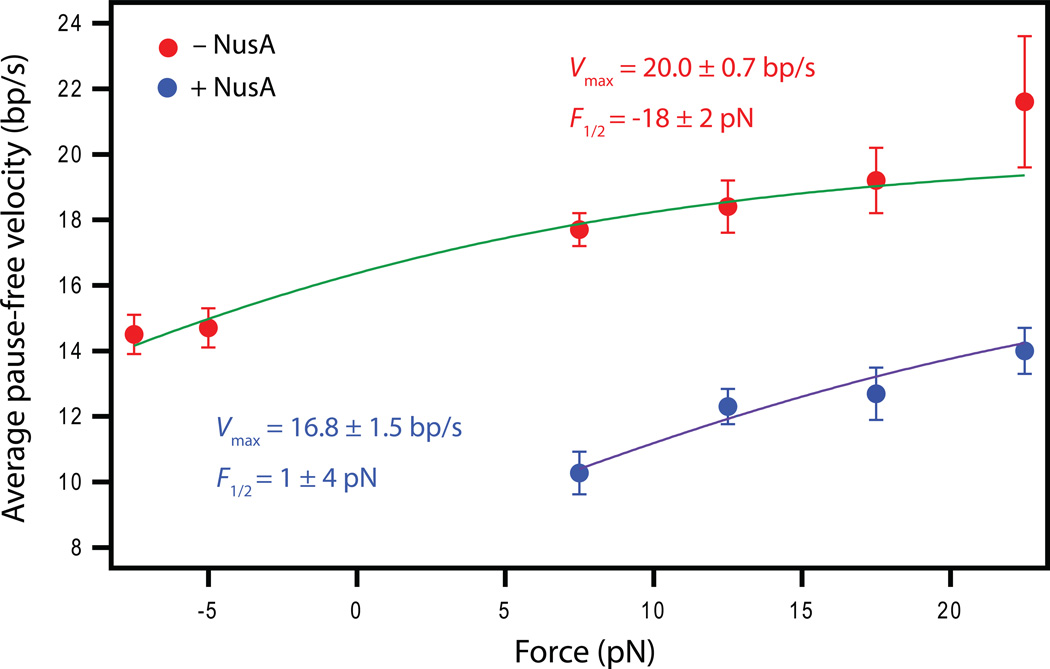
A generalized kinetic scheme for RNAP transcription (Figure 2A), based on previous work (Abbondanzieri et al., 2005b; Bar-Nahum et al., 2005), consists of a main pathway for translocation embodying a Brownian ratchet-type mechanism, a condensation step for nucleotide incorporation into the RNA, and a transition leading to various off-pathway, paused states. For our purposes, this cycle may be further reduced to the simplified reaction scheme shown in Figure 2B. We first studied the active (pause-free) elongation rate, V(F), as a function of the applied force in the presence or absence of NusA. The pause-free elongation velocity is proportional to kn (F), since V(F) = kn (F)·d, where d is the RNAP step size, corresponding to the axial distance subtended by a single base pair (0.34 nm). We found that the pause-free elongation rate was reduced by nearly a factor of two upon the addition of NusA under otherwise identical loads (Figure 3). The experimental force-velocity relationship was well described by a two-state Boltzmann relation (Wang et al., 1998):
| (1) |
where the distance parameter, δ, was fixed at 0.34 nm (1 bp), based on the Brownian ratchet model for translocation (Abbondanzieri et al., 2005a). Vmax, the peak velocity, is reached under large assisting loads, where the translocation step is no longer rate-limiting, and F½ is the force where velocity is half-maximal (additional discussion in Supplemental Material, Figure S1). A two-parameter fit of the pause-free velocity data in the absence of NusA gave Vmax = 20.0 ± 0.7 bp/s and F1/2 = −18 ± 2 pN (Figure 3). In the presence of saturating levels of NusA (0.5 µM), Vmax (16.8 ± 1.5 bp/s) decreased only modestly, but F½ (1 ± 4 pN) increased considerably. The dramatic change in F½ suggests that the primary effect of NusA on elongation is functionally equivalent to applying a load in the hindering direction of FNusA = 19 ± 6 pN, which raises the effective energy barrier against forward translocation, ΔEbarrier, by an amount FNusA ·δ ≈ 1.5 ± 0.5 kBT.
Figure 2. Kinetic Schemes for Transcriptional Elongation and Pausing.
(A) Generalized kinetic scheme for elongation and pausing, with the main RNA synthesis pathway displayed along the top (green boxes). In the Brownian ratchet model, RNAP fluctuates between pre- and post-translocated states prior to the reversible binding of NTP, followed by a less reversible condensation reaction and pyrophosphate release, which rectify forward motion. Alternatively, NTP binding may precede translocation in the presence of a secondary binding site for NTP (Abbondanzieri et al., 2005a). The displacement associated with translocation is δ. The elemental pause state (middle row, red box) branches off the elongation pathway from an intermediate state (here, the pre-translocated state) and may be additionally stabilized to generate longer-lived pause states (bottom row, purple boxes) through backtracking (displacement δ′) or hairpin-interaction mechanisms. (See also Figure S3.)
(B) A simplified kinetic scheme generated from (A), showing a single load-dependent rate on the main elongation pathway with associated distance parameter δ, and load-dependent rates for entry and exit from stabilized pauses, along with associated distance parameters, δ2 and δ−2. (The possible load dependence of transitions in and out of the elemental pause state is considered in the main text).
Figure 3. Single-molecule force-velocity relationships.
Data (mean ± SEM) for the pause-free elongation velocity as a function of applied load in the presence (blue) or absence (red) of 0.5 µM NusA at 1 mM NTPs, along with fits (solid lines) to Boltzmann relations (see text). The distance parameter was fixed at 1 bp. Positive forces correspond to assisting loads. (See also Figure S1.)
NusA raises the probability of entering ubiquitous pauses
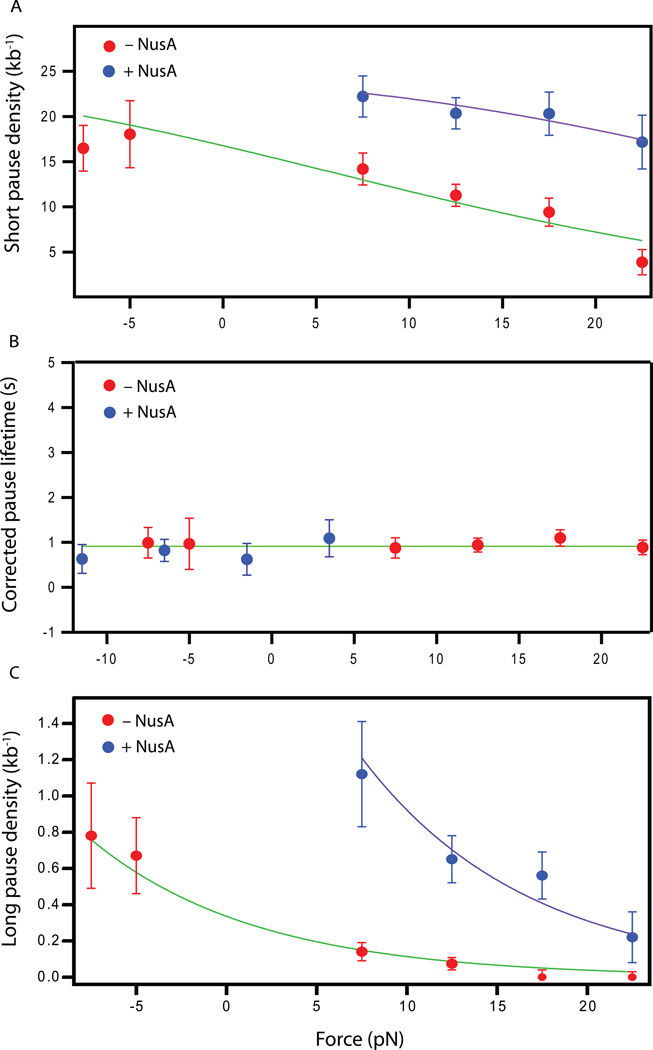
We next examined the effect of NusA and external load on short-lifetime (ubiquitous) pauses (<20 s). Pauses lasting longer than 1 s could be reliably detected, and the distributions of pause lifetimes at each force, in either the presence or absence of NusA, were well fit by a sum of two exponentials (Figure S2A–D). The numbers of pauses were corrected for missing events by adding the number actually scored to the estimated number of undetected pauses (between 0–1 s), obtained by extrapolating each double-exponential distribution to 0 s. We found that the pause density of short-lifetime pauses (defined as the number of short pauses per kilobase DNA traversed) was significantly increased upon the addition of NusA, consistent with the effect of a hindering force (Figure 4A, Figure S2A–D).
Figure 4. Pausing statistics as a function of force.
(A) Pause density as a function of applied load in the presence (blue) or absence (red) of 0.5 µM NusA at 1 mM NTPs, along with global fits to Equation (2).
(B) Corrected (intrinsic) lifetimes (mean ± SEM) for ubiquitous pauses in the presence (blue) and absence (red) of NusA, along with the fit (green line) to a constant lifetime, τ = 0.9 ± 0.1 s.
(C) Long-lifetime pause density vs. applied force in the presence (blue) or absence (red) of NusA, fit to single exponentials (green, purple lines). The distance parameter was globally fit, yielding (δ1 + δ2) = 1.3 ± 0.2 bp. (See also Figure S2.)
Because pausing competes directly with active elongation (Figure 2), the probability of a pause at a given template position (the pause efficiency) is supplied by the branching ratio kp/(kp +kn), where kp is the rate of entry into the pause state and kn is the elongation rate (Figure 2B). The average efficiency across the entire template is proportional to the pause density, ρ, measured by these experiments. However, the vast majority of pauses occurs at only a small fraction of sequence locations, and pauses at the remaining sites are comparatively rare (Herbert et al., 2006). If we assume that pausing at all sites is affected by loads in a similar manner, we can approximate the overall pause density as
| (2) |
where fp is the fraction of sites where pauses can occur (fp would approach 1 bp−1 if all sites on the template generated pauses). The force dependence of ρ was fit by a Boltzmann relation (Figure 4A):
| (3) |
In this particular expression, δ1 represents the effective distance parameter associated with two potentially force-dependent rates, kn and kp. Two distinct candidate pausing mechanisms can lead to this same functional form. The first mechanism is a backtracking model (Mejia et al., 2008), where RNAP moves rearward along the DNA to enter the pause state. Here, kp = kp0 exp(−F δ1/kBT), where kp0 is the pause rate at F = 0, and δ1 is associated with the transition for entry into the ubiquitous pause. In this case, the value of kn at pause sites is only weakly force-dependent and is therefore neglected, which implies that ubiquitous pauses branch off the elongation pathway from the post-translocated state (Discussed in Supplemental Materials). In the alternative mechanism, (most) sequence-dependent ubiquitous pauses do not backtrack (Herbert et al., 2006; Toulokhonov et al., 2007), but instead involve an isomerization of the elongation complex to an elemental pause state, for example, a conformational change in RNAP, fraying of the 3′-end of the RNA-DNA hybrid, or both. In such cases, kp does not involve translocation of the enzyme and is therefore force-independent. Here, ubiquitous pauses branch off the elongation pathway from the pre-translocated state. The rate that directly competes with pausing at any sequence-dependent pause site will have the following force dependence:
| (4) |
where kn0 is the elongation rate at F = 0, and δ1 is the distance parameter associated with the translocation step. Two similar non-backtracking kinetic scenarios that lead to Eq. (4) are discussed in Supplemental Materials (Figure S3). For both candidate mechanisms, A corresponds to kn / kp at F = 0.
A fit of the pause density as a function of force (Eq. 3) to data acquired in the absence of NusA gave fp = 25 ± 7 kb−1 and δ1 = 1.0 ± 0.5 bp, and a global fit to data obtained in the presence and absence of NusA yielded similar values, fp = 25 ± 5 kb−1 and δ1 = 1.0 ± 0.3 bp (Figure 4A). Taking fp to be 25 kb−1 and fixing δ1 at 1 bp, the ratio of values for the fit parameter, A, obtained in the presence and absence of NusA can again be attributed to an equivalent force acting on the polymerase, equal to FNusA = 23 ± 5 pN, corresponding to raising the energy barrier against translocation, ΔEbarrier, by 1.9±0.4 kBT.
The intrinsic lifetime of ubiquitous pauses is unaffected by load, NusA
To distinguish between the alternative candidate mechanisms for ubiquitous pausing, backtracking vs. non-backtracking, we studied the force-dependence of the ubiquitous pause lifetime, which corresponds to 1 / k−p in the kinetic scheme of Figure 2B. We found that the mean lifetime of these pauses was force-dependent and increased upon the addition of NusA (Figure S2A–D). The apparent (measured) lifetime, τ*, is related to the intrinsic (actual) lifetime, τ, through τ* = τ/(1 − ε), where ε is the efficiency of a given pause (Herbert et al., 2006). Here, we estimated the efficiency as ε = ρ/fp. After correcting the apparent lifetimes based on their respective efficiencies, all intrinsic lifetimes were found to be nearly the same, and equal to ~0.9 s, across all forces applied and in either the presence or absence of NusA (Figure 4B). These intrinsic lifetimes are in close agreement with the lifetimes previously reported for the same set of identified pauses (1.1 ± 0.4 s), despite the 4-fold higher GTP levels (1 mM GTP vs. 250 µM) and slightly higher temperature (27°C vs. 21.5°C) used in this study. These findings lend strong support to the notion that ubiquitous pauses represent an elemental state, and that the rate of escape from this state to resume elongation, set by k−p, does not require forward translocation. Therefore, these data disfavor backtracking as an explanation for ubiquitous pausing. The force-dependence of the probability of ubiquitous pauses, and of their apparent lifetimes, is instead attributable to NusA increasing the energy barrier to forward translocation on the main pathway for elongation, which directly affects kn, but not kp or k−p (Figure 2).
NusA and applied force affect the density of long-lived pauses
We next studied how NusA affected long-lived pauses, which may be generated by the stabilization of elemental pauses. We found that, at a given force, the long pause density (operationally defined as the number of pauses exceeding 20 s per kilobase transcribed) was significantly larger in the presence of NusA. Given that long pauses are infrequent (~1 kb−1), the condition ksp << (k−p, kp) is fulfilled. Therefore, the long pause density, ρlong, can be expressed within the scheme of Figure 2B (see Supplemental Materials) as:
| (5) |
A significant fraction of long pauses are known to be caused by backtracking (Shaevitz et al., 2003). In this case, the rate of entry into a long-pause state will be given by ksp = ksp0 exp(− Fδ2/kBT), where δ2 represents the distance parameter associated with energetic barrier against entering the backtracked state. As suggested earlier, both kp and k−p are force-independent, whereas kn will carry the force dependence described by Eq. 4, assuming that the elemental pause state is a precursor to stabilized pauses. Under such circumstances, the long pause density will be described by ρlong = ρlong0 exp(−F(δ1 + δ2)/kBT). The measured long pause densities were well-fit by this single exponential, and separate fits to data obtained in the presence or absence of NusA yielded similar values for (δ1 + δ2) (Figure S2E); a global fit to both datasets returned (δ1 + δ2) = 1.3 ± 0.2 bp (Figure 4C). Similarly, the ratio of values for the parameter ρlong0 in the presence or absence of NusA yielded an equivalent force acting on the polymerase of FNusA = 19 ± 4pN, corresponding to raising the energy barrier against translocation, ΔEbarrier, by 2.0 ± 0.4 kBT.
Modulation of pausing by NusA is sequence-specific
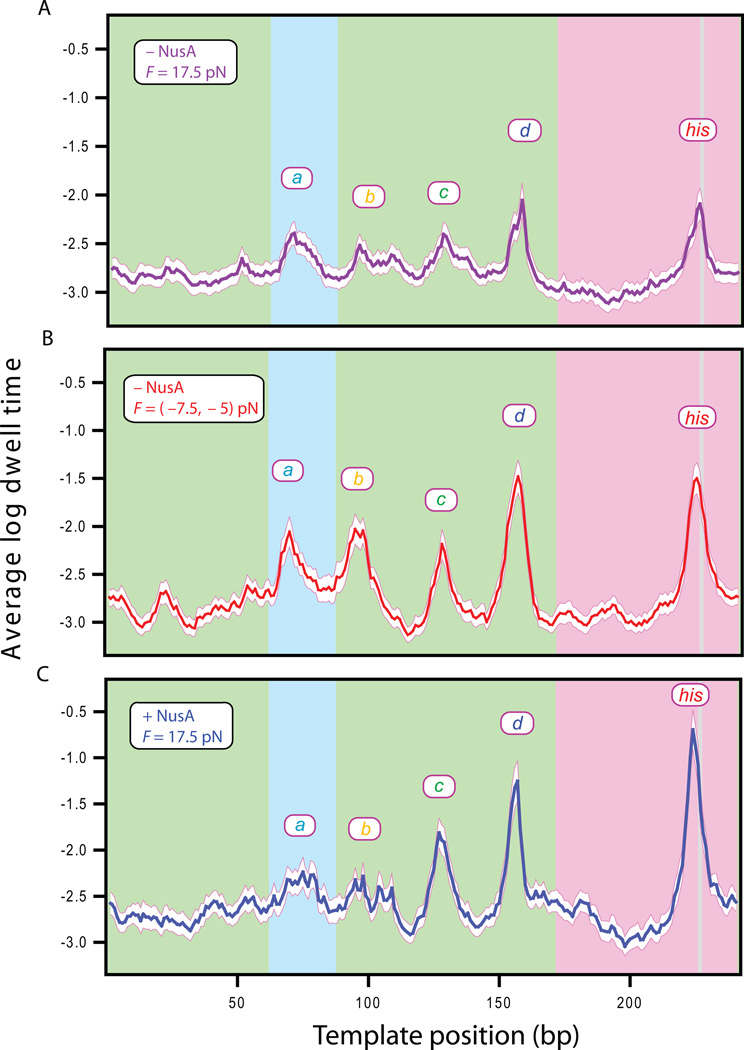
To investigate whether the modulation of pausing and elongation by NusA factor is sequence-specific or uniform across the template, we aligned single-molecule transcription records with the underlying sequence and studied RNAP behavior at identified pause sites (Herbert et al., 2006). Global alignments, where records for all RNAP molecules transcribing the eight tandem repeats were averaged into a single segment, displayed prominent peaks at the various positions in the motif where polymerase tended to pause, studied under three different sets of conditions (Figures 5A–C).
Figure 5. Sequence-specific pauses are influenced by NusA and load.
Display of the mean logarithm of the dwell time for RNAP versus template position (± bootstrapped std. dev., white), computed from aligned data from all eight repeats of the sequence motif carrying the his pause. (A) Data acquired in the absence of NusA for 17.5 pN assisting loads (purple line), (B) in the absence of NusA under 7.5 pN or 5 pN hindering loads (red line), and (C) in the presence of 0.5 µM NusA under 17.5 pN assisting load (blue line). Background color indicates origin of the underlying sequences: rpoB gene (green), restriction sites used for cloning (blue), regulatory pause region (pink), his pause site (light gray). Major pause sites are labeled.
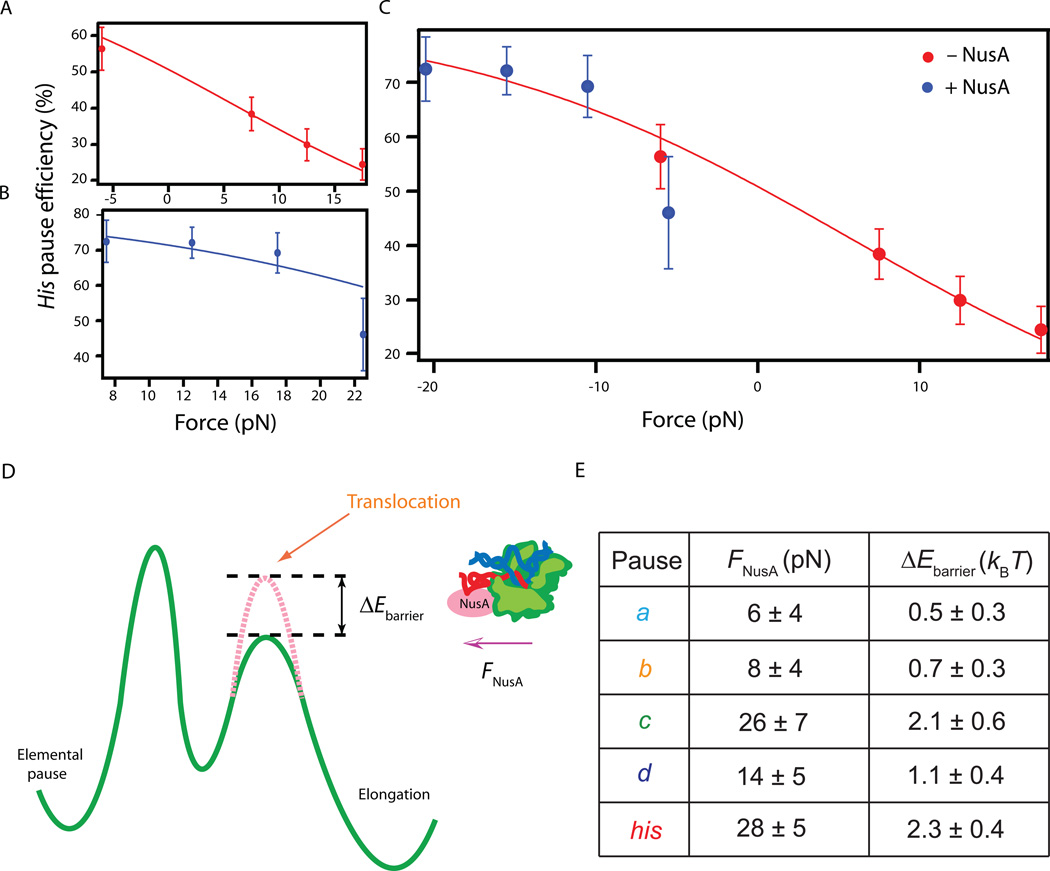
In the absence of NusA under all force conditions, a similar pattern of pausing was observed at the five pause sites identified previously (the a, b, c, d and the his pauses). The heights of these pause peaks varied systematically with the applied load: pausing was enhanced, more-or-less uniformly, as the assisting load was reduced. Pausing was further enhanced when a hindering load was substituted for the assisting load (Figures 5B,C). These findings are fully consistent with the force dependence discussed above for the probability and apparent lifetime for all pauses (Figures 4A,4C,S2A–D). In the presence of NusA under all force conditions, no additional pause sites besides the (a–d) and the his pauses were identified. Moreover, the durations of these sequence-dependent pauses were all enhanced compared to those scored in the absence of NusA under otherwise identical force conditions, although the degree of enhancement varied from site to site. To quantify the increase in the height of the energy barrier against forward translocation produced by NusA factor at various pause sites (Figure 6D), we compared the efficiency of pausing as a function of load in the presence or absence of NusA. As an example, the efficiency of the his pause is shown in Figures 6A–C. The pause efficiency (PE) was fit by the Boltzmann relation (see Eq. (3)):
| (6) |
where APE is ratio of elongation rate (kn) to the rate of pause entry (kp) at F = 0, andδ1 is the distance parameter for translocation, fixed here at 1 bp. The value of PEmax, the pause efficiency at the largest hindering loads, is somewhat less than unity in practice because pauses briefer than 1 s are not detected with 100% reliability; PEmax was therefore obtained by globally fitting efficiency as a function of force in the presence or absence of NusA. The equivalent hindering load produced by NusA and the increase in energy barrier height for all pause sites were determined as described earlier, with results summarized in Figure 6E. Fits returned values of ΔEbarrier for ranging from 0.5–2.3 kBT, suggesting that NusA acts generally to increase the energy barrier against elongation at all template positions (i.e., it acts as an effective hindering load), but that this effect varied with the underlying sequence. Interestingly, the strongest effect of NusA was exerted at the his pause site, which raised the elongation barrier ~3-fold more than at the a or b pause sites.
Figure 6. Effects of NusA on individual pauses and the energetic model.
(A)–(C) Pause efficiencies (PE) for the his pause (mean ± SEM) as a function of applied load, with fits to Equation (3). (A) In the absence of NusA (red), the negative force point (−6 pN) corresponds to pooled data taken under 7.5 pN and 5 pN hindering loads. (B) Data and Boltzmann relation fit (blue) for 0.5 µM NusA. (C) Superposed data in the presence and absence of NusA, showing the fit to a common Boltzmann relation (red line). Here, forces applied in the presence of NusA were first reduced by FNusA obtained from the parameter fit in (B).
(D) Energy diagram illustrating the transcriptional modulation by NusA through a change in barrier height.
(E) The computed increase in the energy barrier to forward translocation produced by NusA for all five identified pause sites and the corresponding hindering load equivalent, obtained from fits of the PE at individual pause sites under various loads in the presence or absence of NusA.
Modulation of transcription by NusA137
To gain further insight into the modulation of elongation by NusA, we studied a mutant consisting of the isolated N-terminal domain (NTD), NusA137. Previous studies have shown that this domain binds RNAP during the elongation phase with somewhat lower affinity than full-length NusA and that the NTD alone can account for the pause-enhancing properties of NusA at the his pause (Ha et al., 2010; Mah et al., 1999). Under assisting loads (17.5 pN) in the presence of a saturating concentration of NusA137 (5 µM), we found that the pause-free elongation rate was suppressed by ~23%, dropping from 18.4 ± 1.0 bp/s to 14.2 ± 0.9 bp/s (Figure 7A). This reduction was only slightly less than that achieved by full-length NusA, which lowered pause-free elongation rate to 12.7 ± 0.8 bp/s (~31%). The short pause density increased considerably upon addition NusA137, but again, not as much as for full-length NusA (Figure 7B). An analogous trend was also observed for long pauses (Figure 7C). All together, these results suggest that the N-terminal domain of NusA is likely to play a central role in the modulation of both elongation and pausing, although it appears that the NTD cannot fully compensate for full-length NusA, even when present at ten-fold higher concentrations. To address sequence-specific effects, individual records were aligned as described previously, and the efficiencies of individual pauses were determined in the presence of either NusA or NusA137 (Figure 7D). As with the full-length construct, the efficiencies all five pauses increased in the presence of NusA137. The pause enhancements at some sequences, e.g., the c and the his sites, were greater for NusA than for NusA137, whereas for the a and b sites, no significant differences between NusA and NusA137 were found.
Figure 7. Modulation of elongation and pausing by NusA137 (NTD).
(A) Average pause-free elongation rates (mean ± SEM) at 17.5 pN assisting load in the presence of 0.5 µM NusA or 5 µM NusA137, and in the absence of NusA..
(B) Short-pause density (mean ± SEM) at 17.5 pN under the same conditions as (A).
(C) Long-lifetime (≥20 s) pause density (mean ± SEM) under the same conditions as (A).
(D) Pause efficiencies (mean ± SEM) of five individual pause sites in the presence of full- length NusA (green), NusA137 (red), or in the absence of either protein (blue). The efficiency at a given pause site was determined by dividing the number of pause events scored (≥ 1 s) within ±5 bp of site by the number of times the site was encountered.
Discussion
The mechanism of sequence-dependent pausing
The mechanisms responsible for transcriptional pausing remain controversial, and competing models have been advanced. In this study, we supply new experimental evidence that addresses some of the models in contention. Broadly speaking, pausing models differ in their overall reaction pathways and kinetics, and may be categorized based on these differences. One category places sequence-dependent pauses directly on the pathway for elongation, where pause states are entered obligatorily, prior to nucleotide condensation by the pre-translocated enzyme (Bai et al., 2007; Bai et al., 2004; von Hippel, 1998). For schemes in this category, the pausing efficiency is necessarily 100%, because the pause state cannot be bypassed. The finding that pause efficiencies vary widely, ranging from ~20% to ~80% in a sequence-dependent fashion, argues against pauses being placed on-pathway [our data, see also (Chan and Landick, 1989; Herbert et al., 2006; Herbert et al., 2010; Kassavetis and Chamberlin, 1981)].
By contrast, the other categories of reaction scheme represent pausing as off-pathway, branching from one or more states of the elongation cycle prior to nucleotide condensation. Off-pathway transitions can occur, in principle, by any of a variety of candidate mechanisms, including enzyme isomerizations, thermal fluctuations, displacements, changes in the nucleic acid structure, or via the influence of transcription co-factors. One particular off-pathway model, inspired by single-molecule studies of eukaryotic RNA polymerase II (Pol II), posits that all pauses are generated by direct enzyme backtracking from the elongation competent state (Depken et al., 2009; Galburt et al., 2007), leading to a (potentially force-biased) random walk of the enzyme over the lattice of upstream template positions until the RNA terminus is restored to the active site, upon which elongation can resume. Long pauses (>25 s duration) by E. coli RNAP induced by nucleotide misincorporation, in particular, have been directly observed to display random-walk behavior (Abbondanzieri et al., 2005a). The backtracking-only model for pausing is attractive in its simplicity, and it accounts in straightforward way for the biphasic, long-tailed distribution observed for pause lifetimes, which can be reasonably fit by a power-law function (Depken et al., 2009; Galburt et al., 2007), as opposed to using multiple exponentials (Neuman et al., 2003; Shaevitz et al., 2003). However, in its present form, a backtracking-only model is not compatible with our experimental observations of the force-dependence of the density and lifetimes for ubiquitous pauses, and the ability of a backtracking-only model to account properly for the observed sequence dependence of pausing remains to be explored.
A third category of reaction scheme, motivated by experiments on prokaryotic RNAP from E. coli, invokes the existence of brief, elemental pause states that are off-pathway and not backtracked, but which may subsequently become stabilized by a variety of mechanisms, including backtracking or hairpin interactions (Ederth et al., 2006; Herbert et al., 2008; Herbert et al., 2006). In previous single-molecule experiments, we found that the intrinsic lifetimes of ubiquitous (elemental, not stabilized) pauses were similar at all pause sequences, including the his and ops elements, in addition to four nearby pause sites characterized to bp-resolution (Herbert et al., 2006). In these experimental conditions, the his and ops pauses appear to reach the stabilized state at most only infrequently. This may be attributable to the lower NTP concentrations and higher temperatures (37 °C) employed in conventional ensemble experiments. Here, we explored the detailed force-dependence of the lifetimes for all four of the ubiquitous pauses, as well as for the his pause. We find once again that the intrinsic pause lifetimes for these elemental pauses are similar in duration, and moreover, that their lifetimes are largely unaffected by external force. These characteristics are fully consistent with a model in which sequence-dependent pauses do not backtrack, but instead represent an elemental state that may, for example, reflect a conformational change at or near the active site of the enzyme (Artsimovitch and Landick, 2000; Herbert et al., 2006; Toulokhonov et al., 2007).
In contrast to the intrinsic lifetime of elemental pauses, both the density and the apparent lifetime of elemental pauses are strongly modulated by assisting load, which tilts the energy landscape for transcription to favor forward translocation (Figure 2A, 6D and S2A–D). Our result, therefore, shows that modulating the translocation step can play a key role in regulating the overall transcription rate, which ultimately regulates the cellular RNA level. Furthermore, quantification of the observed load dependencies provides direct evidence leading to a model that involves kinetic competition between elongation and entry into a pause state which branches directly from the pre-translocated position (Figure S3). Mechanical stabilization of the post-translocated state reduces the branching probability of entering a pause state while simultaneously increasing the chance that the enzyme will resume elongation rather than re-enter the pause state. A branch from the pre-translocated state is also supported by results from previous cross-linking and pyrophosphorolysis experiments (Toulokhonov and Landick, 2003; Toulokhonov et al., 2007). Because the rates involved in this kinetic competition are temperature dependent, it is anticipated that pause efficiencies, lifetimes, and their force sensitivities will likewise be temperature dependent (Abbondanzieri et al., 2005b; Mejia et al., 2008).
Finally, we note that recovery from a pause state to the main elongation pathway need not follow a direct reversal of the sequence of transitions leading to the pause state, although the kinetic analysis here assumes such a scenario for simplicity. Recent findings suggest that at least some pauses which do not backtrack may revert to the elongation pathway after the translocation and NTP binding steps (Kireeva and Kashlev, 2009; Landick, 2009; Toulokhonov et al., 2007). In such a scenario, stabilizing the pre-translocated state will not merely change the pause efficiency, but will also increase the pause lifetime to a degree determined by how rate-limiting the translocation step is during pause recovery. The conclusion that both hindering loads and NusA factor increase the pause density and apparent lifetime by stabilizing the pre-translocated state is qualitatively consistent with such a kinetic scheme as well.
A kinetic model of NusA modulation of elongation and pausing
In the proposed kinetic scheme (Figure 6D), the effects of NusA on active elongation and pausing may be explained quantitatively by an increase of ~1.5 kBT in the energy barrier for forward translocation. Given that the translocation step during each elongation cycle represents the distance subtended by a single base pair, this energy is equivalent to supplying an average hindering load of ~19 pN over that distance. Put in terms of a Brownian ratchet model (Abbondanzieri et al., 2005a), this implies that the action of NusA biases the relative occupancy of the pre- and post-translocated states, further stabilizing (destabilizing) the pre-translocated (post-translocated) state: this is consistent with ensemble experiments suggesting that NusA favors backward translocation (Bar-Nahum et al., 2005).
The force-dependent elongation rate for RNAP in the absence of NusA agrees with previous single-molecule experiments (Abbondanzieri et al., 2005a; Bai et al., 2007). In the presence of NusA, the pause-free velocity was also found to be load-dependent, but we found only a small decrease in Vmax, upon the addition of NusA, suggesting that NusA does not significantly affect the biochemical rates of either NTP condensation or pyrophosphate release. Rather, it would appear that NusA regulates on-pathway elongation in some mechanochemical fashion.
Previous structural and biochemical studies suggested that NusA binds to the flap-tip of the β-subunit of RNAP, the dock domain of the β´ subunit, and the exiting RNA, with its N-terminal domain capping the RNA exit channel (Ha et al., 2010; Toulokhonov et al., 2001; Yang et al., 2009). The effective hindering load produced by NusA might be explained, in principle, by an interaction between the positive surface charge of NusA (Worbs et al., 2001) and nascent RNA, since anchoring of the NusA NTD to the RNAP exit channel would create a continuous NusA-RNA interaction that would preclude RNA looping during translocation. It is also conceivable that the binding of NusA to RNAP may induce allosteric effects in the active site or trigger loop that affect translocation without significantly changing the catalysis rates for active (on-pathway) elongation steps. It might therefore be productive in future single-molecule experiments to explore NusA effects on elongation for RNAP molecules carrying mutations in the trigger loop or bridge helix.
Sequence-specific effects of NusA and their probable mechanisms
Previous experiments have suggested that NusA may exert different effects at different pause sites (Artsimovitch and Landick, 2000; Sigmund and Morgan, 1988; Theissen et al., 1990). The data here show that an assisting force of sufficient magnitude (6–28 pN) can largely neutralize the effect of NusA at different, sequence-specific pause sites, allowing us to conclude that NusA raises the energy barrier against forward translocation in a sequence-specific manner (Figure 6E). The sequence-specific effects of NusA we observed are unlikely to be mediated solely by stable RNA structures, since pauses a–d lack the ability to form obvious, stable RNA hairpins near RNAP (−11 to −16 bp relative to the 3′-end of RNA) and yet are affected to different extents by NusA. Rather differences in the DNA template sequence, the length of the DNA-RNA hybrid (Kyzer et al., 2007) or conceivably small, comparatively unstable RNA structures. We found that NusA NTD (NusA137), which binds to RNAP in or near the RNA-exit channel, also modulates elongation in a sequence-specific manner similar to that of full-length NusA (Figure 7A–C), although the effects of NusA NTD were slightly weaker than those of full-length NusA (Figure 7D). These results are consistent with the effect of NusA NTD on the his pause in ensemble experiments when RNA is present upstream of the pause hairpin (Ha et al., 2010).
Transcriptional regulation by NusA
Unlike some transcription factors, such as RfaH and GreA/B, that are transiently recruited by RNAP at specific sites or for different configurations of the transcription complex (Artsimovitch and Landick, 2002), NusA continuously moderates elongation and pausing (Mooney et al., 2009). A major effect of NusA is attributable to increasing the energy barrier to forward translocation. Remarkably, this is the opposite effect of the other universal elongation factor in bacteria, NusG, which also associates with most elongation complexes (Mooney et al., 2009), but appears instead to promote forward translocation (Herbert et al., 2010). Thus, in vivo, elongating RNAP appears constantly subject to the counter-posing effects of two transcriptional regulators, one that augments pausing and one that diminishes it, both via effects on translocation. Given that NusA and NusG do not compete for binding to elongation complexes, but exhibit selective effects on pauses (Burns et al., 1998), an important challenge for future research will be to determine how differences in the RNA and DNA sequences encountered by RNAP dictate how the opposing effects of these two regulators are differentially balanced.
Experimental Procedures
Transcription templates for optical trapping assays
Plasmid pALB, used for the hindering-load assay, was created from plasmid pBW, which was used to generate the his tandem repeat template employed in previous experiments (Herbert et al., 2006). The AlwNI restriction site was moved from a location ~1000 bp downstream of the rrnBT1 terminator to a location ~1000 bp upstream of the T7A1 promoter using two rounds of site-directed mutagenesis (Qiagen Quickchange). Linear, labeled templates were constructed from the pALB and pBW1 plasmids by first digesting each plasmid at the unique AlwNI site. The resulting 3′ ends were labeled using DIG-ddUTP plus terminal transferase (Roche DIG Oligonucleotide 3′-End Labeling Kit). Subsequent digestion of pBW1 at a unique SapI site resulted in a transcription template suitable for study under assisting load, by removing the digoxigenin label site at the transcriptionally downstream end and leaving a single digoxigenin label site at the upstream end for tethering to anti-digoxigenin antibody-coated polystyrene beads. Digestion of pALB at the unique Eco109I site resulted in a template for hindering load, removing the upstream end and leaving a single label on the downstream end.
Optical trapping assay
Sample preparation for the DNA-pulling dumbbell assay was performed as described (Herbert et al., 2010). All experiments carried out in the presence of 1mM rNTPs in transcription buffer (50 mM HEPES, pH 8.0, 130 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT) and an oxygen scavenging system (Yildiz et al., 2003), and the specified concentrations of NusA and NusA137 at a temperature of 27 °C. Controlled forces were applied by an optical force clamp based on active feedback (Abbondanzieri et al., 2005a). NusA and NusA137 were purified as described (Kyzer et al., 2007).
Data analysis
The contour length of the DNA suspended between the beads was calculated as described (Abbondanzieri et al., 2005a). Template position was determined by subtracting the contour length of the segment of the DNA template located between the transcription initiation site and the 730 nm bead (2,607 bp for assisting load and 5,565 bp for hindering load, based on a DNA helix pitch of 0.338 nm/bp) from the measured DNA contour length during transcriptional elongation. Pauses were identified and aligned by computer as previously described (Herbert et al., 2006). Pause-free elongation velocities of single RNAP molecules were determined by fitting the velocity distribution to a sum of two Gaussians, as previously described (Dalal et al., 2006; Neuman et al., 2003). Statistical uncertainties in the pause efficiency, PE, of individual pauses were determined from , where N is the number of records. Igor Pro software (Wavemetrics) and programs written in C were used for data analysis.
Supplementary Material
Acknowledgements
The authors thank J. Gelles, K. Herbert, and members of the Block lab for stimulating discussions, and P. Anthony for critical reading of the manuscript. This work was supported by grants to S.M.B. and R.L. from the NIH/NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental data include text, experimental procedures, and three figures, and can be found with this article online at www.cell.com.
References
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005a;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005b;89:L61–L63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci U S A. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- Chan CL, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–20804. [PubMed] [Google Scholar]
- Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23:231–239. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Mooney RA, Isaksson LA, Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 2006;356:1163–1179. doi: 10.1016/j.jmb.2005.11.080. [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9:563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DI, Baron LS. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974;58:141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- Greenblatt J, McLimont M, Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981;292:215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401:708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Chamberlin MJ. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981;256:2777–2786. [PubMed] [Google Scholar]
- Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997a;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3' end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997b;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Landick R. Transcriptional pausing without backtracking. Proc Natl Acad Sci U S A. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci U S A. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Landick R, Block SM. Single-molecule studies of RNA polymerase: one singular sensation, every little step it takes. Mol Cell. 2011;41:249–262. doi: 10.1016/j.molcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, Li J, Davidson AR, Greenblatt J. Functional importance of regions in Escherichia coli elongation factor NusA that interact with RNA polymerase, the bacteriophage lambda N protein and RNA. Mol Microbiol. 1999;34:523–537. doi: 10.1046/j.1365-2958.1999.01618.x. [DOI] [PubMed] [Google Scholar]
- Mejia YX, Mao H, Forde NR, Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J Mol Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J Mol Biol. 2001;311:265–282. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F, Curtiss IR, Ingraham JL, Lin ECC, Low KB, Magasanik B, Rfznikopp WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; pp. 822–848. [Google Scholar]
- Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, Chamberlin MJ. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987;195:809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund CD, Morgan EA. Nus A protein affects transcriptional pausing and termination in vitro by binding to different sites on the transcription complex. Biochemistry. 1988;27:5622–5627. doi: 10.1021/bi00415a034. [DOI] [PubMed] [Google Scholar]
- Theissen G, Pardon B, Wagner R. A quantitative assessment for transcriptional pausing of DNA-dependent RNA polymerases in vitro. Anal Biochem. 1990;189:254–261. doi: 10.1016/0003-2697(90)90117-r. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Vogel U, Jensen KF. NusA Is Required for Ribosomal Antitermination and for Modulation of the Transcription Elongation Rate of both Antiterminated RNA and mRNA. J. Biol. Chem. 1997 doi: 10.1074/jbc.272.19.12265. [DOI] [PubMed] [Google Scholar]
- von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, Hankamer B, Lewis RJ, Lewis PJ. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.