Abstract
Despite the fact that approximately half of all HIV patients acquire infection through penile exposure, there have been no recent studies of penile SIV transmission in rhesus macaques and the nature of the virus variants transmitted, target cells, and pathways of virus dissemination to systemic lymphoid tissues are not known. Single genome amplification (SGA) and sequencing of HIV-1 RNA in plasma of acutely infected humans allows the identification and enumeration of transmitted/founder viruses responsible for productive systemic infection. Studies using the SGA strategy have shown that intrarectal and intravaginal SIV transmission to macaques recapitulates key features of human HIV transmission. To date, no studies have used the SGA assay to identify transmitted/founder virus(es) in macaques infected after penile SIV exposure. Here we report that SIV can be transmitted by penile SIV exposure. However, similar exposure to a high-dose inoculum infects only about half the animals, which is about 50% less efficient transmission than occurs after vaginal SIV challenge. In addition, only a single SIV env variant established the systemic infection in all five animals that became infected after penile exposure, a result that is consistent with low incidence and few transmitted HIV variants in heterosexually infected men. Our results suggest that the penile transmission of SIVmac251 in rhesus macaques recapitulates the key features of penile HIV-1 transmission and may provide insight into host or viral factors that permit penile transmission and dissemination. Furthermore, this SIV challenge exposure route will be useful in testing vaccines and other prophylactic approaches.
Introduction
HIV is primarily transmitted by heterosexual contact and approximately equal numbers of men and women are infected with the virus worldwide. Numerous studies to define the biology of HIV transmission have been undertaken with the goal of identifying stages of the infection process, from mucosal exposure through dissemination to distal lymphoid tissues establishment of systemic infection, to identify targets for interventions. To a large extent this effort has focused on understanding rectal transmission in men and vaginal transmission in women,1–22 but heterosexual men are regularly infected through penile HIV exposure. Despite the fact that approximately half of HIV patients acquire infection through penile exposure,23–30 there have been no preclinical studies of AIDS vaccines using penile inoculation to challenge immunized animals and nothing is known about the anti-HIV immune effector mechanisms that may be present on the mucosal surfaces of the penis, which presumably serves as the portal of entry for HIV transmission to males. The foreskin is thought to be particularly important in HIV transmission to males because the presence of an intact foreskin is associated with an approximately 50% increased risk of HIV acquisition.30–36 The foreskin and glans of the human penis have a complete population of immune cells including potential HIV target cells.37–39 Although the foreskin is clearly a critical target tissue for HIV transmission, and circumcision can reduce HIV acquisition,31–33 other tissues of the penis must account for the remaining transmissions in 50% of HIV-infected men.
Strategies to prevent penile HIV transmission should be directed at the viral variants responsible for establishing productive infection in men via this route of exposure. Identifying the individual HIV-1 variants that are transmitted and establish systemic infection is possible using single genome amplification (SGA) and direct sequencing, a technique that provides an accurate characterization of the transmitted/founder viruses involved in the initial establishment of systemic infection following mucosal transmission.40,41 Data generated using the SGA approach and recent empirical data together with mathematical models have shown that immediately after transmission, in the absence of selective pressure, a founder HIV-1 lineage typically diversifies by a Poisson distribution of random nucleotide substitutions leading to a star-like phylogeny of viral variants in the HIV quasispecies with no or few shared mutations.42,43 For each low-diversity HIV-1 lineage in an individual, the consensus of the sequences within that lineage represents the inferred ancestral sequence. If sampled prior to the adaptive immune response or the onset of other selection pressures, mutations occur randomly and thus the consensus sequence of each low diversity lineage represents a transmitted or founder virus sequence.43
Using the SGA sequencing approach and mathematical modeling, it has now been reported in at least eight patient cohorts representing HIV-1 subtypes A, B, C, and D that most (60–90%) mucosal infections originate from single variant transmissions.41,43–47 The remaining 10–40% of infections are initiated by a limited number of transmitted/founder HIV-1 variants. Therefore, for each individual infected, the potential viral diversity in the period of acute infection is limited to a single or few HIV-1 lineages. This genetic bottleneck is less pronounced in individuals engaged in behaviors that may plausibly facilitate transmission (anal-receptive intercourse or intravenous drug use)43,48,49 and in patients with sexually transmitted infections.41,44–47 Importantly, acute infection with ”heterogeneous” HIV populations has been linked to more rapid disease progression.50–52 The study of the number of transmitted viral variants and their overall diversity can thus have important implications for developing both prophylactic vaccines and antiviral therapy.
Simian immunodeficiency virus (SIV) models of mucosal HIV transmission in rhesus macaques are valuable for examining the transmitted or early founder populations that establish productive infection because the genetic composition of the virus stock used for mucosal inoculation can be readily defined and compared to the viruses transmitted following mucosal challenge. Furthermore, because the timing of SIV transmission is known,4,6,9,53–55 mathematical modeling is not needed to infer the timing of virus transmission in macaques. SGA approaches can be used to define mucosal challenge conditions for NHP models that recapitulate typical HIV-1 transmissions in humans, resulting in transmission of one, or only a few viral variants, providing animal models for evaluation of vaccines and other prophylactic interventions that recapitulate the virology of HIV transmission. Using the SGA sequencing approach, one to five transmitted/founder viruses were found in rhesus macaques infected intrarectally (i.r.) either SIVmac251 or SIVsmE660.17 In a separate titration study, a dilution of challenge stock was found that reproducibly infected animals with one or only a few viruses following intrarectal challenge.56 Finally, rhesus macaques vaginally challenged with SIVmac251 became systemically infected with 1–10 founder viruses.11 To date, no studies have used the SGA sequencing approach to enumerate and identify transmitted/founder viruses after penile SIV transmission.
Here we report that SIV can be transmitted to rhesus macaques by penile SIV exposure. However, two exposures to a high-dose inoculum on the same day infected only about half the inoculated animals. In addition, only a single SIV env variant established the systemic infection in all five animals that became infected after penile inoculation, a result that is consistent with low incidence and few transmitted HIV variants in heterosexually infected men. Our results suggest that the penile transmission of SIVmac251 in rhesus macaques recapitulates the key features of penile HIV-1 transmission57,58 and may provide insight into host or viral factors that permit penile transmission and dissemination. Furthermore, this inoculation strategy could be useful in testing vaccines and other prophylactic approaches.
Materials and Methods
Animals
The mature male rhesus macaques (Macaca mulatta) used in these studies were housed at the California National Primate Research Center in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care. These experiments were approved by the Institutional Animal Use and Care Committee of the University of California Davis. All animals were negative for antibodies to HIV-2, SIV, type-D retrovirus, and simian T cell lymphotropic virus type 1 at the time the study was initiated. When necessary, animals were anesthetized with ketamine hydrochloride (10 mg/kg; Parke-Davis, Morris Plains, NJ) or 0.7 mg/kg tiletamine HCl and zolazepan (Telazol, Fort Dodge Animal Health, Fort Dodge, IA) injected intramuscularly.
Penile SIVmac251 inoculation
A cell-free stock of SIVmac251 (UCD-6/04) was produced in Staphylococcus enterotoxin A-stimulated rhesus monkey peripheral blood mononuclear cells (PBMCs) and used for these studies. This SIVmac251 stock (UCD-6/04) contains approximately 109 viral RNA (vRNA) copies/ml and 105 50% tissue culture infection dose (TCID50)/ml when titered on CEMX174 cells. On the day of challenge, serial 10-fold dilutions of the SIV stock were made in phosphate-buffered saline (PBS) (Table 1); 2 ml of virus inoculum was used for all the penile exposures in this study. Animals were exposed repeatedly, at weekly intervals with the more dilute inoculum, and to two exposures to the undiluted SIVmac251 stock on the same day. Animals that did not become infected after the first inoculation with undiluted virus were inoculated using the same procedure to more times at 4 weeks intervals. For the challenge, a small cup was fashioned from a closed end of a disposable 15-ml conical centrifuge tube and 2 ml of virus inoculum was placed into the cup. The penis was extended, the foreskin was retracted, and the glans and shaft were inserted into the inoculum and held there for 5 min. Then the penis was allowed to retract into the foreskin and the animal was placed into its cage in dorsal recumbency and allowed to recover from anesthesia. Thus although no effort was made to specifically inoculate the foreskin, we visually confirmed that some of the inoculum did contact the area when the glans retracted into the foreskin. Five of the 11 animals in this study were repeatedly exposed with several doses of the virus, but the inoculation series was stopped when vRNA was detected in plasma collected 10–14 days after each challenge.
Table 1.
Summary of Animals, Penile SIVmac251 Inoculations, and Virologic Outcomes
| Animal no. | Agea | SIV dose | No. of inoculations | Initial vRNA levelb(log10 copies/ml plasma) | Day of first vRNA+plasmab | No. of founder SGA sequences analyzed | No. of founder variants identified |
|---|---|---|---|---|---|---|---|
| 36527c | 4y6m | 14 | — | — | — | — | |
| 36240d | 4y7m | 14 | — | — | — | — | |
| 36744e | 4y5m | 10 TCID50 | 14 | — | — | — | — |
| 36483f | 4y6m | 14 | — | — | — | — | |
| 36838g | 4y4m | 14 | — | — | — | — | |
| 36527c | 4y6m | 7 | — | — | — | — | |
| 36240d | 4y7m | 103 TCID50 | 7 | — | — | — | — |
| 36317 | 4y8m | 14 | — | — | — | — | |
| 36364 | 4y6m | 14 | — | — | — | — | |
| 36744e | 4y5m | 7 | — | — | — | — | |
| 36483f | 4y6m | 104 | 7 | — | — | — | — |
| 36838g | 4y4m | TCID50 | 1 | 2.54 | 10 | 16 | 1 |
| 36193 | 4y8m | 2h | 3.81 | 10 | 19 | 1 | |
| 36336 | 4y9m | 2h | 4.19 | 10 | 20 | 1 | |
| 37439 | 4y8m | 105 | 2h | 2.86 | 10 | 20 | 1 |
| 36744e | 4y5m | TCID50 | 6i | — | — | — | — |
| 36330 | 4y8m | 6i | 2.42 | 7 | 19 | 1 |
Age at first inoculation.
Above the cutoff of 1.7 log10 copies/ml plasma.
These animals were inoculated with the least concentrated virus inoculum first and most concentrated virus inoculum last.
Animals were inoculated twice in 1 day with a 4 h interval between inoculations.
Animals were inoculated twice in 1 day with a 4 h interval between inoculations on 3 separate days, with 4 weeks between each of the inoculation days.
vRNA, viral RNA; SIV, simian immunodeficiency virus; SGA, single genome amplification.
Viral RNA isolation and cDNA synthesis
Plasma and virus stock were thawed at room temperature and RNA was isolated from 0.5 ml plasma using the QIAamp Ultrasens Viral Kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's protocol and eluted in 50 μl. The vRNA was reverse transcribed into cDNA using SuperscriptIII reagents (Invitrogen, Carlsbad, CA) with 2 μl 50 μM dT23VN (or gene-specific primer for SGA analysis: SIVEnvR1 5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′), 2 μl 10 mM dNTPs, and 22 μl viral RNA. This mixture was heated to 65°C for 5 min followed by incubation on ice for 2 min. A master mix of the following was then added: 8 μl of 5×First-Strand Buffer, 2 μl of 0.1 M DTT, 2 μl of RNaseOUT Recombinant RNase Inhibitor (40 units/μl), and 2 μl of SuperScriptIII RT (200 units/μl) and Incubated at 25°C for 5 min, 50°C for 60 min, and 70°C for 15 min followed by adding 1 μl Escherichia coli RNase H (5 U/μl) and incubated at 37°C for 20 min. cDNA was stored at −20°C to 80°C until amplification.
Quantitative viral RNA analysis
Reverse transcriptase polymerase chain reaction (RT-PCR) was used to detect and quantify SIVgag, RNA levels in plasma samples. Samples were tested in replicates of four reactions carried out in 96-well optical plates (Applied Biosystems, Foster City, CA) in a 25 μl reaction volume containing 5 μl cDNA and 20 μl Mastermix (Applied Biosystems) using the ABI 7900 robotic thermocycler. All sequences were amplified for 2 min at 50°C and 10 min at 95°C followed by 50 cycles of 15 s at 95°C and 1 min at 60°C. The following primer pairs and probes were used: SIVgag forward primer 1, 5′-3′ GGG AGA TGG GCG TGA GAA A, reverse primer, CGT TGG GTC GTA GCC TAA TTT T, and probe TCA TCT GCT TTC TTC CCT GAC AAG ACG GA. The copy number of SIVgag was calculated based on standard curves for a viral gene plasmid spanning a concentration range from 0.1 to 108 copies. Although amplification from wells with 0.1 copy of SIVgag was inconsistent, the assays consistently detected SIVgag in wells containing one or more copies of the plasmid. The results were analyzed with SDS 7900 system software version 2.3 (Applied Biosystems). The results for each sample are reported as log10 vRNA copies per ml of plasma RNA.
In a series of preliminary studies conducted separately to evaluate the specificity and determine the background of the PCR assay, RNA isolated from plasma from six rhesus macaques that had never been exposed to SIV was subjected to amplification. There was no amplification of SIVgag from any of these samples. Thus, 50 SIVgag RNA copies/ml plasma (or 1.7 log10 SIVgag copies/ml plasma) was used as the cutoff for determining if a sample from an SIV-inoculated monkey was positive.
Single genome amplification of SIVmac251 env
The entire env gene was sequenced using a limiting dilution PCR so only one amplifiable molecule is present in each reaction. cDNA was serially diluted and distributed among independent PCR reactions to identify a dilution where amplification occurred in <30% of the total number of reactions. PCR amplification was performed in the presence of 1× buffer, 2 mM MgSO4, 0.2 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 0.025 U/μl Platinum Taq High Fidelity polymerase (Invitrogen) in a 20-μl reaction. First round PCR was performed with sense primer SIVEnvF1 5′-CCT CCC CCT CCA GGA CTA GC-3′ and antisense primer SIVEnvR1 5′-TGT AAT AAA TCC CTT CCA GTC CCC CC-3′ under the following conditions: 1 cycle of 94°C for 2 min, 35 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min, followed by a final extension of 68°C for 10 min. Next, 1 μl from first-round PCR product was added to a second-round PCR reaction that included the sense primer SIVEnvF2 5′-TAT AAT AGA CAT GGA GAC ACC CTT GAG GGA GC-3′ and antisense primer SIVEnvR2 5′-ATG AGA CAT RTC TAT TGC CAA TTT GTA-3′ performed under the same conditions used for first-round PCR, but with a total of 45 cycles. Correct sized amplicons were identified by agarose gel electrophoresis and directly sequenced with second-round PCR primers and six SIV-specific primers using BigDye Terminator technology. To confirm PCR amplification from a single template, chromatograms were manually examined for multiple peaks, indicative of the presence of amplicons resulting from PCR-generated recombination events, Taq polymerase errors, or multiple variant templates. Confirmation that sequences were derived from individual template molecules ensures proportional representation of individual env sequences circulating in vivo. Sequences containing two or more ambiguous sites were excluded from analysis.
Sequence analysis
Sequences were aligned using Clustal W60 and hand edited using Jalview61 or MacClade 4.08 to improve alignment quality. All trees were constructed with Phylip62 using the neighbor-joining method63 with the Kimura two-parameter distance matrix.64 Within-subject env diversity was analyzed in three ways, as described in detail previously,43 but fell only into “homogeneous” diversity. Briefly, diversity was determined by visually inspecting sequences by using neighbor-joining phylogenies and the Highlighter tool (www.hiv.lanl.gov). Also, since the population grows exponentially during the early phase of infection, distribution of pairwise Hamming distances (HD) within each sample were examined for star-like phylogeny and a single Poisson distribution of changes indicating infection from a single viral variant.65 Lastly, mathematical modeling was used to test predictions of expected maximum HD against assumptions of infection variant phylogenies.65 All sequences were deposited in GenBank with accession numbers JF298213–JF298241.
Results
Outcomes of penile SIV inoculation
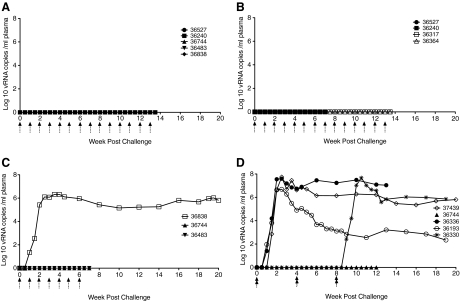
Since no infectivity data were available for penile SIV transmission in rhesus macaques, this study was performed by sequentially escalating the concentration of the virus inoculum from 10 to 105 TCID50. None of five animals inoculated 14 times with 10 TCID50 of the SIVmac251 stock became plasma vRNA+ after penile inoculation (Table 1, Fig. 1A). Neither of the two animals inoculated seven times with 103 TCID50 of the SIVmac251 stock became systemically infected after penile inoculation (Table 1, Fig. 1B) and none of the two animals inoculated 14 times with 103 TCID50 of the SIVmac251 stock became systemically infected after penile inoculation (Table 1, Fig. 1B). One animal inoculated with 104 TCID50 of the SIVmac251 stock became systemically infected after one penile inoculation (Table 1, Fig. 1C), but neither of the two animals inoculated seven times with 104 TCID50 became systemically infected (Table 1, Fig. 1C). Three of the five animals inoculated twice in a single day (4 h interval between inoculations) with the undiluted SIVmac251 stock (105 TCID50) became systemically infected (Table 1, Fig. 1D). One of the two animals inoculated twice in a single day (4 h interval between inoculations) with 105 TCID50 of the SIVmac251 stock every 4 weeks for 8 weeks became systemically infected after penile inoculation with plasma vRNA levels consistent with other infected animals (Table 1, Fig. 1D). Once infected, the pattern of plasma viremia in animals infected by penile SIV inoculation was similar to plasma vRNA in female rhesus macaques infected after vaginal SIV inoculation.10 SIV RNA was first detected in plasma collected at 7–10 days postinfection (PI), however, vRNA peaked at 14–21 days PI (Table 1, Fig. 1), which is delayed compared to vaginal inoculation.10 In both males (Fig. 1) and females,10 a variable set point plasma vRNA level is established at 6–8 weeks PI after genital inoculation.
FIG. 1.
Plasma viral RNA (vRNA) levels after penile SIVmac251 inoculation. (A) With a 10 TCID50 dose, none of the five animals inoculated 14 times became plasma vRNA+ after penile inoculation. (B) At a dose of 103 TCID50, none of the two animals (36527, 36240) inoculated seven times became plasma vRNA+ and none of the two animals (36317, 36364) inoculated 14 times became viremic. (C) One animal (36838) inoculated once with 104 TCID50 of the SIVmac251 stock became plasma vRNA+, but neither of the two animals inoculated seven times became plasma vRNA+. (D) With a 105 TCID50 dose of the SIVmac251 stock, three (37349, 36336, 36193) of the five animals inoculated twice in a single day became plasma vRNA+ and one of two animals became plasma vRNA+ after being inoculated twice in a single day a total of three times over an 8-week period. The arrows under the x-axis indicate the timing of the inoculations. The double arrows in D indicate two inoculations with a 4-h interval between inoculations. The individual animal's symbols are noted in the key and data from animals are graphed up to the day of their last inoculation if they remained uninfected and up to the day of necropsy if they became infected.
Diversity of viral env in the SIVmac251 stock
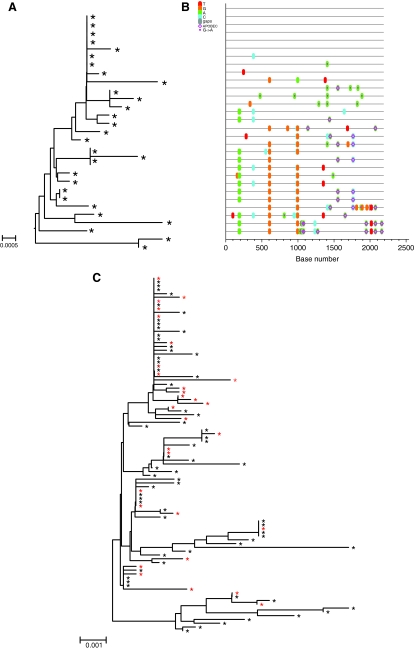
Phylogenetic relationships among the env sequences in the SIVmac251 (UCD-2/02) stock that was produced from the same seed stock that was used produce the SIVmac251 (UCD-6/04) stock for this study have been described.11 To characterize the SIVmac251 (UCD-6/04) stock used in the present study, a total of 36 env nucleotide sequences were derived by SGA. Sequence diversity was analyzed by pairwise sequence comparison, neighbor-joining phylogenetic tree reconstruction (Fig. 2A), and visualization with the Highlighter tool (http://www.hiv.lanl.gov) (Fig. 2B). Overall, the maximum nucleotide diversity of the env sequences from the SIVmac251 stock (UCD-6/04) used for this study was 1.0%, which is consistent with the (UCD-2/02) SIVmac251 stock11 and other SIVmac251 stocks.44 It is worth noting, although perhaps not surprising, that the env variants in the (UCD-6/04) SIVmac251 stock are closely related to the env variants in the (UCD-2/02) SIVmac251 stock (Fig. 2C).
FIG. 2.
Analysis of single genome amplification (SGA)-derived env sequences from the SIVmac251 (UCD 6/04) challenge stock. (A) The neighbor-joining tree (the scale bar represents 0.001 nucleotide substitutions per site) and (B) highlighter plot. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). (C) The neighbor-joining tree including all SGA-derived sequences from the SIVmac251 (UCD 6/04) challenge stock used for the current study and the SIVmac251 (UCD 2/02) challenge stock used in a published study of founder variants after vaginal inoculation.11 Black stars indicate the variants that were amplified from the (UCD 2/02) SIVmac251 stock and red stars indicate the variants that were amplified from the (UCD 6/04) SIVmac251 stock. (The scale bar in A represents 0.0005 nucleotide substitutions per site and the scale bar in C represents 0.001 nucleotide substitutions per site.) The APOBEC symbol indicates the location of APOBEC signatures in hypermutated sequences. The A→G symbol indicates the location of sequences of G to A mutations.
Diversity and divergence of SIVenv in plasma of animals after penile SIV inoculation
A total of 94 SGA-derived 2.7-kb env sequences were obtained from the first vRNA+ plasma sample of five rhesus macaques (median of 20 sequences; range 16–20). SGAs were derived from the first plasma sample that had vRNA levels above the assay cutoff of 1.7 log10 copies/ ml plasma. These samples were collected 7 or 10 days after penile exposure to SIVmac251 (Table 1). Although many of the animals were exposed multiple times, we assumed that the exposure responsible for transmitting the infection occurred 7–10 days before the initial detection of viral RNA in plasma during the ramp-up phase of infection. This estimate of 7–10 days from the transmitting exposure was confirmed using the Poisson diversity tool (www.hiv.lanl.gov). The viral loads in these samples ranged from 2.42 to 4.19 log10 copies/ml of plasma (Table 1). Although no sequences were overtly APOBEC hypermutated as assessed using the Hypermut tool (www.hiv.lanl.gov), several sequences were enriched for G-to-A mutations and excluded from calculations of diversity and divergence from the SIVmac251 consensus reference sequence. The mean sequence diversity of each animal during acute infection ranged from 0.01 to 0.03% with an average for all animals of 0.02%. The maximum sequence diversity for each animal ranged from 0.07 to 0.15% with an average maximum diversity of 0.09%. For each infected animal, a single low-diversity lineage was detected in the ramp-up phase plasma.
In addition to ramp-up sequences, 76 SGA-derived 2.7-kB env sequences were obtained from set point vRNA+ plasma samples of the same five rhesus macaques (median of 17 sequences per animal; range 9–17). These sequences were derived from plasma samples that had vRNA levels above 5.0 log10 copies/ml plasma. These samples were collected 12–19 weeks after the presumed transmitting penile SIVmac251 exposure (Fig. 1). The mean sequence diversity of in these samples ranged from 0.1 to 0.2% with a mean of 0.18%. The maximum sequence diversity for each animal in these samples ranged from 0.3 to 0.6% with an average maximum diversity of 0.4%. Overall, the total diversity correlates to the amount of time since infection.
Enumeration of transmitted/founder populations in animals after penile SIV inoculation
To enumerate the total number of transmitted/founder variants, the number of distinct low-diversity viral env lineages present in each sample was estimated. Low-diversity monophyletic lineages are defined as a unique series of identical or nearly identical nucleotide sequences, differing only by the number of individual polymorphisms that could arise randomly through RT error within 7 days of transmission. In this study, we define a variant as an SGA-derived genetic sequence with up to two individual nucleotide polymorphism differences from another variant assuming no recombination or selection pressure, and a gamma distributed rate of mutation across sites. The maximum number of nucleotide mutations likely to occur by random error within the time frame from virus exposure has been determined mathematically. A 2.2- to 2.7-kb fragment of SIV env will accumulate a maximum of two nucleotide polymorphisms in 1 week of SIV infection. Thus if a plasma SGA sequence had more than two nucleotide substitutions relative to the consensus sequence of each lineage, then the variant is assumed to have been transmitted as distinct founder virus.
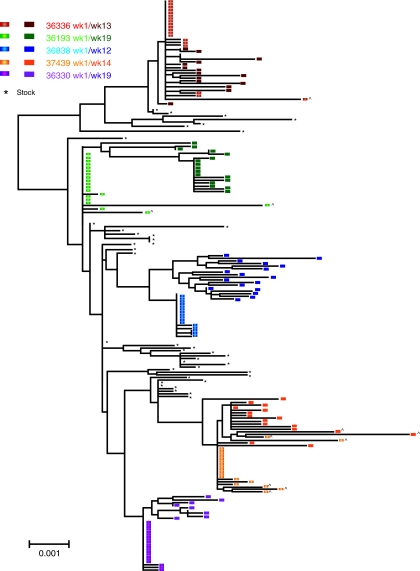
Based on these criteria, a single SIV env variant was detected in the ramp-up plasma of all five SIV-infected animals (Fig. 3). A composite neighbor-joining tree, illustrating the phylogenetic relationship of all SIV env sequences derived from the SIV stock and all five animals, demonstrates that the SIV env variants transmitted to the animals were widely distributed among the env variants in the stock with no overrepresentation of any specific env lineage in the stock (Fig. 3). Furthermore, the set point phase variants in plasma clearly arose from the founder variants detected at week 1 PI (Fig. 3) with no evidence of the emergence of novel variants that could have been missed in the week 1 PI plasma. This result supports the conclusion that each of the five animals was infected with a single SIVmac251 env variant that then gave rise to all subsequent viral variants detected at the set point stage of infection.
FIG. 3.
Composite neighbor-joining tree of SGA env sequences in the SIVmac251 virus stock and plasma SGA SIVenv sequences from five male rhesus macaques infected after penile SIV inoculation. Variants from individual animals are represented as different colors within the phylogenetic tree as denoted in the key. The sequences amplified from week 1 plasma (7 or 10 days PI) are denoted by an elliptical symbol and are lighter shaded whereas set point plasma sequences are denoted by square symbols and are darker shaded. Black asterisk (*) indicates sequences from the (UCD 6/04) SIVmac251 stock. G-to-A mutations in the sequences are indicated with (^). The scale bar represents 0.001 nucleotide substitutions per site.
Discussion
This study demonstrates that SIV can be transmitted to male rhesus macaques by immersing the glans and shaft of the penis in a suspension of cell-free virions. Although penile transmission of SIV is possible, it is not as efficient as vaginal SIV transmission. For example, when exposed to an SIVmac251 inoculum containing 103 TCID50, four males resisted infection despite 7–14 penile exposures, while seven of eight female macaques became infected after 14 vaginal exposures and all eight were infected by 16 challenges.10 Furthermore, 21 of 21 female rhesus macaques became infected after two vaginal challenges (separated by a 4 h interval) in a single day with 105 TCID50 of SIVmac251,9 while only three of five male macaques became infected after penile exposure to the same inoculum and schedule (Table 1, Fig. 1). The relative inefficiency of penile SIV transmission is consistent with the epidemiologic studies in stable HIV-discordant couples that concluded that women are more susceptible to acquiring HIV per heterosexual act than men.27–29,66 The reasons for the difference in gender susceptibility remain to be determined, but as shown here these differences persist even if the doses of virus to which the genders are exposed are the same. It seems likely that differences in the relative surface area of the genital mucosa (the surface area of the vaginal mucosa is much greater than the surface area of the glans and foreskin), the relative density of susceptible target cells in the genital mucosa of men and women, and the time that the inoculum is in contact with the genital mucosa contribute to the differences in the efficiency of HIV transmission between the genders.
In heterosexual men, the presence of an intact foreskin is associated with an increased risk of HIV acquisition30,34–36,67,68 and the efficiency of penile SIV transmission would likely increase if an effort was made to directly inoculate the foreskin mucosa. In addition, HIV transmission to men is dramatically increased by the presence of genital ulcers69; thus experimentally inducing penile inflammation prior to SIV inoculation may be another strategy to enhance penile SIV transmission efficiency while also modeling populations at greatest risk of acquiring HIV.
Transmission of HIV is relatively inefficient compared to other sexually transmitted infections (STI), which is reflected in the fact that in most cases a single variant or only a few variants from the viral quasispecies in an HIV-infected person successfully establishes systemic infection.70–73 As systemic infection with a single variant occurred in five of five animals after penile SIVmac251 transmission, this route of experimental SIV inoculation accurately reflects the virology of HIV transmission. In fact, when infection is transmitted, single SIV env variants establish systemic infections more consistently after penile SIVmac251 exposure compared to vaginal SIVmac251 challenge. After vaginal challenge of randomly selected mature cycling multiparous rhesus macaques with a complex SIVmac251 quasispecies11,17 some animals became infected with one variant, but in about half of the animals, systemic infection was established by five or more unique variants.11 The transmission of multiple variants to some female rhesus may be due to the changes in physiology of the female reproductive tract during the menstrual cycle or the presence of vaginal inflammation at the time of inoculation. Although ulcerative STIs have not been described in macaques, female rhesus monkeys commonly have bacterial vaginosis74,75 that causes genital inflammation.76 Alternatively, the same factors (discussed above) that may make women relatively more susceptible to HIV infection may also increase the number of variants that are transmitted by vaginal SIV inoculation. It remains to be seen if experimentally induced inflammation alters the number of variants transmitted by penile SIV inoculation.
The genetic bottleneck that results in a single viral variant establishing an infection after HIV transmission43,52,70,71,77–79 could be due to a the presence of only a limited number of viral variants in secretions of the infected partner, selective transmission of viral variants, or selective amplification of viral variants that cross the mucosal barrier. Although there could be a reduction in overall diversity in mucosal secretions compared to plasma, this penile infection study is not consistent with first explanation since all five animals inoculated with the genetically diverse SIVmac251 quasispecies became infected with a unique, single virus variant. It should be noted that analyzing 20 SGA-derived sequences provides a 95% confidence estimate that variants representing greater than 15% of the population are not be missed.43 Thus while the most likely conclusion is that all five animals were infected with a single variant we cannot exclude the possibility an individual animal has more than one variant in the plasma sample tested.
Despite this caveat, this is now the fourth study to reach the conclusion that rhesus macaques became infected with a single env variant after mucosal SIVmac251 exposure; the other studies tested animals after vaginal and rectal SIVmac 251 inoculation.11,17,56 Further, because all variants at set-point were derived from the variants we identified at ramp-up, all biologically relevant variants seem to have been found with our approach. Determining the relative importance of selective transmission versus selective amplification of a single variant prior to systemic dissemination will be important for understanding the challenges associated with preventing HIV transmission through vaccination. In some HIV clades, the transmitted HIV env variants share features that presumably enhance transmission to a seronegative host, but they also make the transmitted variants relatively susceptible to antibody neutralization.80 In this case preexisting antibodies may be able to efficiently block infection by the founder viruses. On the other hand, if the neutralization-sensitive variants appear as the founder population in blood by outcompeting less fit viruses for limited target cells in the first few days after transmission, then a neutralization-resistant variants may emerge as the primary founder viruses in the presence of established vaccine-induced neutralizing antibodies.
Finally, although infection is established relatively inefficiently by penile SIV inoculation, these data are consistent with the epidemiology of HIV transmission in human populations and represent the first use of this route in NHP models. Even after penile inoculations with a high virus dose the pattern of viremia and the single variant founder SIV populations that establish infection accurately reflect the virology of HIV infection. Given the consistency of infection, this route of SIV inoculation can be used to model penile HIV transmission to better understand virus transmission and dissemination. Furthermore, this route of virus challenge will be very useful for accurately modeling HIV transmission in the preclinical testing of HIV vaccines.
Acknowledgments
The authors thank the Primate Services Unit at the CNPRC and Joseph Dutra, Ding Lu, and Lili Guo for excellent technical assistance. This work was supported by NIH Grants RR00169 and AI8227, National Cancer Institute contract no. HHSN266200400088C, and a gift from the James B. Pendleton Charitable Trust.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Miller C. Vogel P. Dandekar S, et al. Localization of SIV in the reproductive tract of rhesus macaques: Relationship to the distribution of T cells and macrophages. Paper presented at the VII International Conference on AIDS; Florence, Italy. 1991. [Google Scholar]
- 2.Miller CJ. Use of the SIV/rhesus macaque model of the heterosexual transmission of HIV in vaccine research. Vaccine Res. 1992;1:295–301. [Google Scholar]
- 3.Miller CJ. Mucosal transmission of SIV. In: Desrosiers RC, editor; Letvin NL, editor. Current Topics in Microbiology and Immunology. Vol. 188. Springer-Verlag; Berlin: 1994. pp. 107–122. [DOI] [PubMed] [Google Scholar]
- 4.Miller CJ. Alexander NJ. Sutjipto S, et al. Genital mucosal transmission of simian immunodeficiency virus: Animal model for heterosexual transmission of human immunodeficiency virus. J Virol. 1989;63:4277–4284. doi: 10.1128/jvi.63.10.4277-4284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller CJ. Alexander NJ. Vogel P, et al. Mechanism of genital transmission of SIV: A hypothesis based on transmission studies and the location of SIV in the genital tract of chronically infected female rhesus macaques. J Med Primatol. 1992;21:64–68. [PubMed] [Google Scholar]
- 6.Miller CJ. Marthas M. Torten J, et al. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J Virol. 1994;68:6391–6400. doi: 10.1128/jvi.68.10.6391-6400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CJ. McChesney MB. Lu X, et al. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic SIVmac239. J Virol. 1997;71(3):1911–1921. doi: 10.1128/jvi.71.3.1911-1921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CJ. Shattock RJ. Target cells in vaginal HIV transmission. Microbes Infect. 2003;5:59–67. doi: 10.1016/s1286-4579(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 9.Miller CJ. Li Q. Abel K, et al. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005;79(14):9217–9227. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma ZM. Abel K. Rourke T, et al. A period of transient viremia and occult infection precedes persistent viremia and antiviral immune responses during multiple low-dose intravaginal simian immunodeficiency virus inoculations. J Virol. 2004;78:14048–14052. doi: 10.1128/JVI.78.24.14048-14052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone M. Keele BF. Ma ZM, et al. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J Virol. 2010;84(14):7083–7095. doi: 10.1128/JVI.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J. Gardner MB. Miller CJ. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol. 2000;74:6087–6095. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couedel-Courteille A. Butor C. Juillard V, et al. Dissemination of SIV after rectal infection preferentially involves paracolic germinal centers. Virology. 1999;260:277–294. doi: 10.1006/viro.1999.9809. [DOI] [PubMed] [Google Scholar]
- 14.Cranage MP. Baskerville A. Ashworth LAE, et al. Intrarectal challenge of macaques vaccinated with formalin-inactivated simian immunodeficiency virus. Lancet. 1992;339:273–274. doi: 10.1016/0140-6736(92)91335-6. [DOI] [PubMed] [Google Scholar]
- 15.Cranage MP. Whatmore AM. Sharpe SA, et al. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology. 1997;229(1):143–154. doi: 10.1006/viro.1996.8419. [DOI] [PubMed] [Google Scholar]
- 16.Crowley-Nowick PA. Bell MC. Brockwell R, et al. Rectal immunization for induction of specific antibody in the genital tract of women. J Clin Immunol. 1997;17(5):370–379. doi: 10.1023/a:1027312223474. [DOI] [PubMed] [Google Scholar]
- 17.Keele BF. Li H. Learn GH, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206(5):1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlowski PA. Cu-Uvin S. Neutra MR, et al. Comparison of the oral, rectal, and vaginal immunization routes for induction of antibodies in rectal and genital tract secretions of women. Infect Immun. 1997;65:1387–1394. doi: 10.1128/iai.65.4.1387-1394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlowski PA. Williams SB. Lynch RM, et al. Differential induction of mucosal and systemic antibody responses in women after nasal, rectal, or vaginal immunization: Influence of the menstrual cycle. J Immunol. 2002;169:566–574. doi: 10.4049/jimmunol.169.1.566. [DOI] [PubMed] [Google Scholar]
- 20.Kuller L. Beneviste RE. Clark EA, et al. Successful intrarectal inoculation of two macaque species with SIVmne CL E11S. Paper presented at the 10th Annual Symposium on Nonhuman Primate Models for AIDS; San Juan, Puerto Rico. 1992. [Google Scholar]
- 21.Kuller L. Benveniste RE. Tsai CC, et al. Intrarectal inoculation of macaques by the simian immunodeficiency virus, SIVmne E11S: CD4+ depletion and AIDS. J Med Primatol. 1994;23(7):397–409. doi: 10.1111/j.1600-0684.1994.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 22.Polacino P. Stallard V. Montefiori DC, et al. Protection of macaques against intrarectal infection by a combination immunization regimen with recombinant simian immunodeficiency virus SIVmne gp160 vaccines. J Virol. 1999;73(4):3134–3146. doi: 10.1128/jvi.73.4.3134-3146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall HI. Espinoza L. Benbow N, et al. Epidemiology of HIV infection in large urban areas in the United States. PLoS One. 2010;5(9):e12756. doi: 10.1371/journal.pone.0012756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abu-Raddad LJ. Hilmi N. Mumtaz G, et al. Epidemiology of HIV infection in the Middle East and North Africa. AIDS. 2010;24(Suppl 2):S5–23. doi: 10.1097/01.aids.0000386729.56683.33. [DOI] [PubMed] [Google Scholar]
- 25.Wilson CM. Wright PF. Safrit JT, et al. Epidemiology of HIV infection and risk in adolescents and youth. J Acquir Immune Defic Syndr. 2010;54(Suppl 1):S5–6. doi: 10.1097/QAI.0b013e3181e243a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan S. Dunbar MS. Minnis AM, et al. Poverty, gender inequities, and women's risk of human immunodeficiency virus/AIDS. Ann NY Acad Sci. 2008;1136:101–110. doi: 10.1196/annals.1425.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padian N. Marquis L. Francis DP, et al. Male to female transmission of human immunodeficiency virus. J Am Med Assoc. 1987;258:788–790. [PubMed] [Google Scholar]
- 28.Padian N. Shiboski S. Glass S, et al. Heterosexual transmission of human immunodeficiency virus (HIV) in Northern California: Results of a ten year study. Am J Epidemiol. 1997;146:350–357. doi: 10.1093/oxfordjournals.aje.a009276. [DOI] [PubMed] [Google Scholar]
- 29.Padian NS. Shiboski SC. Jewell NP. Female to male transmission of human immunodeficiency virus. J Am Med Assoc. 1991;266:1664–1667. [PubMed] [Google Scholar]
- 30.Turner AN. Morrison CS. Padian NS, et al. Men's circumcision status and women's risk of HIV acquisition in Zimbabwe and Uganda. AIDS. 2007;21(13):1779–1789. doi: 10.1097/QAD.0b013e32827b144c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auvert B. Taljaard D. Lagarde E, et al. Randomized, controlled intervention trial of male circumcision for reduction of HIV infection risk: The ANRS 1265 Trial. PLoS Med. 2005;2(11):e298. doi: 10.1371/journal.pmed.0020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey RC. Moses S. Parker CB, et al. Male circumcision for HIV prevention in young men in Kisumu, Kenya: A randomised controlled trial. Lancet. 2007;369(9562):643–656. doi: 10.1016/S0140-6736(07)60312-2. [DOI] [PubMed] [Google Scholar]
- 33.Gray RH. Kigozi G. Serwadda D, et al. Male circumcision for HIV prevention in men in Rakai, Uganda: A randomised trial. Lancet. 2007;369(9562):657–666. doi: 10.1016/S0140-6736(07)60313-4. [DOI] [PubMed] [Google Scholar]
- 34.Hallett TB. Singh K. Smith JA, et al. Understanding the impact of male circumcision interventions on the spread of HIV in southern Africa. PLoS One. 2008;3(5):e2212. doi: 10.1371/journal.pone.0002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreiss JK. Hopkins SG. The association between circumcision status and human immunodeficiency virus infection among homosexual men. J Infect Dis. 1993;168:1404–1408. doi: 10.1093/infdis/168.6.1404. [DOI] [PubMed] [Google Scholar]
- 36.Sidler D. Smith J. Rode H. Neonatal circumcision does not reduce HIV/AIDS infection rates. S Afr Med J. 2008;98(10):762–764. , 766. [PubMed] [Google Scholar]
- 37.Hirbod T. Bailey RC. Agot K, et al. Abundant expression of HIV target cells and C-type lectin receptors in the foreskin tissue of young Kenyan men. Am J Pathol. 2010;176(6):2798–2805. doi: 10.2353/ajpath.2010.090926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoombe SG. Short RV. Potential HIV-1 target cells in the human penis. AIDS. 2006;20(11):1491–1495. doi: 10.1097/01.aids.0000237364.11123.98. [DOI] [PubMed] [Google Scholar]
- 39.Donoval BA. Landay AL. Moses S, et al. HIV-1 target cells in foreskins of African men with varying histories of sexually transmitted infections. Am J Clin Pathol. 2006;125(3):386–391. [PubMed] [Google Scholar]
- 40.Palmer S. Kearney M. Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salazar-Gonzalez JF. Bailes E. Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HY. Giorgi EE. Keele BF, et al. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261(2):341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrahams MR. Anderson JA. Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83(8):3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haaland RE. Hawkins PA. Salazar-Gonzalez J, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kearney M. Maldarelli F. Shao W, et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83(6):2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salazar-Gonzalez JF. Salazar MG. Keele BF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Bar KJ. Wang S, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6(5):e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bar KJ. Li H. Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84(12):6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagar M. Lavreys L. Baeten JM, et al. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J Virol. 2003;77(23):12921–12926. doi: 10.1128/JVI.77.23.12921-12926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganeshan S. Dickover RE. Korber BT, et al. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu T. Mo H. Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 53.Greenier JL. Miller CJ. Lu D, et al. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J Virol. 2001;75(8):3753–3765. doi: 10.1128/JVI.75.8.3753-3765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lifson JD. Nowak MA. Goldstein S, et al. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowak MA. Lloyd AL. Vasquez GM, et al. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 1997;71(10):7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J. Keele BF. Li H, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol. 2010;84(19):10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shattock RJ. Moore JP. Inhibiting sexual transmission of HIV-1 infection. Nat Rev Micro. 2003;1(1):25–34. doi: 10.1038/nrmicro729. [DOI] [PubMed] [Google Scholar]
- 58.Wawer MJ. Gray RH. Sewankambo NK, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191(9):1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 59.Marthas ML. Lu D. Penedo MC, et al. Titration of an SIVmac251 stock by vaginal inoculation of Indian and Chinese origin rhesus macaques: Transmission efficiency, viral loads, and antibody responses. AIDS Res Hum Retroviruses. 2001;17:1455–1466. doi: 10.1089/088922201753197123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clamp M. Cuff J. Searle SM, et al. The Jalview Java alignment editor. Bioinformatics. 2004;20(3):426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 62.Felsenstein J. Confidence on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 63.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 64.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 65.Giorgi EE. Funkhouser B. Athreya G, et al. Estimating time since infection in early homogeneous HIV-1 samples using a poisson model. BMC Bioinform. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicolosi A. Correa Leite ML. Musicco M, et al. The efficiency of male-to-female and female-to-male sexual transmission of the human immunodeficiency virus: A study of 730 stable couples. Italian Study Group on HIV Heterosexual Transmission. Epidemiology. 1994;5(6):570–575. doi: 10.1097/00001648-199411000-00003. [DOI] [PubMed] [Google Scholar]
- 67.Gray RH. Serwadda D. Kong X, et al. Male circumcision decreases acquisition and increases clearance of high-risk human papillomavirus in HIV-negative men: A randomized trial in Rakai, Uganda. J Infect Dis. 2010;201(10):1455–1462. doi: 10.1086/652184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tobian AA. Serwadda D. Quinn TC, et al. Male circumcision for the prevention of HSV-2 and HPV infections and syphilis. N Engl J Med. 2009;360(13):1298–1309. doi: 10.1056/NEJMoa0802556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gray RH. Wawer MJ. Brookmeyer R, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 70.Yuste E. Sanchez-Palomino S. Casado C, et al. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novella IS. Quer J. Domingo E, et al. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J Virol. 1999;73(2):1668–1671. doi: 10.1128/jvi.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duarte EA. Novella IS. Weaver SC, et al. RNA virus quasispecies: Significance for viral disease and epidemiology. Infect Agents Dis. 1994;3:201–214. [PubMed] [Google Scholar]
- 73.Fischer W. Ganusov VV. Giorgi EE, et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One. 2010;5(8):e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spear GT. Gilbert D. Landay AL, et al. Pyrosequencing of genital microbiota in HIV-seropositive and -seronegative women shows Lactobacillus iners as the predominant Lactobacillus species. Appl Environ Microbiol. 2011;77(1):378–381. doi: 10.1128/AEM.00973-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doyle L. Young CL. Jang SS, et al. Normal vaginal aerobic and anaerobic bacterial flora of the rhesus macaque (Macaca mulatta) J Med Primatol. 1991;20(8):409–413. [PubMed] [Google Scholar]
- 76.Thurman AR. Doncel GF. Innate immunity and inflammatory response to Trichomonas vaginalis and bacterial vaginosis: Relationship to HIV acquisition. Am J Reprod Immunol. 2011;65(2):89–98. doi: 10.1111/j.1600-0897.2010.00902.x. [DOI] [PubMed] [Google Scholar]
- 77.Domingo E. Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 78.Zhu T. Wang N. Carr A, et al. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: Evidence for virus compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolinsky SM. Wike CM. Korber BT, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255(5048):1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 80.Derdeyn CA. Decker JM. Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]