Abstract
Although there is considerable evidence that PrPSc is the infectious form of the prion protein, it has recently been proposed that a transmembrane variant called CtmPrP is the direct cause of prion-associated neurodegeneration. We report here, using a mutant form of PrP that is synthesized exclusively with the CtmPrP topology, that CtmPrP is retained in the endoplasmic reticulum and is degraded by the proteasome. We also demonstrate that CtmPrP contains an uncleaved, N-terminal signal peptide as well as a C-terminal glycolipid anchor. These results provide insight into general mechanisms that control the topology of membrane proteins during their synthesis in the endoplasmic reticulum, and they also suggest possible cellular pathways by which CtmPrP may cause disease.
INTRODUCTION
Prion diseases are fatal neurodegenerative disorders characterized by dementia, ataxia, and cerebral spongiosis. A recent epidemic of bovine spongiform encephalopathy in the United Kingdom and the likely transmission of this disease to human beings has focused public attention on the origin and transmission of prion disorders (Collinge, 1999). Infectious, inherited, and sporadic forms of these diseases are all due to conformational conversion of a normal cell-surface glycoprotein called PrPC 1, expressed in neurons and glia, to a protease-resistant isoform denoted PrPSc (Harris, 1999; Prusiner, 1999). A great deal of evidence has accumulated indicating that PrPSc is infectious in the absence of nucleic acids, and that it is the principal component of infectious prion particles. It is also commonly assumed that PrPSc is the primary cause of neurodegeneration, based on the spatial and temporal correlation between the accumulation of this isoform and the degree of neuronal damage during the course of prion diseases (DeArmond and Ironside, 1999).
Recently, however, an alternative topological variant of PrP called CtmPrP has been proposed as a key intermediate in infectious and inherited forms of prion disease. Whereas most molecules of PrP are anchored to the cell membrane exclusively by a C-terminal glycosyl-phosphatidylinositol (GPI) anchor (Lehmann and Harris, 1995), CtmPrP spans the membrane once via a conserved, hydrophobic segment encompassing residues 111–134, with the C terminus on the exofacial surface (Hegde et al., 1998a). A third topological variant, denoted NtmPrP, spans the membrane via the same hydrophobic domain, but in the opposite orientation (N terminus on the exofacial surface) (Hegde et al., 1998a). Transmembrane forms of PrP were originally observed after in vitro translation of PrP mRNA on rabbit reticulocyte or wheat germ ribosomes in the presence of canine pancreatic microsomes (Hay et al., 1987a,b; De Fea et al., 1994). There is evidence that the relative proportions of the three topological variants are determined by as yet unidentified accessory proteins present during the translation process, as well as by a region of nine hydrophilic acids (the “stop transfer effector”) adjacent to the transmembrane domain of PrP (Lopez et al., 1990; Yost et al., 1990; Hegde et al., 1998b). However, the precise mechanism by which transmembrane forms are produced is still unclear.
Several pieces of evidence have implicated CtmPrP in the pathogenesis of prion diseases. First, transgenic mice were created that express PrP molecules carrying mutations in or near the transmembrane domain that favor formation of CtmPrP (Hegde et al., 1998a, 1999). Mice that produced CtmPrP above a threshold level developed a spontaneous neurological disease with scrapie-like features, but without detectable PrPSc. In addition, when these mice were inoculated with scrapie prions, the amount of PrPSc that accumulated was inversely related to the amount of CtmPrP present, indicating that CtmPrP rather than PrPSc may be the proximate cause of neurodegeneration (Hegde et al., 1999). Finally, after scrapie inoculation of mice that carried a wild-type hamster PrP transgene that served as a reporter of CtmPrP formation, CtmPrP was found to accumulate during the course of the infection (Hegde et al., 1999). Taken together, these data have been interpreted to suggest that CtmPrP is the direct cause of neurodegeneration in familial and infectious prion diseases, and that PrPSc acts indirectly by increasing the amount of CtmPrP.
It remains unknown, however, how CtmPrP might affect cellular physiology and result in neuronal death and central nervous system pathology. Part of the difficulty in addressing this issue has been that it has not been possible to produce CtmPrP in cells in the absence of the other topological variants. Thus, even basic questions such as the subcellular localization of this isoform remain unanswered.
We report here on the identification of a mutant form of PrP that is synthesized exclusively with the CtmPrP topology, and our use of cells expressing this protein to investigate the cell biology and metabolism of CtmPrP. In addition, we have discovered that CtmPrP contains an uncleaved, N-terminal signal peptide as well as a C-terminal GPI anchor, observations that provide clues to how this form is generated during the translation process.
MATERIALS AND METHODS
PrP Plasmids and mRNA Synthesis
All mouse PrP cDNAs were cloned into the vector pcDNA3 (Invitrogen, Carlsbad, CA), and carried an epitope tag for monoclonal antibody 3F4 created by changing residues 108 and 111 to methionine. The following mutations were introduced into the wild-type PrP cDNA by using the polymerase chain reaction as described previously (Lehmann and Harris, 1995): L9R, A116V, 3AV (A→V at 112, 114, and 117), and L9R/3AV. The FLAG epitope (DYKDDDDK) was inserted between residues 22 and 23 by using the polymerase chain reaction. Plasmids were linearized with XbaI and gel-purified. In vitro transcriptions were performed with the mMessage mMachine T7 kit (Ambion, Austin, TX).
In Vitro Translation
Messenger RNAs were translated in the presence of [35S]methionine (Promix; Amersham Pharmacia Biotech, Pitcataway, NJ) by using rabbit reticulocyte lysate (Promega, Madison, WI) as directed by the manufacturer, except that the final lysate concentration was 50%. Translation reactions also contained microsomal membranes prepared from canine pancreas (Promega) or from murine BW5174.3 thymoma cells (Vidugiriene and Menon, 1995). To detect protease-protected products, 5-μl aliquots of translation reactions were incubated in a final volume of 50 μl with 100 μg/ml proteinase K (PK) (Roche Molecular Biochemicals, Indianapolis, IN) in 50 mM Tris-HCl (pH 7.5) and 1 mM CaCl2 for 60 min at 4°C, followed by addition of 5 mM phenylmethylsulfonyl fluoride (PMSF) to terminate digestion. Some digestion reactions also contained 0.5% Triton-X 100 to solubilize membranes. Samples were then analyzed by SDS-PAGE and autoradiography.
Immunoprecipitation
Aliquots of the translation reactions were boiled in the presence of 1% SDS for 5 min to denature the proteins, and were then diluted with 10 volumes of RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) containing 1 mM CalCl2 plus protease inhibitors (1 μg/ml pepstatin A, 1 μg/ml leupeptin, 5 mM PMSF). Anti-PrP antibody P45-66 (Lehmann and Harris, 1995), or anti-FLAG monoclonal antibodies M1 or M2 (Sigma, St. Louis, MO), were added and samples incubated on ice for 90 min. Protein A-Sepharose beads (for P45-66 and M1) or protein G-agarose beads (for M2) were added, and samples were rotated at 4°C for 30 min. Beads were collected by low-speed centrifugation and washed three times with RIPA buffer plus 1 mM CaCl2, after which proteins were eluted with sample buffer, and analyzed by SDS-PAGE.
Transfected Cells
Baby hamster kidney (BHK) and Chinese hamster ovary (CHO) cells were maintained in α-Eagle's minimum essential medium supplemented with 10% fetal calf serum, nonessential amino acids, and penicillin/streptomycin. Transfections were performed with Lipofectamine (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. Cells were harvested 24 h after transfection by scraping or by brief trypsinization, rinsed twice with phosphate-buffered saline (PBS), and resuspended in 0.25 M sucrose, 10 mM HEPES (pH 7.4), 1 μg/ml pepstatin A, 1 μg/ml leupeptin. Cells were disrupted by 10 passages through silastic tubing (0.3 mm i.d.) connecting two syringes with 27-gauge needles. A postnuclear supernatant was prepared by centrifugation at 2500 × g for 2 min. PK protection assays were performed by incubating postnuclear supernatants in 50 mM Tris-HCl (pH 7.5), 250 μg/ml PK, and in some cases 0.5% Triton X-100. After 60 min at 22°C, digestion was terminated by addition of 5 mM PMSF and samples were treated with peptide-N-(acetyl-β-glucosaminyl)asparagine amidase (PNGase F) (New England Biolabs, Beverly, MA) and analyzed by Western blotting with anti-PrP antibody 3F4 (Bolton et al., 1991).
To test the glycosidase sensitivity of PrP, lysates of transfected cells prepared in 0.5% Triton X-100, 0.5% deoxycholate, 50 mM Tris-HCl (pH 7.5) were treated with endoglycosidase H (New England Biolabs) or PNGase F according to the manufacturer's directions before methanol precipitation and Western blotting with 3F4.
To test the effect of proteasome inhibitors, transfected cells were treated for 18 h with 20 μM of Z-Ile-Glu(OtBu)-Ala-Leu-CHO (proteasome inhibitor 1 [PSI 1]; Calbiochem, San Diego, CA). Cells were lysed in 0.5% SDS, 50 mM Tris-HCl (pH 7.5), and proteins were analyzed by Western blotting with 3F4 and anti-actin antibodies.
GPI Anchor Analysis
Transiently transfected cells were labeled for 16 h with either 3.5 mCi/ml [3H]palmitic acid (American Radiolabeled Chemicals, St. Louis, MO.) or 250 μCi/ml [35S]methionine. After lysis in 0.5% SDS, 50 mM Tris-HCl (pH 7.5), samples were diluted with 5 volumes of 0.5% Triton X-100, 50 mM Tris-HCl (pH 7.5) and incubated for 16 h at 37°C with PNGase F in the presence or absence of phosphatidylinositol-specific phospholipase C (PIPLC) (1 unit/ml) from Bacillus thuringiensis. PrP was then immunoprecipitated with 3F4 antibody and analyzed by SDS-PAGE.
Immunofluorescence Microscopy
Transiently transfected cells grown on glass coverslips were fixed for 1 h in 4% paraformaldehyde in PBS, and then permeabilized for 2 min in 0.5% Triton X-100 in PBS. After treatment for 30 min in 2% goat serum/PBS (blocking solution), cells were incubated for 1 h with primary antibodies in blocking solution (P45-66 and mouse anti-protein disulfide isomerase [Stressgen]), washed, and then incubated for 1 h with fluorescently labeled secondary antibodies in blocking solution (Alexa-488–coupled anti-rabbit IgG and Alexa-594–coupled anti-mouse IgG; Molecular Probes, Eugene, OR). Coverslips were then mounted in 50% glycerol/PBS and viewed with a Zeiss Axioplan fluorescence microscope equipped with a Bio-Rad MRC1024 laser confocal scanning system. To selectively visualize surface PrP, living cells were stained with 3F4 antibody in Opti-MEM (Life Technologies) plus 2% goat serum, washed, fixed in 4% paraformaldehyde, and then incubated with Alexa-488–coupled anti-mouse IgG.
RESULTS
CtmPrP Contains an Uncleaved, N-Terminal Signal Peptide
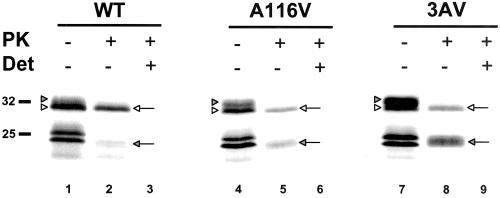
When PrP mRNA is translated in vitro by using rabbit reticulocyte lysate supplemented with canine pancreatic microsomes, products of ∼32 and ∼25 kDa are synthesized, corresponding to core-glycosylated and untranslocated/unglycosylated PrP, respectively (Figure 1, lanes 1, 4, and 7). Incubating microsomes with PK to cleave off the cytoplasmically exposed domains of newly synthesized PrP molecules resulted in the appearance of two protease-protected species (lanes 2, 5, and 8): a 32-kDa form (SecPrP) that corresponds to intact, fully translocated chains, and a 24-kDa fragment that corresponds to the transmembrane and lumenal domains of CtmPrP. The latter fragment is distinct from untranslocated/unglycosylated PrP, which has a slightly larger molecular size, and is not present in lanes 2, 5, and 8 because it is completely degraded by the protease. As reported previously (Hegde et al., 1998a, 1999), the presence of either of two mutations (A116V or A112V/A114V/A117V, referred to as 3AV) in the transmembrane domain significantly increased the proportion of CtmPrP (lanes 5 and 8). No PrP was detected after PK treatment in the presence of Triton X-100 detergent, which disrupts the microsomal membrane (lanes 3, 6, and 9), confirming that the 32- and 24-kDa fragments do not represent intrinsically protease-resistant portions of the molecule.
Figure 1.
Mutations in the transmembrane region increase the proportion of CtmPrP, and reveal that this form is slightly larger than SecPrP. mRNA encoding wild-type (WT), A116V, or 3AV PrP was translated in rabbit reticulocyte lysate supplemented with canine pancreatic microsomes. Aliquots of the reaction were then incubated with (lanes 2, 3, 5, 6, 8, and 9) or without (lanes 1, 4, and 7) PK in the presence (lanes 3, 6, and 9) or absence (lanes 1, 2, 4, 5, 7, and 8) of Triton X-100 (Det). Samples were then analyzed by SDS-PAGE and autoradiography. Note the presence of a closely spaced doublet of glycosylated PrP in lanes 1, 4, and 7, corresponding to SecPrP (white arrowheads) and CtmPrP (shaded arrowheads). The protease-protected forms of SecPrP and CtmPrP are indicated by the white and shaded arrows, respectively, in lanes 2, 5, and 8. Molecular size markers are given in kilodaltons.
We noticed the presence of a 33-kDa glycosylated product that was present before PK digestion and that could be resolved from the 32-kDa band corresponding to SecPrP (Figure 1, lanes 1, 4, and 7). The amount of this 33-kDa species correlated with the amount of the 24-kDa CtmPrP fragment produced after PK digestion: it was present in largest amounts for 3AV, at intermediate levels for A116V, and in smallest amounts for wild-type PrP. This observation suggested that the 33-kDa species corresponded to full-length CtmPrP, which then gave rise to a 24-kDa protected fragment after PK digestion. To explain why full-length CtmPrP might be slightly larger than full-length SecPrP, we hypothesized that it might contain an uncleaved, N-terminal signal peptide.
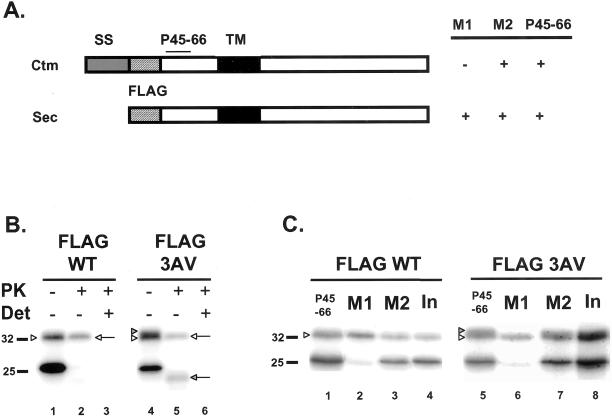
To test this hypothesis, we constructed wild-type and 3AV versions of PrP that contained an eight amino acid FLAG epitope (DYKDDDDK) inserted at the signal peptide cleavage site, between residues 22 and 23 (Figure 2A). An antibody designated M1 is available that recognizes the FLAG epitope only if it displays a free N terminus (Prickett et al., 1989), so that FLAG-tagged PrP will only react with M1 if the signal peptide has been cleaved. As positive controls, we used antibody M2, which recognizes the FLAG epitope regardless of its sequence context, as well as antibody P45-66, which reacts with the octapeptide repeat region of PrP (Lehmann and Harris, 1995). For these experiments we used microsomes derived from BW5174.3 murine thymoma cells because they are efficient at attaching GPI anchors to newly synthesized polypeptide chains, in contrast to microsomes from canine pancreas (Vidugiriene and Menon, 1995; Stewart and Harris, 2001). In addition, introduction of the FLAG epitope has relatively little effect on the proportions of SecPrP and CtmPrP translated with thymoma microsomes, whereas it increases the amount of CtmPrP produced with pancreatic microsomes (data not shown). It had been previously shown that introduction of a FLAG epitope at the signal peptide cleavage site of PrP did not interfere with signal peptide removal, oligosaccharide addition, or the ability of the protein to be converted to PrPSc in transgenic mice (Telling et al., 1997).
Figure 2.
Immunoprecipitation of FLAG-tagged PrP reveals that CtmPrP contains an uncleaved signal peptide. (A) Schematic of the immunoreactivity of FLAG-tagged CtmPrP and SecPrP. SS, signal sequence; TM, transmembrane domain. (B) In vitro translation and PK protection assays of FLAG-tagged wild-type (WT) or 3AV PrP were performed as in Figure 1, except that microsomes were from murine BW5174.3 thymoma cells. The white and shaded arrowheads indicate, respectively, the positions of SecPrP and CtmPrP before protease digestion (lanes 1 and 4); it is not possible to completely separate the FLAG-tagged versions of these two species. The white and shaded arrows indicate, respectively, the protease-protected forms of SecPrP and CtmPrP (lanes 2 and 5). Only SecPrP is visible for wild-type PrP. (C) FLAG-WT and FLAG-3AV PrPs were synthesized by in vitro translation, and were immunoprecipitated with anti-PrP antibody P45-66 (lanes 1 and 5), anti-FLAG antibody M1 (lanes 2 and 6), or anti-FLAG antibody M2 (lanes 3 and 7). Lanes 4 and 8 show samples before immunoprecipitation (Input). Note that SecPrP (white arrowheads) but not CtmPrP (shaded arrowhead) is immunoprecipitated with M1, whereas both forms are immunoprecipitated with P45-66 and M2.
We confirmed that FLAG-tagged PrP, like its untagged counterpart, gave rise to protected products corresponding to SecPrP (32 kDa) and CtmPrP (24 kDa) after PK digestion of microsomes, and that the amount of the CtmPrP product was increased by the presence of the 3AV mutation (Figure 2B, lanes 2 and 5). Before PK digestion, there was a broad band at 32–33 kDa, which was especially apparent for 3AV PrP (Figure 2B, lane 4), and which represents a combination of the 32- and 33-kDa species that we had seen after translation with pancreatic microsomes (Figure 1); these two products are not completely resolved from each other in this experiment, probably because of the small increment in size contributed by the FLAG epitope. When undigested samples were subjected to immunoprecipitation, we found that the M1 antibody reacted with the lower part of the 32–33-kDa band corresponding to SecPrP, but not the upper portion of the band corresponding to what we postulate is full-length CtmPrP (Figure 2C, compare lanes 6 and 8). As expected, the entire band (representing both SecPrP and full-length CtmPrP) was immunoprecipitated by antibodies P45-66 and M2 (Figure 2C, lanes 5 and 7). This result indicates that although the signal peptide has been removed from SecPrP, it is still present in CtmPrP. The 25-kDa unglycosylated form of PrP, which represents untranslocated molecules whose signal peptide would still be present, was not recognized by M1 (Figure 2C, lanes 2 and 6), a result that serves as a control for the specificity of the M1 antibody.
Our conclusion that CtmPrP contains an uncleaved signal peptide was confirmed by labeling in vitro translation products with either [3H]leucine or [35S]methionine. We observed that radiolabeling of the 33-kDa band corresponding to CtmPrP was much greater with [3H]leucine than with [35S]methionine (data not shown), consistent with the fact that the N-terminal signal sequence contains five leucine residues, whereas the rest of the polypeptide chain (excluding the C-terminal GPI signal sequence) contains only two leucine residues.
Creation of a Mutant That Is Synthesized Exclusively as CtmPrP
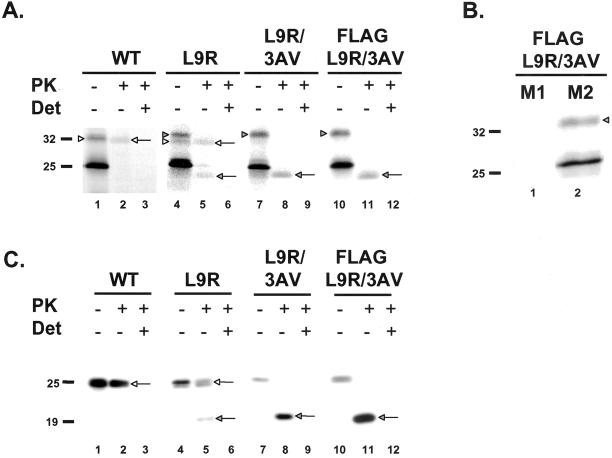
Previously identified mutations that have been found to alter the proportion of CtmPrP are all localized within or adjacent to the transmembrane domain (Hegde et al., 1998a, 1999). Given our conclusion that CtmPrP contains an uncleaved, N-terminal signal peptide, we reasoned that mutations in the signal peptide itself might also affect the amount of CtmPrP. We found that the substitution of a charged residue for a hydrophobic residue within the signal sequence (L9R) markedly increased the proportion of CtmPrP produced after in vitro translation to ∼50% (Figure 3A, lanes 4 and 5). Combining this mutation with one in the transmembrane domain to create L9R/3AV resulted in a protein that was synthesized exclusively as CtmPrP. Before protease treatment, the glycosylated form of this double mutant migrated at 33 kDa (Figure 3A, lane 7), and after PK digestion an equimolar amount of a 24-kDa protected fragment was produced without any SecPrP (lane 8). In experiments where the protected products were immunoprecipitated and enzymatically deglycosylated before SDS-PAGE, a protocol that facilitates detection of NtmPrP, we failed to observe any NtmPrP (not shown). When we translated a FLAG-tagged version of L9R/3AV, we observed that none of the mutant protein was immunoprecipitated with M1 antibody, confirming the conclusion that CtmPrP has an uncleaved signal sequence (Figure 3B, lane 1).
Figure 3.
Mutations in the signal sequence increase the proportion of CtmPrP. (A) In vitro translation and PK protection assays of wild-type and mutant PrPs were performed as in Figure 2. The full-length forms of SecPrP and CtmPrP are indicated by the white and shaded arrowheads, respectively (lanes 1, 4, 7, and 10). The protease-protected forms of SecPrP and CtmPrP are indicated by the white and shaded arrows, respectively (lanes 2, 5, 8, and 11). (B) FLAG-L9R/3AV PrP was synthesized by in vitro translation, and was immunoprecipitated with anti-FLAG antibodies M1 (lane 1) or M2 (lane 2). Neither CtmPrP (shaded arrowhead) nor untranslocated/unglycosylated PrP (25 kDa) are immunoprecipitated by M1, whereas both forms are immunoprecipitated by M2. (C) BHK cells were transiently transfected with plasmids encoding wild-type or mutant PrPs. Postnuclear supernatants prepared from cells 24 h after transfection were incubated with (lanes 2, 3, 5, 6, 8, 9, 11, and 12) or without (lanes 1, 4, 7, and 10) PK in the presence (lanes 3, 6, 9, and 12) or absence (lanes 1, 2, 4, 5, 7, 8, 10, and 11) of Triton-X-100 (Det). Proteins were then solubilized in SDS, deglycosylated with PNGase F, and subjected to Western blotting with 3F4 antibody. The protease-protected forms of SecPrP and CtmPrP are indicated by the white and shaded arrows, respectively.
We found that the L9R mutation altered the topology of PrP in cultured cells as it did after in vitro translation. By carrying out PK protection assays on postnuclear supernatants prepared from transfected BHK cells, we found that the L9R mutation increased the proportion of CtmPrP to ∼50% (Figure 3C, lanes 4 and 5), whereas both untagged and FLAG-tagged versions of L9R/3AV were synthesized entirely as CtmPrP (Figure 3C, lanes 7, 8, 10, and 11). (In these experiments, PrP was deglycosylated with PNGase F before Western blotting.) We also observed that FLAG-L9R/3AV PrP reacted with P46-66 antibody but not with M1 antibody, indicating that CtmPrP has an uncleaved signal peptide when synthesized in BHK cells (not shown). The availability of L9R/3AV PrP provided us for the first time the ability to analyze the properties of CtmPrP in a cellular context in the absence of the other two topological variants (SecPrP and NtmPrP).
CtmPrP Has a GPI Anchor
We tested whether CtmPrP has a GPI anchor by labeling BHK cells expressing FLAG-tagged L9R/3AV PrP with either [35S]methionine or the anchor precursor [3H]palmitate. PrP was then deglycosylated with PNGase F before immunoprecipitation. We found that the mutant protein was labeled with both compounds, and that the palmitate label was removed by treating samples with PIPLC, which cleaves the GPI anchor (Figure 4). Of note, the methionine-labeled protein is shifted upward on the gel by a small amount after phospholipase treatment, a phenomenon that is indicative of loss of the GPI anchor (Narwa and Harris, 1999). This experiment directly demonstrates that CtmPrP carries a C-terminal GPI anchor in addition to a transmembrane anchor.
Figure 4.
CtmPrP has a GPI anchor. Transfected BHK cells expressing FLAG-L9R/3AV PrP were metabolically labeled for 16 h with either [35S]methionine (top) or [3H]palmitate (bottom). Detergent lysates of the cells were treated with PNGase F in the presence (+ lane) or absence (− lane) of PIPLC. PrP was then immunoprecipitated with 3F4 antibody and analyzed by SDS-PAGE and autoradiography. The slightly reduced mobility of PrP after PIPLC treatment (top) is characteristic of PrP without its GPI anchor.
CtmPrP Is Retained in the Endoplasmic Reticulum (ER) and Is Degraded by the Proteasome
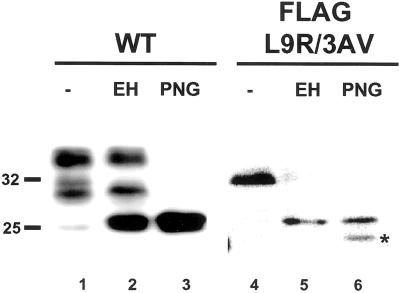
We noticed that FLAG-L9R/3AV PrP synthesized in BHK cells migrated on SDS-PAGE as a sharp band of ∼33 kDa, the same size as core-glycosylated PrP produced by in vitro translation (Figure 5, lane 4). In contrast, wild-type PrP displayed a sharp band of 25 kDa as well as two broad bands of 28–30 and 33–35 kDa, corresponding to unglycosylated, mono-, and diglycosylated forms (lane 1). This observation suggested the possibility that the oligosaccharide chains of the mutant protein were being only partially processed. In support of this idea, we found that FLAG-L9R/3AV PrP was quantitatively deglycosylated by endoglycosidase H, which acts only on high-mannose oligosaccharide chains added in the ER (lane 5). In contrast, only a small amount of wild-type PrP was susceptible to this enzyme (lane 2), probably representing molecules in transit to the cell surface. As expected, both proteins were fully deglycosylated by PNGase F, which cleaves both complex and high-mannose oligosaccharide chains (lanes 3 and 6). These results indicate that FLAG-L9R/3AV PrP, and thus CtmPrP, does not transit beyond the mid-Golgi stack where oligosaccharides become resistant to endoglycosidase H. Similar results were obtained with an untagged version of L9R/3AV (not shown).
Figure 5.
The oligosaccharide chains of CtmPrP are sensitive to digestion with endoglycosidase H. Detergent lysates of transfected BHK cells expressing wild-type or FLAG-L9R/3AV PrP were incubated without enzyme (lanes 1 and 4), with endoglycosidase H (lanes 2 and 5) or with PNGase F (lanes 3 and 6). Proteins were then precipitated with methanol and analyzed by Western blotting by using antibody 3F4. The band in lane 6 indicated by the asterisk is a proteolytic breakdown product.
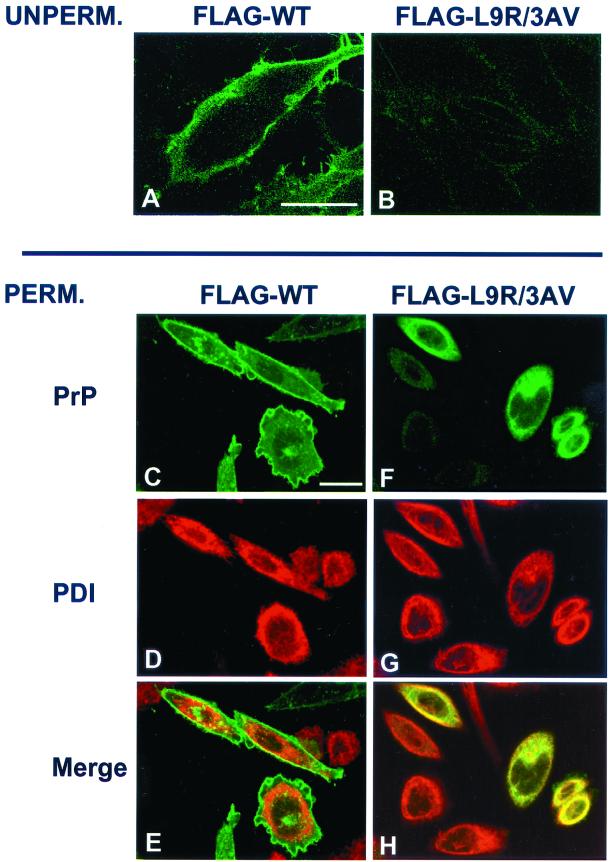
To directly determine the subcellular localization of CtmPrP, we analyzed transfected cells by immunofluorescence microscopy. When we stained unpermeabilized cells we found that, in contrast to FLAG-wild-type PrP, FLAG-L9R/3AV PrP was not detectable on the cell surface (Figure 6, A and B). When detergent-permeabilized cells were analyzed, we observed that FLAG-L9R/3AV PrP was distributed within the cell in a reticular pattern that largely colocalized with the ER marker protein disulfide isomerase (Figure 6, F–H), whereas FLAG-wild-type PrP was largely localized to the cell surface, with a small amount of internal punctate staining that corresponded to the Golgi (Figure 6, C–E). Results similar to those shown in Figure 6 were obtained when non-FLAG versions of wild-type and L9R/3AV PrP were used (not shown). We also observed that wild-type PrP but not L9R/3AV PrP could be labeled on intact cells using a membrane-impermeant biotinylation reagent, further confirming the absence of the mutant protein from the cell surface (data not shown).
Figure 6.
CtmPrP is retained in the ER. (A and B) Transfected BHK cells expressing FLAG-wild-type PrP (A) or FLAG-L9R/3AV PrP (B) were stained with 3F4 antibody without permeabilization to visualize PrP on the cell surface. The faint staining visible in B represents background, because it is also seen on untransfected cells (not shown). (C–H) Transfected CHO cells were fixed, permeabilized, and stained with antibodies to PrP (P45-66) and protein disulfide isomerase. After treatment with secondary antibodies, cells were viewed by laser scanning confocal microscopy to visualize PrP (green, C and F) or protein disulfide isomerase (red, D and G). E and H show a merge of the PrP and protein disulfide isomerase staining patterns. The punctate intracellular staining for PrP seen in C and E colocalizes with a Golgi marker (data not shown). Bars, 25 μm.
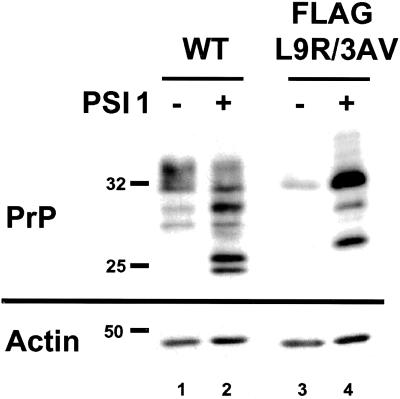
Because proteins that are trapped in the ER are often subject to degradation by the proteasome as part of a quality control process (Ellgaard et al., 1999), we tested whether this was true for FLAG-L9R/3AV PrP. We found that PSI 1 [Z-Ile-Glu(OtBu)-Ala-Leu-CHO], a reversible aldehyde inhibitor of the proteasome, caused a marked accumulation of the mutant protein in transfected cells, indicating that CtmPrP is subject to proteasomal degradation (Figure 7, lanes 3 and 4). Similar results were obtained with lactacystin, an irreversible proteasome inhibitor, and with untagged L9R/3AV (not shown). PSI 1 also caused a change in the amount and glycoform distribution of wild-type PrP (lanes 1 and 2), indicating that SecPrP is also degraded by the proteasome (Drisaldi, Stewart, and Harris, unpublished data).
Figure 7.
CtmPrP is degraded by the proteasome. Transfected BHK cells expressing wild-type or FLAG-L9R/3AV PrP were incubated for 16 h with (lanes 2 and 4) or without (lanes 1 and 3) 20 μM PS 1. Cell lysates were then subjected to Western blotting with 3F4 and anti-actin antibodies. The blot probed with anti-actin antibody confirms equal loading of total protein in all lanes.
DISCUSSION
We describe here several previously unappreciated features of CtmPrP, a novel transmembrane form of PrP that has been postulated to play a critical role in the pathogenesis of prion diseases. Our results provide insight into general mechanisms that control the topology of membrane proteins during their synthesis in the ER, and they also suggest possible cellular pathways by which CtmPrP may cause disease.
CtmPrP has the topology of a type II transmembrane protein (C terminus on the exofacial side of the bilayer), and it obeys the “positive inside” rule in which there is a preponderance of positively charged residues on the cytoplasmic side of the membrane-spanning sequence (von Heijne, 1989). However, CtmPrP is unusual in that it contains an uncleaved, N-terminal signal peptide, a feature we have demonstrated here for the first time. Most type II proteins contain an internal signal-anchor sequence that serves both to initiate translocation and to anchor the polypeptide chain in the lipid bilayer (Denzer et al., 1995; Gafvelin et al., 1997). A few type II proteins have an uncleaved, N-terminal signal sequence, but unlike the case of CtmPrP, this sequence serves as a membrane anchor (Ozols, 1998). The retention of the N-terminal signal peptide on CtmPrP can be rationalized by the fact that the N terminus of the polypeptide chain does not enter the ER lumen where signal peptidase is located. In contrast, the signal sequence is cleaved from SecPrP (this study) and NtmPrP (data not shown), whose N termini lie on the lumenal side of the membrane.
CtmPrP is unusual in one other respect, which is the presence of a C-terminal GPI anchor in addition to the transmembrane anchor. We have demonstrated that CtmPrP contains a GPI anchor by labeling cells with [3H]palmitate (this study), as well as by PIPLC treatment of PrP translated in vitro followed by Triton X-114 phase partitioning (Stewart and Harris, 2001). This dual mode of membrane attachment has been described in only a few other proteins (Hitt et al., 1994; Howell et al., 1994; Koster and Strand, 1994). The presence of a GPI anchor on CtmPrP is consistent with the fact that anchor addition occurs on the lumenal side of the ER membrane after cleavage of a C-terminal segment of the polypeptide chain (Udenfriend and Kodukula, 1995).
Our results, in conjunction with previous work (Hegde and Lingappa, 1999), suggest a model in which the membrane orientation of PrP is determined by competition during the translation process between two conflicting topological determinants in the polypeptide chain: an N-terminal signal sequence (residues 1–22) that directs translocation of the N terminus of the polypeptide chain across the membrane to produce SecPrP or NtmPrP; and a central hydrophobic domain (residues 111–134) that acts as a type II signal-anchor sequence, directing translocation of the C terminus across the membrane to produce CtmPrP. In this model, the effects of mutations on the membrane orientation of PrP (Hegde et al., 1998a, 1999) can be understood in terms of how they affect the relative functional strength of these two topological domains. Mutations within or near the central, hydrophobic domain either enhance (3AV, N107I, K109I/H110I, A116V) or diminish (G122P) the efficacy of the internal signal-anchor sequence, thereby either increasing or decreasing the proportion of CtmPrP. The “stop transfer effector” (residues 103–111), which modulates formation of CtmPrP, is also thought to act by altering the action of the adjacent signal-anchor region (Lopez et al., 1990; Yost et al., 1990). In contrast, the L9R mutation we describe here weakens the translocation activity of the N-terminal signal peptide by introducing a charged residue into the hydrophobic core of the sequence, thus increasing the proportion of CtmPrP. By combining mutations in both the signal and signal-anchor domains (L9R/3AV), we have been able to achieve a synergistic effect that completely shifts the topology of PrP to the CtmPrP form. Competitive interactions between an N-terminal signal sequence and an internal signal-anchor sequence have also been observed in model chimeric proteins (Goder et al., 1999), indicating that the adoption of alternate membrane topologies by a single polypeptide chain is not a unique feature of PrP. The precise mechanisms by which signal and signal-anchor sequences interact during the translation process remain to be investigated, although there is evidence that the two determinants compete within the translocon itself rather than at the level of binding to signal recognition particle (Hegde et al., 1998b; Goder et al., 1999). Consistent with this suggestion, we find that PrP in which the N-terminal signal sequence has been deleted is not translocated into microsomes at all (unpublished data), indicating that the signal-anchor sequence is not by itself competent for binding to the signal recognition particle and targeting to the ER.
The availability of a mutant form of PrP (L9R/3AV) that is synthesized exclusively in the CtmPrP form has allowed us for the first time to investigate the cell biological properties of this variant in isolation. Previously identified mutations in the transmembrane domain increase the proportion of CtmPrP to at most 30–40% of the total PrP chains after in vitro translation (Hegde et al., 1998a, 1999), and to only 2% after transfection of cultured cells (Stewart and Harris, 2001), with the remainder of the molecules being primarily SecPrP. Using the L9R/3AV mutant, we have shown that CtmPrP fails to reach the cell surface after synthesis, and is retained primarily in the ER. This localization may result from recognition of CtmPrP as a misfolded substrate by the ER quality control machinery (Ellgaard et al., 1999). Consistent with this interpretation, we have found that proteasome inhibitors dramatically increase the steady-state levels of CtmPrP, indicating that this form of the protein is normally degraded at least in part by the proteasome. It is now well established that misfolded proteins in the ER are retro-translocated into the cytoplasm where they are degraded by the proteasome, often following ubiquitination (Bonifacino and Weissman, 1998). Whether proteasome-mediated degradation of CtmPrP requires ubiquitination, and whether it involves complete translocation of the polypeptide chain into the cytoplasm, or selective proteolysis of the cytoplasmic domain are questions requiring further investigation. We have also observed that proteasome inhibitors increase the amount of wild-type PrP in transfected cells, suggesting that some wild-type molecules are also subject to ER retention and proteasomal degradation, although most are transported efficiently to the cell surface (our unpublished data).
In contrast to our study, a previous report concluded that CtmPrP transits beyond the ER, based on the endoglycosidase H-resistance of PrP from the brains of transgenic mice that express the 3AV or K109I/H110I mutations (Hegde et al., 1998a). However, only 20–30% of the PrP in these animals is actually CtmPrP, and biochemical characterization of these molecules in the presence of excess SecPrP would have been problematic. Interestingly, Zanusso et al. (1999) found that PrP carrying a stop codon at position 145, a mutation described in a Japanese patient with a Gerstmann-Sträussler-like syndrome, retained the N-terminal signal peptide and was rapidly degraded by the proteasome. Unlike L9R/3AV PrP, however, this mutant was partially secreted. These results suggest that alterations of the C-terminal part of PrP beyond the signal-anchor sequence can produce a topological variant with the characteristics of both CtmPrP and SecPrP.
Our results provide clues to the mechanisms by which CtmPrP might play a role in the pathogenesis of prion diseases. Hegde et al. (1999) have hypothesized that CtmPrP is a component of a common pathway of neurodegeneration underlying both infectious and genetic forms of prion diseases, and that PrPSc is pathogenic because it enhances the formation of CtmPrP (Hegde et al., 1999). This hypothesis is based on indirect evidence that levels of CtmPrP in mouse brain increase during the course of scrapie infection, and on the finding that transgenic mice expressing PrP with CtmPrP-favoring mutations develop a scrapie-like neurological illness without PrPSc. However, the mechanism by which CtmPrP may cause neurodegeneration has remained a mystery. Our observation that CtmPrP is retained in the ER where it is subject to proteasomal degradation suggests that CtmPrP may damage neurons by activating stress-induced signaling pathways that are engaged by the accumulation of misfolded proteins in the ER. Some of these pathways, such as the unfolded protein response that results in up-regulation of ER chaperone synthesis, are adaptive in nature, but others, such as induction of the transcription factor CHOP/GADD153 and phosphorylation of the translation initiation factor eIF-2, can kill cells by an apoptotic mechanism (Chapman et al., 1998; Kaufman, 1999). Whether these or other ER stress pathways are activated by synthesis of CtmPrP will be an important subject for future investigation. In any case, our results link prion diseases for the first time to other inherited, human disorders that are due to retention of misfolded proteins in the ER (Perlmutter, 1999).
Available data indicate that CtmPrP is distinct from PrPSc. We have found that L9R/3AV PrP is detergent-insoluble, possibly because of the presence of the hydrophobic, N-terminal signal peptide on the cytoplasmic domain, but it is not resistant to digestion with even low concentrations of PK in detergent solution (our unpublished data), a cardinal feature of PrPSc from infectious, familial, and sporadic cases of prion disease. Similarly, Hegde et al. (1998a) have not observed a PrP 27-30 fragment after digestion of PrP molecules carrying other CtmPrP-favoring mutations (although small amounts of slightly smaller fragment are produced under mild digestion conditions) (Hegde et al., 1998a). Whether CtmPrP and PrPSc contribute independently to neurodegeneration, or whether they form part of a common biochemical pathway remains to be determined. Expression of L9R/3AV PrP in transgenic mice, which would be predicted to produce a severe neurological illness without PrPSc, may help to further illuminate the role of CtmPrP in prion diseases.
ACKNOWLEDGMENTS
We thank Richard Kascsak for 3F4 antibody. This work was supported by a grant from the National Institutes of Health (R01-NS35496). R.S.S. was supported by a training grant from the National Institutes of Health (T32-NS07129).
Abbreviations used:
- GPI
glycosyl-phosphatidylinositol
- PK
proteinase K
- PIPLC
phosphatidylinositol-specific phospholipase C
- PrP
prion protein
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- SecPrP
secretory form of PrP
- NtmPrP
N-transmembrane form of PrP
- CtmPrP
C-transmembrane form of PrP
REFERENCES
- Bolton DC, Seligman SJ, Bablanian G, Windsor D, Scala LJ, Kim KS, Chen CM, Kascsak RJ, Bendheim PE. Molecular location of a species-specific epitope on the hamster scrapie agent protein. J Virol. 1991;65:3667–3675. doi: 10.1128/jvi.65.7.3667-3675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Collinge J. Variant Creutzfeldt-Jakob disease. Lancet. 1999;354:317–323. doi: 10.1016/S0140-6736(99)05128-4. [DOI] [PubMed] [Google Scholar]
- De Fea KA, Nakahara DH, Calayag MC, Yost CS, Mirels LF, Prusiner SB, Lingappa VR. Determinants of carboxyl-terminal domain translocation during prion protein biogenesis. J Biol Chem. 1994;269:16810–16820. [PubMed] [Google Scholar]
- DeArmond SJ, Ironside JW. Neuropathology of prion diseases. In: Prusiner SB, editor. Prion Biology and Diseases. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. pp. 585–652. [Google Scholar]
- Denzer AJ, Nabholz CE, Spiess M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 1995;14:6311–6317. doi: 10.1002/j.1460-2075.1995.tb00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Gafvelin G, Sakaguchi M, Andersson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DA. Cellular biology of prion diseases. Clin Microbiol Rev. 1999;12:429–444. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Barry RA, Lieberburg I, Prusiner SB, Lingappa VR. Biogenesis and transmembrane orientation of the cellular isoform of the scrapie prion protein. Mol Cell Biol. 1987a;7:914–920. doi: 10.1128/mcb.7.2.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Prusiner SB, Lingappa VR. Evidence for a secretory form of the cellular prion protein. Biochemistry. 1987b;26:8110–8115. doi: 10.1021/bi00399a014. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Lingappa VR. Regulation of protein biogenesis at the endoplasmic reticulum membrane. Trends Cell Biol. 1999;9:132–137. doi: 10.1016/s0962-8924(99)01504-4. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Mastrianni JA, Scott MR, Defea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998a;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Voigt S, Lingappa VR. Regulation of protein topology by trans-acting factors at the endoplasmic reticulum. Mol Cell. 1998b;2:85–91. doi: 10.1016/s1097-2765(00)80116-1. [DOI] [PubMed] [Google Scholar]
- Hitt AL, Lu TH, Luna EJ. Ponticulin is an atypical membrane protein. J Cell Biol. 1994;126:1421–1431. doi: 10.1083/jcb.126.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S, Lanctot C, Boileau G, Crine P. A cleavable N-terminal signal peptide is not a prerequisite for the biosynthesis of glycosylphosphatidylinositol-anchored proteins. J Biol Chem. 1994;269:16993–16996. [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Koster B, Strand M. Schistosoma mansoni: Sm23 is a transmembrane protein that also contains a glycosylphosphatidylinositol anchor. Arch Biochem Biophys. 1994;310:108–117. doi: 10.1006/abbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- Lehmann S, Harris DA. A mutant prion protein displays an aberrant membrane association when expressed in cultured cells. J Biol Chem. 1995;270:24589–24597. doi: 10.1074/jbc.270.41.24589. [DOI] [PubMed] [Google Scholar]
- Lopez CD, Yost CS, Prusiner SB, Myers RM, Lingappa VR. Unusual topogenic sequence directs prion protein biogenesis. Science. 1990;248:226–229. doi: 10.1126/science.1970195. [DOI] [PubMed] [Google Scholar]
- Narwa R, Harris DA. Prion proteins carrying pathogenic mutations are resistant to phospholipase cleavage of their glycolipid anchors. Biochemistry. 1999;38:8770–8777. doi: 10.1021/bi990736c. [DOI] [PubMed] [Google Scholar]
- Ozols J. Determination of lumenal orientation of microsomal 50-kDa esterase/N-deacetylase. Biochemistry. 1998;37:10336–10344. doi: 10.1021/bi9807916. [DOI] [PubMed] [Google Scholar]
- Perlmutter DH. Misfolded proteins in the endoplasmic reticulum. Lab Invest. 1999;79:623–638. [PubMed] [Google Scholar]
- Prickett KS, Amberg DC, Hopp TP. A calcium-dependent antibody for identification and purification of recombinant proteins. Biotechniques. 1989;7:580–589. [PubMed] [Google Scholar]
- Prusiner SB. Prion Biology and Diseases. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- Stewart RS, Harris DA. Most pathogenic mutations do not alter the membrane topology of the prion protein. J Biol Chem. 2001;276:2212–2220. doi: 10.1074/jbc.M006763200. [DOI] [PubMed] [Google Scholar]
- Telling GC, Tremblay P, Torchia M, DeArmond SJ, Cohen FE, Prusiner SB. N-terminally tagged prion protein supports prion propagation in transgenic mice. Protein Sci. 1997;6:825–833. doi: 10.1002/pro.5560060409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J, Menon AK. Soluble constituents of the ER lumen are required for GPI anchoring of a model protein. EMBO J. 1995;14:4686–4694. doi: 10.1002/j.1460-2075.1995.tb00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Yost CS, Lopez CD, Prusiner SB, Myers RM, Lingappa VR. Non-hydrophobic extracytoplasmic determinant of stop transfer in the prion protein. Nature. 1990;343:669–672. doi: 10.1038/343669a0. [DOI] [PubMed] [Google Scholar]
- Zanusso G, Petersen RB, Jin T, Jing Y, Kanoush R, Ferrari S, Gambetti P, Singh N. Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J Biol Chem. 1999;274:23396–23404. doi: 10.1074/jbc.274.33.23396. [DOI] [PubMed] [Google Scholar]