Abstract
Copy-number variants (CNVs) reshape gene structure, modulate gene expression, and contribute to significant phenotypic variation. Previous studies have revealed CNV patterns in natural populations of Drosophila melanogaster and suggested that selection and mutational bias shape genomic patterns of CNV. Although previous CNV studies focused on heterogeneous strains, here, we established a number of second-chromosome substitution lines to uncover CNV characteristics when homozygous. The percentage of genes harboring CNVs is higher than found in previous studies. More CNVs are detected in homozygous than heterozygous substitution strains, suggesting the comparative genomic hybridization arrays underestimate CNV owing to heterozygous masking. We incorporated previous gene expression data collected from some of the same substitution lines to investigate relationships between CNV gene dosage and expression. Most genes present in CNVs show no evidence of increased or diminished transcription, and the fraction of such dosage-insensitive CNVs is greater in heterozygotes. More than 70% of the dosage-sensitive CNVs are recessive with undetectable effects on transcription in heterozygotes. A deficiency of singletons in recessive dosage-sensitive CNVs supports the hypothesis that most CNVs are subject to negative selection. On the other hand, relaxed purifying selection might account for the higher number of protein–protein interactions in dosage-insensitive CNVs than in dosage-sensitive CNVs. Dosage-sensitive CNVs that are upregulated and downregulated coincide with copy-number increases and decreases. Our results help clarify the relation between CNV dosage and gene expression in the D. melanogaster genome.
Keywords: copy-number variation, gene expression, gene dosage sensitivity, recessive CNV, selection
Introduction
Recent analyses of structural genetic variation have highlighted the presence of extensive naturally occurring copy-number variants (CNVs) in organisms as diverse as humans, fruit flies, yeast, and plants (Sebat et al. 2004; Snijders et al. 2005; Perry et al. 2006; Redon et al. 2006; Dopman and Hartl 2007; Emerson et al. 2008; McCarroll et al. 2008; Carreto et al. 2008; DeBolt 2010). About 10% of the human genome harbor CNVs (Redon et al. 2006), with an estimated average of 12 CNVs per individual relative to a reference sequence (Feuk et al. 2006). In humans, a number of studies have indicated links between CNV and disease phenotypes (McCarroll and Altshuler 2007; Zhang et al. 2009; WTCCC 2010), whereas only a handful of studies have been conducted in natural populations of Drosophila melanogaster. Similar to estimates from the human genome, about 5–8% of the D. melanogaster genome were estimated to contain CNVs (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010). The nonrandom distribution CNV patterns in D. melanogaster suggest that selection and mutational biases are primary forces that shape structural variation (Dopman and Hartl 2007; Emerson et al. 2008). Furthermore, the occurrence of CNVs was found to be negatively associated with the abundance of protein–protein interactions (Dopman and Hartl 2007). To date, all the reported CNV analyses in D. melanogaster were based on heterogeneous isofemale strains from natural populations. Many CNVs are presumably heterozygous in these lines, which is problematic because the incidence of CNVs may be underestimated. Hence, a better resolution of CNVs may be expected from studies of homozygous genotypes.
In the human genome, about half of the CNVs detected overlap with protein-coding regions (Sebat et al. 2004) changing gene structure and dosage. Therefore, CNV loci encompassing genes may potentially affect gene expression, which can subsequently shape ecologically, evolutionarily, and medically relevant phenotypes (Stranger et al. 2007; Henrichsen et al. 2009; Schuster-Böckler et al. 2010).
Compensatory mechanisms are commonly invoked in attempts to understand the functional and evolutionary consequences of ploidy and sex determination (Birchler et al. 2007; Vicoso and Bachtrog 2009), but dosage must also be important for CNV loci encompassing individual genes. Indeed, disruption in the stoichiometric balance of proteins belonging to molecular complexes may affect gene expression (Birchler et al. 2005). The effects of aneuploidy result from a change in the relative dosage balance among various regulatory components that arise due to unbalanced alterations in gene copy number (Birchler et al. 2001, 2005). Dosage sensitivity is an essential evolutionary mechanism that influences gene dispensability. Although the underlying causes of dosage sensitivity remain poorly understood, previous reports suggested a complex relationship between haploinsufficiency and duplication sensitivity (Veitia 2002). Complexity may be explained from the balance hypothesis (Birchler et al. 2007) in which multiprotein complexes need to maintain the stoichiometry of their subunits to perform biological functions (Papp et al. 2003). As CNVs harboring duplications and deletions potentially create gene dosage effects, understanding the balance between CNV gene dosage and expression should shed light on the evolution of CNVs and how CNVs affect gene regulation.
It was previously reported that more than 70% of genes in D. melanogaster that are differentially expressed in contrasts between homozygous genotypes lack expression differences when in the heterozygous state (Lemos et al. 2008). This result suggested that recessive alleles with regulatory consequences might be abundant in Drosophila (Lemos et al. 2008). Because gene heterozygosity is prevalent in natural populations (Singh and Rhomberg 1987), the expression of genes encompassed in CNV could also be largely masked in heterozygotes.
Here, we addressed the relevance of CNV in homozygous and heterozygous genotypes to reveal dosage effects of CNVs. We generated six second-chromosome substitution homozygous lines and two heterozygous lines to investigate CNV patterns. We also utilized gene expression data conducted with some of the same substitution lines to infer the association between CNVs and their gene expression. We found that most CNVs appear to have low levels of dosage sensitivity, and they are often recessive in heterozygous state. Nevertheless, increases and decreases in copy number coincide with up- and downregulation in a number of cases. Overall, our work highlights complex relationships between gene dosage and expression.
Materials and Methods
Fly Stocks
Some of the second-chromosome substitution strains (PS1, PS2, PS3, CS) in this study were previously described by Lemos et al. (2008). Strains PS4 and PS5 were established using identical methodology (supplementary fig. S1 in Lemos et al. 2008). Heterozygous strains PS2/CS and PS5/CS are obtained in the F1 generation of homozygous second-chromosome substitution strains and contain two different second chromosomes in an otherwise identical genetic background. A total of eight strains were assayed.
DNA Isolation and Digestion
Genomic DNA was isolated from either 40 adult females or 60 males, using QIAGEN DNeasy blood and tissue kit (Cat. No. 695004). Genomic DNA was then digested with 1.5 μl MspI enzyme to randomly digest the genome into moderately sized fragments (average size ∼ 3.5 kb, Barker et al. 1984). Restriction digests followed the manufacture's recommendations (New England BioLabs, 20,000 U/ml) of 37 °C for 1 h; an equal amount of enzyme was added for an additional 1 h to assure complete digestion. DNA was further cleaned by Phase Lock Gel (Eppendorf) and phenol purification. Five micrograms DNA was used for each sample resulting in 10 μg DNA in each microarray reaction.
Microarray Platform
Array comparative genomic hybridizations (aCGH) were performed with an 18,000-feature DNA microarray. Labeling and hybridization were conducted with the 3DNA Array 900 MPX kit (Genisphere), with a Cy5–Cy3 two-channel dye swap for each reaction. All the DNA copy-number increases and decreases in the other seven sampled strains were estimated relative to PS1. Besides the dye swap, every reaction had at least two replicates (experimental design shown in supplementary fig. S1, Supplementary Material online). Upon hybridization, microarray slides were scanned in an Axon 4000B scanner (Axon Instruments). Gene expression microarrays, experimental designs, and previous results used in this study were obtained from Lemos et al. (2008).
Microarray Analyses
Scanned microarray slides were first analyzed with GenePix Pro 6.0 software (Axon Instruments). Fluorescence Cy5 and Cy3 intensities were then normalized by the Limma library of software R (Version 2.10.1). Two different methods were used to ascertain copy-number increases and decreases: threshold analysis and Bayesian Analysis of Gene Expression Levels (BAGEL). In threshold analysis, probes were suggested as indicating an occurrence of a CNV event if the standard error of the log-intensity ratio was beyond an intensity-ratio threshold. The threshold ratio was established from self–self-hybridizations in the reference strain, by controlling the false-positives to <1% (for more details, see Dopman and Hartl 2007). BAGEL analysis uses Bayesian algorithm to compute the probe signal ratios between samples and the reference strain, with P values indicating the significance (for more details, see Townsend and Hartl 2002; Lemos et al. 2008). Array probes located in transposons or containing repetitive sequences were removed from the analyses. The two methods were in good agreement and the patterns herein described are robust to the choice of method for ascertaining gains and losses. Only the threshold results are shown in great detail in the Results.
Analyses Schemes for Associations between CNVs and Gene Expression
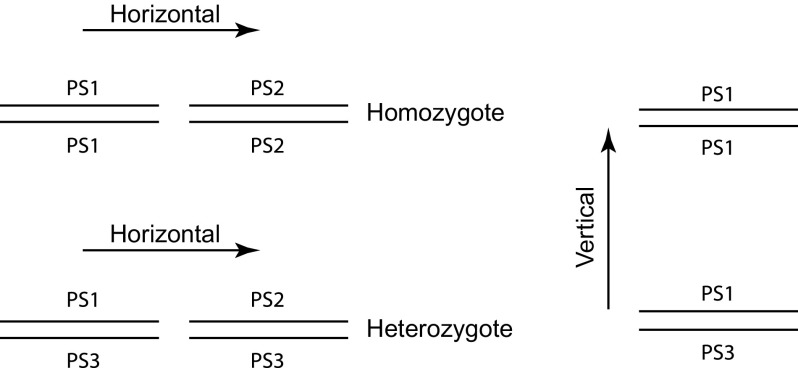
To determine dosage effects of CNVs in homozygotes, “horizontal” comparisons of gene expression levels between homozygous PS2 and PS1 were conducted as illustrated in figure 2. To determine dosage effects of CNVs in heterozygotes, horizontal comparisons of gene expression levels between heterozygous PS1/PS3 and PS2/PS3 were conducted. To determine recessive CNVs (heterozygous masking effect), the results collected from homozygous PS2 and PS1 comparisons were combined with those for PS1/PS3 and PS2/PS3 heterozygotes to infer if the same CNV genes in PS2 are recessive to PS3. To determine if upregulated or downregulated CNVs are matched with copy-number increases or decreases, “vertical” comparisons between PS1/PS1 and PS1/PS3 were conducted as also shown in figure 2, where in this comparison, PS3 harbors CNVs relative to PS1 instead of no differences from PS1 in the horizontal comparisons.
FIG. 2.—
Diagrams of analyses on associations of CNVs and gene expression. PS1, PS2 and PS3 are three second-chromosome substitution strains. The horizontal solid lines represent CNV alleles in each strain. The left panel shows a “horizontal” comparison between PS1/PS1 and PS2/PS2 homozygotes as well as between PS1/PS3 and PS2/PS3 heterozygotes, where PS2 contains CNVs relative to PS1 but PS3 allele is the same as PS1. The right panel shows a “vertical” comparison between PS1/PS1 and PS1/PS3, where PS3 allele harbors CNVs.
Protein Interactions for CNV Genes
The interaction data set from BIOGRID (Stark et al. 2006; http://thebiogrid.org/) was used to detect protein–protein interactions for dosage-sensitive, dosage-insensitive, recessive and nonrecessive CNV genes in different context. Only genes with ≥1 interactions were analyzed.
Results
Populations of D. melanogaster can be polymorphic for as many as 43% of their gene loci, and an average individual typically shows a level of heterozygosity on the order of 10% (Singh and Rhomberg 1987). Several recent studies have accessed CNVs with microarray and sequencing technologies using genetically heterogeneous isofemale strains (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010). However, the contribution of copy-number heterozygosity to estimates of copy-number variation is difficult to evaluate. Therefore, we investigated CNVs in completely homozygous chromosome substitution lines, which differ exclusively in the origin of the second chromosome but are otherwise genetically identical. All the second chromosomes were derived from a single Pennsylvania population (except line CS), whereas other chromosomes were originated from the marker lines used to construct these substitution lines (for details, see supplementary fig. S1 in Lemos et al. 2008). These second-chromosome substitution strains offer two major advantages. First, false positive error rates can be experimentally ascertained because no CNVs are expected to be found from probes located in the third, fourth, and X chromosomes. Second, chromosomes are homozygous within each strain, and so issues of detection associated with identifying CNVs in heterozygotes can be avoided. In this study, we utilized six homozygous and two heterozygous second-chromosome substitution lines, originally established by Lemos et al. (2008), to reveal the CNV patterns.
Variation in gene expression levels contributes to dramatic phenotypic differences between individuals and populations. Gene copy-number differences among individuals and populations can provide a source of gene expression variation (Stranger et al. 2007), although evidence suggests complex relationships between gene copy number and expression (Birchler et al. 2005, 2007). Changes in dosage of individual chromosome or chromosomal segments have more extreme global effects on gene expression than observed in ploidy series (Birchler et al. 2007). The balance between CNV gene dosage and expression levels can address how significant gene copy variation as well as gene structural changes induced by CNVs may affect gene regulation. Here, we addressed the extent of copy-number variation across chromosomes sampled from a single population (except for strain CS) and also combined this CNV data with previously reported gene expression data (Lemos et al. 2008) to investigate the balance between gene dosage and expression.
Validation of Methods Used in the Detection of CNVs
As females have two copies of X-linked genes and males only have one copy, male–female aCGH result in an excess of female signals for X-linked genes that can be used to calibrate the threshold values and detection methods. Indeed, lower signal ratios between male and female X-linked genes are reflected in supplementary figure S2A (Supplementary Material online) (based on threshold analysis; data on a log scale). In addition, only the second chromosomes in the substitution lines may be expected to contain gene copy-number variation, as all other chromosomes are in principle invariant across all strains. Indeed, as shown in supplementary figure S2B (Supplementary Material online), in one of the substitution strains (PS2) relative to the reference strain PS1, the second chromosome contains virtually all of the CNVs detected by microarray hybridizations. Other strains in this study all showed similar patterns (data not shown). With regard to BAGEL analyses, supplementary figure S3 (Supplementary Material online) demonstrates the distributions of probabilities of CNV occurrence in the sample strain PS2 compared with the reference strain PS1. As expected, the distribution of P values for probes located in the second chromosome is notably skewed to low (P < 0.05) or high (P > 0.95), indicating copy-number decrease and increase in PS2, respectively. In contrast, and in agreement with the expectation if the third, fourth, and X chromosomes are invariant, the distribution of P values is uniform for pooled data from all other chromosomes. These observations suggest a substantial level of variation of gene copy numbers on the second chromosome that can be detected with two distinct methods. In the following, we only show results and analyses based on the threshold method.
CNVs in Homozygous Second-Chromosome Substitution Strains
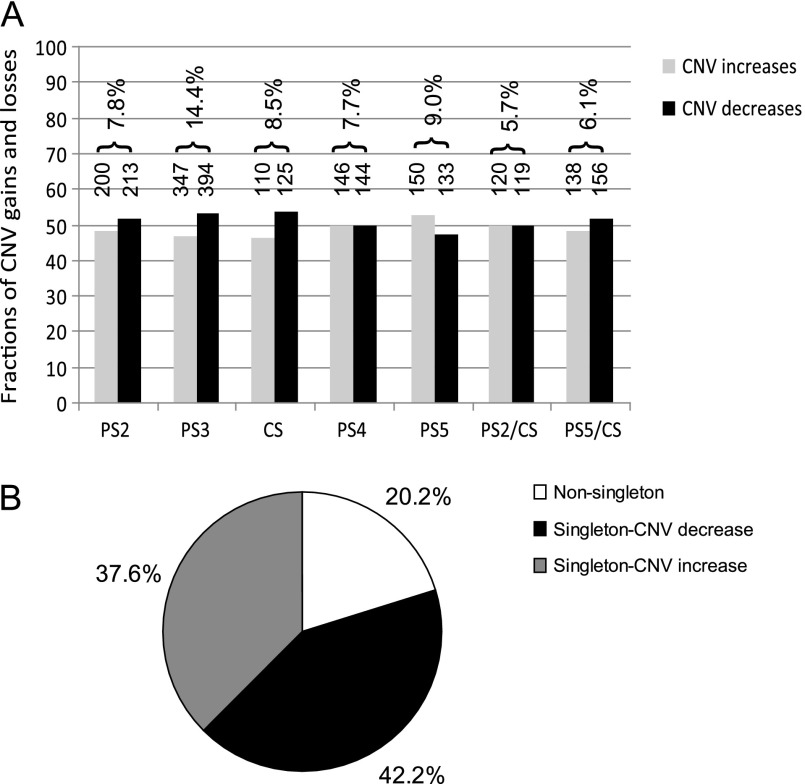
The number and fractions of CNV increases and decreases in five homozygous and two heterozygous second-chromosome substitution strains, relative to the reference strain, are plotted in figure 1. Because sample arrays and their replicates were not all prepared at the same time, batch variation may result in different detection rates of aCGH. The number of CNV increases between a sample strain and PS1 ranged from ∼100 to 350 and the number of CNV decreases ranged from ∼100 to 400 depending on the batch and strain. However, the fraction of CNVs that increased or decreased in number was found to be balanced in each strain (fig. 1A). In some of the strains, such as PS2, PS3, CS, and Heterozygous PS5/CS, there were slightly more copy-number decreases (on average 5%) than increases. In PS5, more increases (5%) were observed. The fractions were within 0.5% variation in PS4 and PS2/CS.
FIG. 1.—
CNV composition in homozygous second chromosomes. (A) Summary of CNV copy-number increases and decreases relative to a reference strain PS1 in the seven second-chromosome substitution lines, homozygous PS2, PS3, CS, PS4, and PS5; heterozygous PS2/CS and PS5/CS. Bars represent the fractions of CNV increases (gray) and decreases (black). The numbers on top of the bars show the number of increases and decreases detected in each strain, respectively. The percentages on top of the numbers indicate the percentage of genes (probes) containing CNVs among all the detected genes (probes) from the second chromosomes. (B) A pie chart demonstrating the fraction of singleton and nonsingleton CNVs derived from the five homozygous lines (CNV allele frequencies) relative to PS1. The black area shows the fraction of singletons that contain decreased copies in one strain relative to the other four strains and the reference strain PS1. The gray area shows the fraction of singletons that contain increased copies in only one strain. The white area shows the fraction of nonsingletons that appear in more than one strain as either increase or decrease.
The percentages of probes containing CNVs among all the detected genes on the second chromosomes are shown on top of the bars in figure 1A. For the five homozygous strains, the average CNV fraction is about 9.5% (range from 7.7% to 14.4%), a finding that is somewhat higher than previous reports of 5–8% in D. melanogaster (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010). The levels of variation detected in heterozygous PS2/CS (5.7%) and PS5/CS (6.1%) were lower than in their homozygotes. Two factors can account for these lower percentages in heterozygotes. First, some duplications and deletions may complement each other in heterozygotes resulting in no variation compared with the reference strain. Second, and most likely in view of the observation that most CNVs are singletons (discussed in the following paragraphs), aCGH arrays may be less sensitive to detecting copy-number variation in heterozygotes. This is because whenever a CNV is unique to only one homozygous strain, the magnitude of fold-change between the homozygous reference strain and the heterozyogous is less extreme.
CNVs can be either clustered at certain regions or dispersed across a whole chromosome. To distinguish CNVs from larger scale segmental duplications, we investigated CNV clustering by checking the fraction of CNVs that can be found in contiguous sets of more than three CNVs along the second chromosome. Minor clustering was found (supplementary table S1, Supplementary Material online). None of those clustered area involved more than six genes. Instead, CNVs were spread across the whole second chromosomes.
CNV Allele Frequency
All CNVs present in the five homozygous strains were assessed for their allele frequencies. All copy-number increases and decreases were evaluated relative to PS1. Therefore, we did not know if a detected copy-number increase or decrease represents a derived or ancestral allele. We make the parsimonious assumption that the minor allele (lower frequency) represents the derived state. For example, a focal probe showing higher copy number across all five “test” strains is most parsimoniously interpreted as a copy-number reduction in PS1. As shown in figure 1B, 37.6% of the copy-number increases as well as 42.2% of the decreases were unique to a single strain (singleton). The singletons discovered in only one strain are more likely to be true deletions and duplications relative to all other five strains including reference strain PS1, otherwise one would need to posit a shared CNV in other five strains. The nonsingleton 20.2% of CNVs were detected more than once among the five strains, among which 7.7% shared copy-number increase only relative to PS1, 7.3% shared copy-number decrease only, and 5.2% showed either increase or decrease in different strains. The fractions of singleton and nonsingleton between copy-number increase and decrease are not significantly different (Fisher's exact test, P = 0.193).
Dosage Effects of CNVs on Expression in Homozygotes
The majority of the array probes used in this study are located in gene regions. In total, 11,934 genes are represented on the array with an average of 1.2 probes per gene (Hild et al. 2003; Dopman and Hartl 2007). Therefore, the same array platform can be used to compare gene copy number and expression variation. How gene expression levels and CNVs correlate with each other is essential to understanding how structural changes induced by CNVs affect gene regulation.
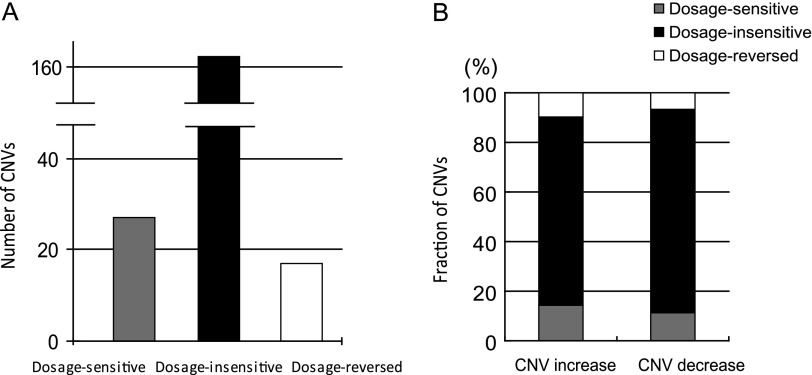
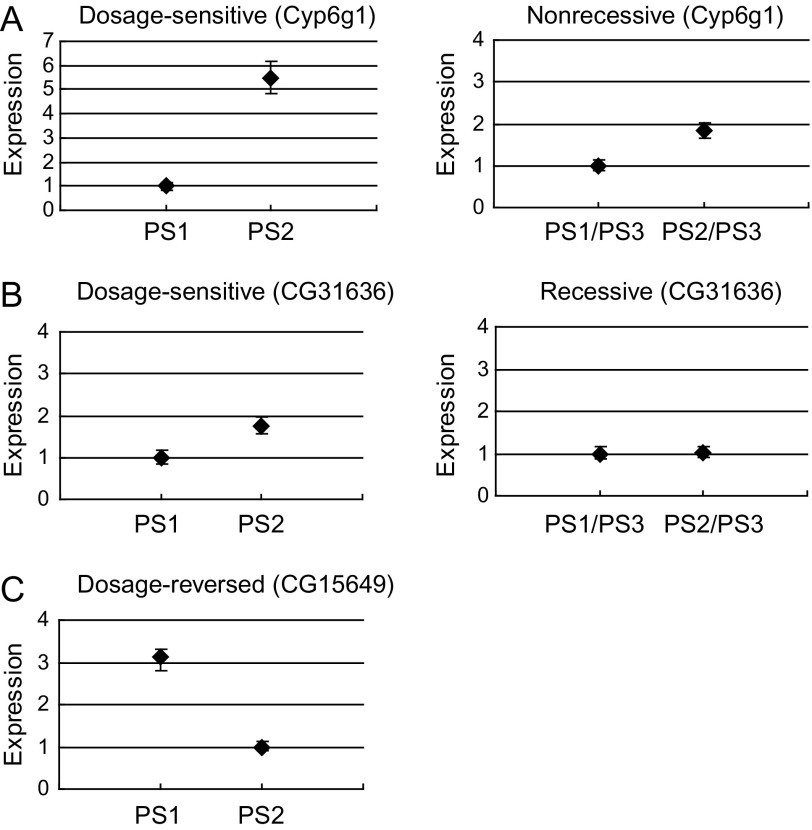
We began by investigating CNVs and their expression levels in homozygous PS2 and homozygous PS1. Shown in figure 2, if a gene in homozygous PS2 showed both an increase in copy number and expression level relative to that of the homozygous reference strain PS1, this focal gene is termed “dosage sensitive.” A gene in PS2 that showed both a decrease in copy number and expression relative to the reference is likewise termed dosage sensitive. Conversely, a gene showing an increase in copy number but a lower expression level than the reference is termed “dosage reversed.” Genes whose expression levels do not change despite alterations in copy number are termed “dosage insensitive.” We observed that 21% of CNVs had matching expression variation, with 27 and 17 CNVs in PS2 showing dosage-sensitive and dosage-reversed expression phenotypes, respectively. On the other hand, 163 CNVs (79%) showed no corresponding expression variation (dosage insensitive) in the homozygous–homozygous comparison between PS1 and PS2 (fig. 3A). The dosage effects for gene copy-number increases and decreases on gene expression levels were similar, as shown in figure 3B. Overall, 14.6% and 11.5% CNVs were dosage sensitive for copy-number increases and decreases, respectively; 9.7% and 6.7% CNVs were dosage reversed for increases and decreases, respectively; and 75.7% and 81.7% CNVs were dosage insensitive for increases and decreases, respectively. There was no significant difference between CNV increase and decrease (Fisher exact test, P = 0.553). The largest fraction of CNVs fell into the dosage-insensitive categories, which is examined further in the Discussion. More importantly, the absolute expression levels for dosage-sensitive genes did not differ from that of dosage-insensitive genes. Two dosage-sensitive CNV genes are shown in figure 4, in which both gene Cyp6g1 and CG31636 had copy-number increases and higher expression levels in PS2 relative to PS1. A dosage-reversed gene CG15649 is also shown which had a higher copy number but lower expression level in PS2 compared with PS1.
FIG. 3.—
Dosage effects of CNVs on expression in homozygotes. (A) In the left column, dosage sensitive indicates that CNVs have either copy-number increases with higher expression levels than reference strain PS1 or else that copy-number decreases with lower expression levels than PS1. In the right column, dosage reversed suggests opposite negative associations. The middle panel shows the number of CNVs that are not sensitive to copy-number dosage effects. (B) The bars indicate the fractions of dosage-sensitive (gray), dosage-insensitive (black) and dosage-reversed CNVs (white) for copy-number increases and decreases separately.
FIG. 4.—
Examples illustrating dosage sensitivity of CNV genes. Expression indicates normalized estimates from BAGEL analysis. For all panels, copy number is higher in PS2 than in PS1. Diamonds represent genes, with credible intervals shown. The expression levels are normalized. (A) Dosage-sensitive gene Cyp6g1 shows a copy-number increase and higher expression level in PS2 relative to PS1. Its expression level is also higher in PS2/PS3 heterozygotes compared with PS1/PS3, suggesting nonrecessive phenotype of Cyp6g1 CNV gene. (B) Dosage-sensitive gene CG31636 shows a copy-number increase and higher expression level in PS2 relative to PS1. Its expression level is not different between PS2/PS3 and PS1/PS3 heterozygotes, suggesting recessive phenotype of CG31636 CNV gene. (C) Dosage-reversed gene CG15649 shows a copy-number increase but lower expression level in PS2 relative to PS1.
CNVs are Largely Recessive (Masked) in Heterozygotes
Are changes in copy number resulting in expression changes in homozygous state recessive in heterozygotes? To address this issue, we considered CNVs in PS2 homozygotes with the expression phenotype manifested in the comparison between homozygous PS1 versus PS2 and investigated if such expression differences were still present when heterozygous PS2/PS3 were contrasted with heterozygous PS1/PS3. For example, for a gene with both increased copy number and higher expression in homozygous PS2 relative to homozygous PS1 (fig. 2, dosage sensitive in homozygotes), the CNV is recessive if PS2/PS3 shows no expression difference from PS1/PS3 heterozygotes. In contrast, the CNV is nonrecessive if a difference in expression observed in the homozygotes is maintained in the contrast between PS2/PS3 and PS1/PS3. One possible cause of the recessivity could be background trans-factors from PS3. Nevertheless, the observation of no expression difference suggests the CNVs are masked in heterozygous background.
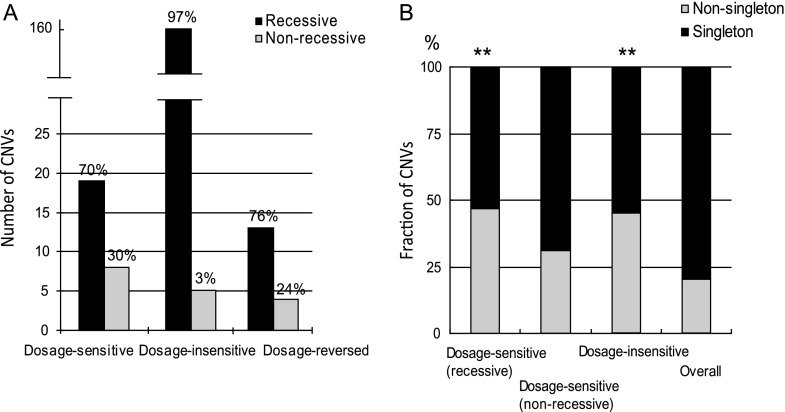
We observed 19 (70%) recessive CNVs and 8 (30%) nonrecessive CNVs. Conversely, only 3% (5 of 163) of all dosage-insensitive CNVs that did not show expression differences in the homozygous PS2 versus PS1 contrast appeared to show expression differences in the heterozygous PS2/PS3 versus PS1/PS3. There were 17 dosage-reversed CNVs in the homozygous PS2 versus PS1 comparison, 76% of which were recessive in the heterozygous PS2/PS3 versus PS1/PS3 contrast (fig. 5A). The absolute expression levels for recessive genes did not differ from that of nonrecessive genes (P = 0.10, Mann–Whitney test). In both of the dosage-sensitive and dosage-reversed groups, there were more recessive CNVs than nonrecessive CNVs. As to the dosage-insensitive CNVs, three CNVs harboring higher copy number showed no expression difference in homozygotes but higher levels in heterozygotes, and the other two CNVs harboring lower copy number showed lower levels in heterozygotes. The overall result suggests that expression differences caused by gene copy-number changes are largely masked in heterozygotes. Examples of nonrecessive and recessive CNV genes are shown in the right panel of figure 4. Gene Cyp6g1 appears to have a copy-number increase and higher expression level in PS2/PS3 relative to PS1/PS3 heterozygotes. In contrast, gene CG31636 has a higher copy number but its expression level in PS2/PS3 does not differ from PS1/PS3, therefore appears recessive.
FIG. 5.—
Reconstruction of recessive and nonrecessive CNVs. (A) Three groups of CNVs (dosage sensitive, dosage insensitive and dosage reversed) from homozygous PS2 were investigated for their expression in heterozygous PS2/PS3 (for details, see Results). The numbers of recessive (black bars) and nonrecessive (gray bars) CNVs in dosage-sensitive, -insensitive and -reversed CNVs of homozygous PS2 are plotted, respectively. Percentages of recessive and nonrecessive CNVs are shown on the top. (B) Fractions of singleton and nonsingleton CNVs in the above groups of CNVs. Both dosage-sensitive and -reversed CNVs respond to copy-number change such that they are grouped together in comparison with the overall allele frequency derived from all five homozygous strains. Black bars indicate singleton CNVs. Gray bars indicate nonsingleton CNVs. Overall indicates singleton and nonsingleton data collected from five homozygous strains. Asterisks indicate P < 0.001 in the comparison between either dosage sensitive (recessive) or dosage insensitive and overall.
For the recessive and nonrecessive CNVs identified in homozygous PS2 that were already categorized into dosage-sensitive, -insensitive, and -reversed groups, the CNVs were investigated for their allele frequencies (singleton or nonsingleton). Because both dosage-sensitive and dosage-reversed CNVs respond to copy-number changes, they were grouped together to compare with the overall allele frequency derived from all five homozygous strains. The group of dosage-insensitive CNVs corresponds to “recessive” in yielding no expression difference between PS2/PS3 and PS1/PS3. Shown in figure 5B, the fraction of nonsingleton CNVs is significantly increased (Fisher exact test, P < 0.001) for both recessive dosage-sensitive CNVs and dosage-insensitive CNVs. However, we did not observe a significant difference (Fisher exact test, P = 0.137) for the fraction of singletons and nonsingletons between nonrecessive dosage-sensitive CNVs and overall CNVs.
Dosage Effects of CNVs on Expression in Heterozygotes
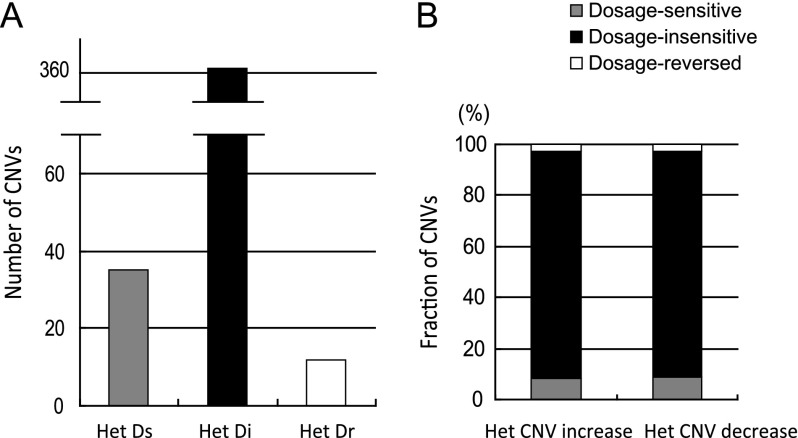
We also reconstructed heterozygous CNVs to directly correlate heterozygous gene expression levels in PS2/PS3 and PS2/CS heterozygotes. In particular, for genes with variable copy number between homozygous PS2 and PS1, we asked what happens to the expression of genes in the heterozygous state. This analysis differs from determining recessive CNVs classified as dosage sensitive, dosage insensitive, or dosage reversed in homozygous PS2. Gene expression may change in the heterozygous background. In this analysis, horizontal comparisons (shown in fig. 2) of gene expression levels between heterozygous PS1/PS3 and PS2/PS3 (as well as PS1/CS and PS2/CS) were directly conducted and the CNVs classified in regard to dosage sensitivity in the heterozygous background. A total of 415 CNVs were pooled from PS2/PS3 and PS2/CS heterozygotes in contrast to PS1/PS3. Thirty-six CNVs showed dosage-sensitive effects, whereas 11 showed dosage-reversed effects. The remaining CNVs were dosage insensitive. Nearly 90% of the total CNVs fell into the dosage-insensitive group. The number of CNV increases and decreases is plotted separately for three groups in figure 6A, along with their corresponding fractions shown in figure 6B. The fractions show no significant differences (chi-square test, P = 0.989).
FIG. 6.—
Reconstruction of CNVs in heterozygotes and their effects on gene expression. (A) As shown in figure 2, heterozygous PS2/PS3 expression can be compared with PS1/PS3 to infer CNVs' dosage effects on heterozygotes. The heterozygotes data were pooled from PS2/PS3 and PS2/CS together (for details, see Results) and plotted. The graph shows the number of three groups of CNVs that are dosage sensitive, dosage insensitive and dosage reversed. Het Ds: heterozygous dosage sensitive; Het Di: heterozygous dosage insensitive; Het Dr: heterozygous dosage reversed. (B) The fractions of dosage sensitive (gray), dosage insensitive (black) and dosage reversed (white) for CNV increases and decreases in heterozygotes are plotted separately.
Protein Interactions for CNV Genes
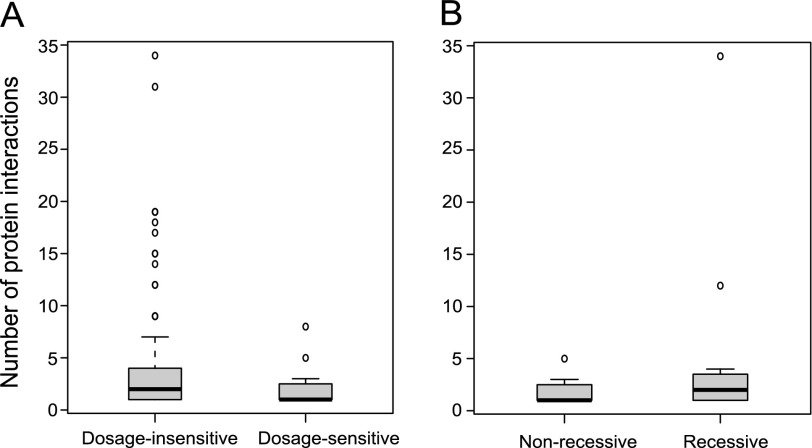
Dopman and Hartl (2007) reported that the occurrence of CNV is negatively correlated with the degree of protein interaction network. Natural selection plays critical roles in shaping CNV patterns, and dosage-sensitive CNVs might be expected to have greater functional consequences and fewer protein–protein interactions than dosage-insensitive ones. As shown in figure 7A, the dosage-sensitive CNVs have a significantly lower number of protein–protein interactions than that of dosage-insensitive CNVs (dosage sensitive: one protein interactions [median]; dosage insensitive: two protein interactions [median]; P = 0.04, Mann–Whitney test). The same pattern holds true for another measure of centrality: betweenness (dosage sensitive, betweenness = 0 [median]; dosage insensitive, betweenness = 1002 [median]; P = 0.02, Mann–Whitney test).
FIG. 7.—
The degree of protein interactions for CNV genes. (A) The number of protein interactions for dosage-sensitive and dosage-insensitive CNV genes is plotted. Dosage-sensitive genes have significantly more protein interactions. (B) The number of protein interactions for recessive and nonrecessive CNV genes is plotted. Recessive genes appear to have more protein interactions than nonrecessive genes. However, the difference is not significant. Bold horizontal bars are the median value, the box is the interquartile range, and the whiskers indicate the 95% confidence interval.
Similarly, one would expect to see a higher number of interactions in recessive CNVs than that of nonrecessive CNVs. Although the trend showed the prediction, the difference was not significant (recessive: two protein interactions [median]; nonrecessive: one protein interactions [median]; P = 0.22, Mann–Whitney test), possibly due to the relatively smaller sample size (fig. 7B). The same pattern holds true for another measure of centrality: betweenness (recessive, betweenness = 311 [median]; nonrecessive, betweenness = 0 [median]; P = 0.34, Mann–Whitney test].
Upregulation and Downregulation in CNVs
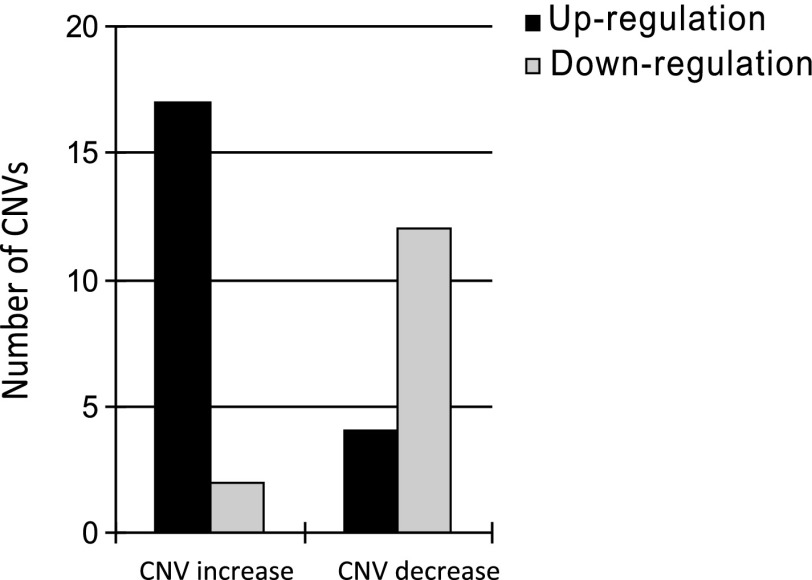
To infer if upregulated or downregulated CNVs match with their copy-number changes, we employed a slightly different analysis. As illustrated in figure 2, PS1/PS1 and PS1/PS3 (or PS1/CS) were vertically compared in which PS3 (or CS) contained CNVs. A number of CNVs involving upregulation and downregulation were detected. These CNVs were then sorted based upon copy-number increases or decreases. Shown in figure 8, 17 CNVs containing copy-number increases in PS3 and CS were upregulated (higher copy number and higher expression) relative to PS1 in heterozygous PS1/PS3 or PS1/CS, whereas only two CNVs with increases were downregulated (lower expression). In contrast, more downregulation events were discovered in CNVs with copy-number decreases. Twelve downregulations (lower copy number and lower expression) were found for PS3 and CS, whereas only four upregulated CNVs (higher expression) were found for PS3 and CS. It appears that up- and downregulated CNVs are positively associated with gene copy-number changes.
FIG. 8.—
Upregulated and downregulated CNVs are matched with copy-number increase and decrease. As shown in figure 2, heterozygous PS1/PS3 expression can be compared with homozygous PS1 to infer if CNVs are upregulated or downregulated in heterozygotes (here, PS3 contains CNVs). The black bars indicate the number of CNVs in which PS3 (as well as CS, for details, see Results) CNV allele is upregulated relative to PS1 in expression. The gray bars indicate downregulation of PS3 (or CS) allele relative to PS1. The left two columns show the CNVs with copy-number increases and the right two columns show the CNVs with copy-number decreases.
Discussion
CNV Pattern in Homozygous Genotypes
Studies of copy-number variation in natural populations of D. melanogaster had previously been conducted with heterozygous isofemale strains (Dopman and Hartl 2007; Emerson et al. 2008). In these cases, many low-frequency CNVs are heterozygous and may remain undetected. Here, we established a number of second-chromosome substitution strains derived from a single Pennsylvania population (except strain CS) to evaluate CNV occurrence in completely homozygous genotypes. The results indicate extensive copy-number variation on the second chromosome of these flies. Indeed, within the context of our own experimental design, the fraction of protein-coding genes harboring CNVs that can be detected in homozygotes is higher than that of heterozygotes. Furthermore, the faction of CNVs detected in homozygous genotypes is also higher than that reported in previous studies with heterozygous genotypes. These findings suggest that the aCGH microarray analysis underestimates CNVs in heterozygous genotypes because of a diminished power of detecting CNVs.
Also, previous studies found more duplications than deletions in fruit flies (Emerson et al. 2008). One possibility is that duplicating a region may confer milder phenotypes than deleting it, such that purifying selection may be stronger against deletions in CNV genes. Also, deletions in heterozygous state may potentially reduce the detection power of CNVs in aCGH arrays. In another study examining mammalian genomes, the results suggested a strong bias against duplications for genes whose protein products belong to complexes, with less than a quarter of the CNVs scored as gains (Schuster-Böckler et al. 2010). In contrast, our results found that the frequencies of copy-number increase and decrease are exceptionally close in these substitution lines, suggesting a highly variable CNV composition across species and within species.
CNV Dosage Sensitivity and Effects on Expression
CNVs can have drastic phenotypic consequences as a result of altering gene dosage, disrupting coding sequences, or perturbing gene regulation. The degree of penetrance (the fraction of a genotype that shows the associated phenotype) of CNV encompassed genes is essential to understanding the impact of CNVs on expression and potentially their association with genetic disorders (Beckmann et al. 2007). We found that 13% of homozygous CNVs were dosage sensitive, meaning that gene expression levels positively associate with copy-number increase or decrease. Conversely, we discovered that 8% of CNVs were dosage reversed which exhibited negative associations between expression and copy number in homozygotes. The two categories were 9% and 3%, respectively, for CNVs in heterozygous states.
Dosage-reversed CNVs were also discovered in human genomes. In the case of copy-number duplications, 10% of the CNVs in human genome were found to be dosage reversed (Stranger et al. 2007; Beckmann et al. 2007). Schuster-Böckler et al. (2010) also reported a complex relationship between copy number and expression level in human heterozygous CNVs. For example, more than 10% of the CNVs exhibited dosage-reversed expression pattern in their study. In addition to genes exhibiting changes in both copy number and expression, the remaining 79% of the CNVs in homozygotes or 89% of the CNVs in heterozygotes in our study were not responsive to gene copy-number changes (dosage insensitive). Similarly, around 65% of CNVs were dosage insensitive in the studies conducted by Schuster-Böckler et al. (2010). All the above findings strongly suggest an extremely complex relationship between gene copy number and expression.
Young duplicated genes typically exhibit increased expression divergence (Farre and Alba 2010). Under certain conditions, gene duplications may induce reduced transcripts or even gene silencing. On the contrary, deletion of a transcriptional repressor could serve to elevate gene expression (Stranger et al. 2007). Both factors could contribute to the discovery of CNVs whose expression phenotype is dosage reversed. On the other hand, dosage-insensitive CNVs could arise if gene promoter regions were not duplicated or deleted along with the CNV regions. Also partial duplication or deletion of genes may not significantly affect gene expression levels. Nevertheless, the presence of detectable gene expression implies that at least one copy of the gene is present. Therefore, a deletion occurred in one of the other copy or copies did not significantly change the expression.
CNVs can alter gene doses without abolishing gene function or changing phenotype. As shown in the results, the majority of CNVs were found to be dosage insensitive, particularly in heterozygous CNVs. Therefore, CNVs appear to be less likely to contain dosage-sensitive genes, indicating that negative selection acts on the shaping of CNVs. Previously, CNV genes encoding protein complexes were found to be significantly underrepresented (Dopman and Hartl 2007; Schuster-Böckler et al. 2010). Hence, selection facilitates the formation and spread of CNV patterns due to functional constraints.
The observations between low or no change in gene expression and change of gene copy number suggest that cells may attempt to compensate changes in gene copy number on expression by modifying transcription. Dosage compensation has been widely addressed in plants, worms, mammals, and fruit flies (Charlesworth 1996; Birchler et al. 2005, 2007; Vicoso and Bachtrog 2009; Prestel et al. 2010). The molecular mechanism of dosage compensation involves chromatin structure remodeling (Bachtrog et al. 2010; Prestel et al. 2010). Transcription factors, chromatin proteins, and signal-transduction genes were found to be predominantly responsible for dosage effects (Birchler et al. 2001, 2005). However, the mechanisms by which CNVs affect dosage compensation are not well understood. CNVs dosage effects on gene expression may be dependent on local chromatin modifications or regulatory genes in the dosage compensation cascades. Note that some CNVs change dosage status from homozygotes to heterozygotes (e.g., dosage-sensitive CNVs in homozygotes become insensitive or vice versa), again suggesting a complex relationship between gene dosage and expression.
CNVs are Largely Recessive in Heterozygous State
Previous studies reported that 70% of differentially expressed genes in homozygotes were masked in heterozygous state (Lemos et al. 2008). CNVs encompassed genes appeared to show similar patterns in our studies. More than 70% of the CNVs that were sensitive to copy-number changes in contrasts between homozygous individuals appeared to be recessive when in the heterozygote (fig. 5A). This finding suggests a buffered response to structural changes induced by CNVs and implies that heterozygous masking effect may protect genes from harmful consequences. In the case of gene duplications, apparent masking in heterozygotes may reflect the reduced power to detect transcript abundances of 3:2 in heterozygotes versus 4:2 in homozygotes. Silencing by unpaired DNA might be another mechanism operating in heterozygous CNVs (Shiu and Metzenberg 2002). The unpaired copy of a gene might reduce the expression of other homozygous copies in the genome.
Consistent with a previous report (Dopman and Hartl 2007), we found that only 20% of the CNVs were nonsingletons in the population. Interestingly, the fraction of nonsingletons for recessive dosage-sensitive CNVs is increased significantly (47%) compared with that of overall CNVs (fig. 5B). The increase suggests that selection typically prevents the spread of CNVs in natural populations, however, with a higher tolerance if the CNVs are recessive in their effects on expression. Consistent with this hypothesis, the fractions of singletons and nonsingletons for nonrecessive dosage-sensitive CNVs did not differ from that of overall CNVs (fig. 5B). Another category in which the fraction of nonsingleton increased significantly consists of dosage-insensitive CNVs, most likely due to the low penetrance of CNVs having little or no effects on phenotypes.
Selection May Constrain Protein Interactions for CNV Genes
It is known that protein-coding changes may impair the ability of a protein to form dependable network interactions (Fraser et al. 2002). CNVs were reported to negatively correlate with the degree of protein interaction network (Dopman and Hartl 2007), indicating selection is likely to shape the CNV distribution. Here, we also found that dosage-sensitive CNVs have fewer protein–protein interactions than dosage-insensitive CNVs (fig. 7A). Dosage-insensitive genes are less stringent to structural changes such that their mutational influences in the protein network are kept minimal. In contrast, stronger selection on central nodes may result in dosage-sensitive genes showing a lessened number of protein–protein interactions and betweenness. Similarly, recessive CNV genes were expected to have more protein interactions than nonrecessive ones. However, possibly due to a relatively small sample size, they did not show statistically significant difference in our study although the trend appeared consistent with the expectation (fig. 7B).
Up- and Downregulated CNVs Coincide with Copy-Number Increase and Decrease
CNVs that are upregulated or downregulated in expression were found positively associated with their copy-number changes (fig. 8). The fraction of upregulated or downregulated CNVs is higher than that of dosage-sensitive CNVs in heterozygotes discussed above (∼27% vs. ∼10%). Some trans-effects (or background effects from PS3) may be involved in determining dosage effects in heterozygotes. In the case of up- and downregulated CNVs (fig. 8), one may expect that the fraction of singletons would increase relative to that of overall CNVs because selection is presumably against gene copy-number changes. However, the fraction of singletons and nonsingletons did not differ from that of overall CNVs (data not shown).
Conclusions
This study has revealed several important features of CNVs in a number of second-chromosome substitution lines from a single natural population of D. melanogaster, particularly with respect to the balance between CNV encompassed gene dosage and expression. The fraction of CNVs among homozygotes appeared to be higher than in heterozygotes, indicating underestimation by aCGH arrays. We found many cases of CNV genes that are sensitive to copy-number changes. However, the majority of genes show no significant change in expression with copy number. More than 70% of the CNVs are recessive in expression in heterozygotes. Selection appears to prevent CNVs from spreading in the population as indicated by allele frequencies, recessive CNVs, and protein interaction data. With this CNV and expression association study in D. melanogaster, we have achieved an understanding of CNV dosage effect to some extent. Many questions still remain unsolved such as CNV distributions on other chromosomes besides the second, what mechanisms cells employ to modulate CNV dosage and reach a balance. Despite the critical role of CNVs in shaping genotypes and phenotypes, the majority of the identified CNVs have not yet been finely resolved to the nucleotide level. Large CNV genotype data sets from different populations are required to extensively study the roles of CNV in genome evolution. Next-generation sequencing or a combination of aCGH array and sequencing tools will enable us to dissect this relationship further to greater resolution.
Supplementary Material
Supplementary figures S1–S3 and table S1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe).
CNV raw data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27632).
Acknowledgments
We thank Tim Sackton for his critical comments on the manuscript and Hsiao-Han Chang for her assistance on part of the data analysis. This research was supported by National Institutes of Health grant GM065169 and GM084236 to D.L.H.
References
- Bachtrog D, Toda NR, Lockton S. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr Biol. 2010;20:1476–1481. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Schafer M, White R. Restriction sites containing CpG show a higher frequency of polymorphism in human DNA. Cell. 1984;36:131–138. doi: 10.1016/0092-8674(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Biological consequences of dosage dependent gene regulatory systems. Biochim Biophys Acta. 2007;1769:422–428. doi: 10.1016/j.bbaexp.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreto L, et al. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol. 2010;2:83–101. doi: 10.1093/gbe/evq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S. Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol. 2010;2:441–453. doi: 10.1093/gbe/evq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- Farre D, Alba MM. Heterogeneous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol. 2010;27:325–335. doi: 10.1093/molbev/msp242. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- Hild M, et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci U S A. 2008;105:14471–14476. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Altshuler DM. Human disease. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, et al. Intergrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Perry GH, et al. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci U S A. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B, Conrad D, Bateman A. Dosage sensitivity shapes the evolution of copy-number varied regions. PLoS One. 2010;5:e9474. doi: 10.1371/journal.pone.0009474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shiu PKT, Metzenberg RL. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics. 2002;161:1483–1495. doi: 10.1093/genetics/161.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS, Rhomberg LR. A comprehensive study of genic variation in natural Populations of Drosophila melanogaster. II. Estimates of heterozygosity and patterns of geographic differentiation. Genetics. 1987;117:255–271. doi: 10.1093/genetics/117.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, et al. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0071. RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–84. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 2009;17:585–602. doi: 10.1007/s10577-009-9053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium (WTCCC) Genome-wide association study of CNVs in 16000 cases of eight common diseases and 3000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]