Abstract
Miniature inverted-repeat transposable elements (MITEs) are abundant repeat elements in plant and animal genomes; however, there are few analyses of these elements in fungal genomes. Analysis of the draft genome sequence of the fungal endophyte Epichloë festucae revealed 13 MITE families that make up almost 1% of the E. festucae genome, and relics of putative autonomous parent elements were identified for three families. Sequence and DNA hybridization analyses suggest that at least some of the MITEs identified in the study were active early in the evolution of Epichloë but are not found in closely related genera. Analysis of MITE integration sites showed that these elements have a moderate integration site preference for 5′ genic regions of the E. festucae genome and are particularly enriched near genes for secondary metabolism. Copies of the EFT-3m/Toru element appear to have mediated recombination events that may have abolished synthesis of two fungal alkaloids in different epichloae. This work provides insight into the potential impact of MITEs on epichloae evolution and provides a foundation for analysis in other fungal genomes.
Keywords: Epichloë, transposable element, endophyte, genome evolution, fungi
Introduction
Transposable elements are characterized by their ability to move, or transpose, within genomes and are ubiquitous in all kingdoms of life. Transposons have a substantial impact on genome function and evolution: transposition of these “selfish” elements can lead to mutation by insertion within genes and can alter transcription by removal or addition of cis elements or by epigenetic mechanisms (Kidwell and Lisch 1997; Feschotte 2008). The repeat sequences generated by transposon movement and expansion can also be responsible for local and global genome rearrangements (Fierro and Martin 1999; Mieczkowski et al. 2006).
Transposable elements have been divided into two classes. Type 1 elements, or retroelements, transpose through an RNA intermediate, whereas type 2, or DNA transposons, mostly utilize a “cut and paste” mechanism of transposition. Miniature inverted-repeat transposable elements (MITEs) are nonautonomous DNA (type 2) transposable elements that require the transposase from an autonomous parent element for transposition (Feschotte et al. 2002). Like autonomous DNA transposons, MITEs are characterized by terminal inverted repeats (TIRs) and a target site duplication (TSD). However, unlike autonomous elements, MITEs have no coding capacity, and unlike other deleted elements, MITEs amplify to high copy number and copies are homogeneous in size (usually <500 bp) (Feschotte et al. 2002).
Some MITEs appear to be direct deletion derivatives of autonomous copies (Jiang et al. 2003), whereas in many other cases, MITEs appear to evolve independently by recombination events that lead to a pair of TIRs sufficiently similar to those of an autonomous element to be able to be mobilized by its transposase (Jiang et al. 2004). It has long been a puzzle as to how these deleted elements are able to amplify to a much higher copy number than their parents. A recent landmark study showed that the Stowaway MITE in rice does not contain a repressor element present in the autonomous Mariner elements (Yang et al. 2009). In addition, the Stowaway MITE has an enhancer of transposition that further facilitates its ability to amplify over Mariner. Although MITEs arising from simple deletion are unlikely to contain an enhancer, deletion of repressor elements may be a common mode of copy number amplification of these elements.
In higher eukaryotes, MITEs can make up a large proportion of the genome repeat content, especially in plants where a substantial proportion of the genome can consist of these elements (Santiago et al. 2002; Jiang et al. 2004; Juretic et al. 2004; Benjak et al. 2009). In plants thus far examined, MITEs have an integration site bias for genic regions of the genome (e.g., Bureau and Wessler 1994a, 1994b; Mao et al. 2000) and thus likely influence expression of associated genes. MITEs in fungi have received little attention, with just two families being characterized: Guest in Neurospora crassa (Yeadon and Catcheside 1995; Ramussen et al. 2004) and mimp in Fusarium oxysporum (Hua-Van et al. 2000; Dufresne et al. 2007; Bergemann et al. 2008). However, recently, a number of uncharacterized MITEs have been reported in fungal genome sequences (Martin et al. 2008; Spanu et al. 2010).
We previously identified five MITE-like elements present within secondary metabolite gene clusters in epichloid fungi (Epichloë and Neotyphodium species: Ascomycota, Sordariomycetes, Hypocreales, Clavicipitaceae). These fungi are endophytic symbionts of grasses, producing alkaloids that protect the host plant from herbivory by insects and grazing animals. Annotation of the EAS gene cluster for ergot alkaloid synthesis in Neotyphodium lolii identified two MITEs, Toru and Rima (Fleetwood et al. 2007). Examination of the LTM gene cluster for lolitrem B biosynthesis revealed three futher MITEs, labeled EFT-14, EFT-24, and EFT-25 (Young et al. 2009). The presence of five putative MITEs in such a restricted sequence analysis led to the hypothesis that these elements are abundant components of epichloae genomes.
Here we describe the presence of 13 families of degenerate MITEs in the 34.4-Mb draft genome sequence of Epichloë festucae E2368. We show that at least some of these families were present in the common ancestor of the epichloae lineage, that overall MITEs show a bias for integration within 5′ regions of genes, and are particularly enriched near secondary metabolism genes. We further describe the probable impact of EFT-3m elements on rearrangements and deletions at two secondary metabolite gene loci, highlighting the possibly large impact of these elements on genome evolution of epichloid fungi.
Materials and Methods
Fungal Strains
Strains used for computational, sequence, and Southern blot analysis are described in table 1. Fungi were grown in potato dextrose broth or agar at 22 °C.
Table 1.
Fungal Strains Used in This Study
| Fungal Strain | Parentagea | ATCC# or Reference |
| Epichloë clarkii E426 | n/a | ATCC 200741(Moon et al. 2004) |
| E. festucae E2368 | n/a | C. Schardl, University of Kentucky, KY |
| E. festucae Fl1 | n/a | ATCC MYA-3407 (Young et al. 2005; Moon et al. 1999) |
| E. sylvatica E503 | n/a | ATCC 200751 (Moon et al. 2004) |
| E. typhina E8 | n/a | ATCC 200736 (Chung et al. 1997) |
| Neotyphodium coenophialum e19 | Efe × ETC × LAE | ATCC 90664 (Tsai et al. 1992) |
| N. lolii Lp19 | Efe | (Christensen et al. 1993) |
| N. lolii AR1 | Efe | (Moon et al. 1999) |
| N. lolii Lp14 (AKA AR37) | Efe | (Christensen et al. 1993) |
| Neotyphodium sp. Lp1 | Efe × ETC | (Christensen et al. 1993) |
| N. uncinatum e167 | Ebr × ETC | (Blankenship et al. 2001) |
| Claviceps cynodontis Haskell | n/a | (Marek et al. 2006) |
| Fusarium graminearum | n/a | Unnamed, K. Craven, Noble Foundation, OK |
| Phymatotrichopsis omnivora OKAlf8 | n/a | (Marek et al. 2009) |
Note.—n/a, not applicable.
Closest sexual ancestors to asexual species (Moon et al. 2004). Ebr, E. bromicola; Ef, E. festucae; ETC, E. typhina complex (=E. typhina, E. sylvatica, E. clarkii); LAE, Lolium-associated endophyte (closest extant species = E. baconii).
Identification of MITEs in the E. festucae Genome Sequence
MITEs were computationally mined from the E. festucae genome in two parts: 1) identification of seed MITE sequences used to create libraries of hidden Markov models (HMMs) representing distinct MITE families and subfamilies and 2) searching of HMM libraries against the genome to comprehensively identify and classify MITE instances, including degraded, nested, or autonomous elements, and to support analysis of insertion sites. Bioinformatics analyses were implemented using a combination of various software and custom Perl scripts.
The E. festucae E2368 genome contigs (version 200606) were first masked for the following classes of repeats to prevent repetitive regions resulting in spurious MITE candidates: simple repeats and known fungal repeats (RepeatMasker 3.2.8, http://www.repeatmasker.org, cited 2011 Oct 7), microsatellites (Sputnik, http://espressosoftware.com/sputnik/index.html, cited 2011 Oct 7), tandem repeats TRF 4.00 (Benson 1999), and low complexity regions (dust; Morgulis et al. 2006). Seed MITEs were identified from the masked genome using two successive rounds of Vmatch (Kurtz et al. 2001) to identify TIRs (TIR length range 10–65 bp, ≥80% identity, maximum inter-TIR distance 650 bp). TIRs identified by the first Vmatch round were used to demarcate approximate MITE-containing regions; in the second round, each region was submitted to an individual Vmatch search using the same criteria after masking inter-TIR regions with Ns, which focused the palindrome search to the terminal ends of the regions. The additional search round served two purposes: 1) refinement of the seed MITE boundaries because we observed that Vmatch sometimes extended match regions in the second round and 2) collapsing of multiple palindromes within the same region to a single seed MITE at each locus. Seed MITE sequences were extracted from the genome and clustered into putative families using an all-against-all basic alignment search tool (Blast) with sensitive discontiguous Blast parameters, which forced matches to be seeded within TIRs (-e 1 × 10−10 -b 10000 -v 10000 -U T -F “m D” -r 1 -q -1 -G 2 -E 2 -W 9 -m 9), followed by clustering using the Markov Cluster algorithm (MCL; van Dongen 2000) using Blast similarity scores (normalized bit scores) as the similarity criterion. Clusters with <10 members were discarded, and clusters remaining were deemed to represent putative MITE families. Seed MITE sequences within each family were aligned using TCoffee (Notredame et al. 2000) and visualized using JalView (Waterhouse et al. 2009). To focus on conservation within TIRs and improve alignment in these regions, additional alignments containing seed MITEs with masked inter-TIR regions (replaced by 5 Ns) were generated.
To identify subfamily structure, we attempted sequence-based clustering within families using MCL or hierarchical agglomerative clustering (hclust in the R statistical package; http://www.R-project.org, cited 2011 Oct 7) but did not recover the manually identified subfamily structure of the Toru family, most likely due to degeneracy of subfamily members. Therefore, we manually partitioned families into putative subfamilies based on element lengths. The subfamilies were then aligned using Muscle (Edgar 2004) for visualization in JalView. Flanking regions (50 bp either side) were extracted and used to create separate alignments to examine similarity within their genome contexts. Those subfamilies with marginal conservation or conserved flanking regions were discarded.
To comprehensively identify MITE instances in the genome, libraries of subfamily HMMs were constructed from the seed MITE subfamily alignments using hmmbuild from the HMMER2 package (Eddy 1998). Two libraries were created: “global” containing global HMMs aimed at finding complete elements and “local” containing local HMMs aimed at finding deleted instances (fragments of elements which have most likely arisen from deletions occurring within full-length elements over time). We included deleted instances in our analysis and also instances that did not contain both putative TSD sequences. This means there may be a low level of false-positive instances for some families; however, we were willing to accept this in order to perform a comprehensive analysis of the highly degenerate MITEs in the E. festucae genome. The libraries were compared with the masked E. festucae genome contigs using hmmpfam (from HMMER2), explicitly searching both forward and reverse strands in separate searches; positive, nonzero scoring hits were flagged as candidate MITE instances. Multiple, overlapping MITEs at a single genomic locus were reduced to a single representative by selecting the instance with the best score (note that a global match always trumped a local match). We used hmmalign (from HMMER2) to build subfamily alignments using the appropriate model to guide the alignment so that it best represented any structural characteristics of that subfamily. Alignments including flanking sequences were used to manually correct element boundaries. These corrected elements represented the final collection of MITE instances, used for subsequent analyses. In addition, subfamily consensus sequences were identified using hmmemit (from HMMER2), and TIR coordinates on consensus were found using Vmatch.
The collection of MITE instances were postprocessed to identify nested MITEs and possible parent autonomous elements because criteria used in the original Vmatch search would not identify TIRs separated by extraordinary distances, which could arise due to displacement of TIRs by nested elements or an autonomous state prior to any significant deletion of the inter-TIR region. Proximal MITE TIRs were linked (assumed to be derived from a single element) if they belonged to the same subfamily and lay within 4 kb in the correct orientation relative to one another.
A track showing MITE positions in the E. festucae Gbrowse is available at the E. festucae Genome Project webpage (http://www.endophyte.uky.edu/, cited 2011 Oct 7).
Integration Site Analysis
To identify whether MITEs are preferentially located near genes, insertion sites were compared with locations of open reading frames (ORFs) extracted from messenger RNA models (version 2, available on request), first converting genome contig coordinates of MITEs to positions on scaffolded supercontigs (E. festucae genome assembly version 200606). Some spurious computationally generated ORFs were removed by manual curation. An instance was classified as “near” an ORF if it was within 500 bp of an ORF boundary, excluding ranges occupied by other proximal MITEs to simulate the state of the genome prior to any insertion events and avoid penalizing MITEs clustered near ORFs. Counts of MITEs observed near an ORF, near an ORF 5′ end, and near an ORF 3′ end were submitted to two-tailed binomial tests to determine the significance of these observations compared with random insertion. The null probability (P) for random insertion was derived by dividing the number of genome positions in near regions by the total number of positions in the genome, excluding positions spanned by MITEs and other ORFs in both cases. Proximity of MITE insertion sites to 41 manually annotated nonribosomal peptide synthetase (NRPS) and polyketide synthetase (PKS) genes was examined in a similar way. Fungal secondary metabolite genes are usually found in tight gene clusters and thus in the absence of other annotated secondary metabolism genes “near” was defined as within 10 kb of an NRPS/PKS ORF, as an approximation of any secondary metabolite gene cluster regions.
DNA Extraction and Southern Blot Analysis
Genomic DNA from Epichloë spp., Neotyphodium spp., N. crassa, Fusarium graminearum, Claviceps cynodontis, and Phymatotrichopsis omnivora was isolated from freeze-dried mycelium using ZR Fungal/Bacterial DNA kit (Zymo Research), Plant DNeasy kit (Qiagen, Hilden, Germany), or a published method (Byrd et al. 1990). Genomic DNA (2 μg) was digested overnight at 37 °C with 48 units of EcoRI (Promega). Digested genomic DNA was separated overnight in 0.7% agarose gel and transferred overnight to nylon membranes (Zeta probe blotting membrane, BioRad) by capillary transfer. Membranes were UV cross-linked (120,000 μJ/cm2) in UV stratalinker 2400 (Stratagene). The EFT-14 MITE element was amplified from E. festucae genomic DNA using primers EFT-14F, 5′-GTGAGACAGATATATCAGGCACA-3′, and EFT-14R, 5′-GATTTAAGACGGATTGGAATGATG-3′. Sequence-specific polymerase chain reaction (PCR) was carried out in a reaction volume of 50 μl containing 5 ng E. festucae genomic DNA, 1× green reaction buffer (Promega), 200 μM of each deoxyribonucleotide triphosphate, 200 nM of each primer, and 1 U GoTaq (Promega). Thermocycling conditions were 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 55 °C for 30 s, 72 °C for 1 min, and then a final extension at 72 °C for 10 min. PCR products were purified using a PCR purification kit (Qiagen). Probe labeling and hybridization were performed using Amersham gene images AlkPhos direct labeling and detection system (GE Healthcare). Hybridizations were carried out overnight at 50 °C in AlkPhos Direct hybridization buffer. Posthybridization washes were performed according to the manufacturer’s instructions (GE Healthcare) using CDP-Star (GE Healthcare) for chemiluminescent signal generation. Blots were exposed to Biomax XAR film (Kodak) for 30 min to 24 h depending on signal strength.
PCR and DNA Sequencing
For targeted sequencing of repeat regions, PCR products were amplified using either Pfx50 (Invitrogen) for the N. lolii AR1 and Neotyphodium sp. Lp1 easA–easG region or Triplemaster (Eppendorf) for the N. lolii Lp14 perA 3′ region. PCR products were either sequenced directly or cloned into p-GEMT and sequenced by M13 and custom primers. Dye terminator sequencing was performed using BigDye v3.1 (Applied Biosystems) and separated on either an ABI3130XL or ABI3730 capillary sequencer (Applied Biosystems) at the University of Auckland Centre for Genomics and Proteomics or the Massey University Alan Wilson Centre Genome Service, respectively.
Sequences obtained in this study are available at GenBank under accessions JF494831 (AR1 easA–easG) and GU966659 (Lp14 perA).
Results
Identification and Characterization of 13 MITE Families
Five MITEs previously identified in epichloid endophytes were found to be highly degenerate. For this reason, we predicted that existing algorithms for MITE identification, which rely on high similarity between copies (Tu 2001) or known TIR sequences (Santiago et al. 2002; Bergemann et al. 2008), were not suitable for use in E. festucae. We thus developed a computational pipeline utilizing various programs to identify MITEs ab initio.
A list of candidate MITEs was identified from the E. festucae genome by searching for inverted-repeat sequences using Vmatch (supplementary data, Supplementary Material online). Candidates were then sorted into 79 putative MITE families based on similarity in inverted-repeat regions. Alignments of clustered MITE sequences were examined manually, and 66 of these were discarded due to poor overall similarity or due to extensive similarity in the sequences flanking the inverted repeats, indicative of the inverted repeat being within a larger repeat element. We thus examined 13 putative MITE families further. These were named EFT-[number]m, with “m” standing for MITE. Previously identified MITEs, Toru and Rima, were labeled EFT-3m/Toru and EFT-5m/Rima, respectively. Characteristics of the MITE families are described in table 2; consensus sequences are available as supplementary data (Supplementary Material online). Mean pairwise identity within each family varied but was in most cases low, varying between 49% and 90%. Accurate genomic copy numbers of elements were obtained, including counting of degraded and deleted elements which were likely to be missed by the Vmatch search, by searching a MITE library of HMMs representing MITE families/subfamilies against the E. festucae genome. Copy number varied, with the highest copy subfamily, EFT-3A, containing 97 full-length elements. Two families contained fewer than 10 full-length copies; however, we considered them as MITEs alongside the high copy elements as in each case there were substantially more deleted instances in the genome sequence and fungal genomes are relatively small in comparison to the plants for which the 10-copy criterion was considered applicable for MITE categorization (Feschotte et al. 2002). Combining all families, a total of 1,249 MITE instances were identified (415 full length and 834 deleted).
Table 2.
Characteristics of MITEs in the Epichloë festucae Genome
| Family | Mean Length (range) | Mean Pairwise %ID | TSD | TIR Length (%ID) | #Full Copies (#Deleted)a | Putative Superfamily | Parent Element? |
| EFT-3mA/Toru | 135 (109–177) | 75 | AT | 29 (96) | 97 (47) | Unknown | No |
| EFT-3mB | 256 (218–389) | 67 | AT | 29 (96) | 24 (63) | Unknown | No |
| EFT-5m/Rima | 294 (259–356) | 62 | TAb | 61 (86) | 7 (14) | Tc1/mariner | No |
| EFT-8m | 401 (364–567) | 76 | TWY | 34 (91) | 12 (98) | Pif/Harbinger | Yes |
| EFT-9m | 246 (139–535) | 70 | TA | 34 (88) | 61 (101) | Tc1/mariner | No |
| EFT-11m | 406 (393–410) | 86 | 8 bp | 10 (100) | 29 (106) | hAT | No |
| EFT-14m | 291 (193–434) | 68 | TA | 36 (91) | 63 (116) | Tc1/mariner | No |
| EFT-24m | 380 (297–653) | 49 | 9 bp | 115 (81) | 31 (89) | Mutator | No |
| EFT-25m | 81 (77–86) | 90 | TA | 24 (95) | 14 (27) | Tc1/mariner | Yes |
| EFT-26m | 364 (287–470) | 69 | 9 bp | 116 (93) | 13 (83) | Mutator | Yes |
| EFT-27mA | 148 (130–188) | 84 | AT | 38 (97) | 14 (20) | Unknown | No |
| EFT-27mB | 267 (254–402) | 67 | AT | 40 (87) | 19 (14) | Unknown | No |
| EFT-28m | 259 (207–292) | 65 | ATc | 35 (91) | 10 (30) | Unknown | No |
| EFT-29md | 83 (57–108) | 83 | TA | 25 (88) | 16 (13) | Tc1/mariner | No |
| EFT-30m | 524 (509–540) | 80 | TA | 43 (95) | 5 (13) | Tc1/mariner | No |
Note.—ID, identity.
Copy number data are from publicly available genome assembly (EF201006); other data are from the version 200606 assembly that all other analysis was performed on.
Putative—degenerate at ends.
Only two with putative AT TSD although frequent AT at one end.
Closely related to EFT-25, internal 30–40 bp dissimilar, and TIRs imperfect.
To further categorize the MITE elements, we attempted to identify subfamilies within the family designations. The high level of degeneracy precluded identification of relationships from multiple sequence alignments; we thus, used a size-based criterion to identify subfamily relationships. Based on manual size separation and analysis of alignments, two families were identified that contained two subfamilies each, EFT-3m and EFT-27m. EFT-3mA and EFT-3mB differed only in size, whereas EFT-27mA and EFT-27mB shared only the 34 bp of the TIR with no similarity between intervening sequences.
In the absence of transposase sequences, superfamilies were predicted for each family based on TIR and putative TSD characteristics (Wicker et al. 2007) (table 2). Six families belonged to the Tc1/mariner superfamily, consistent with Tc1/mariner being a common type 2 superfamily in fungi. Other MITE families were characterized as Mutator, Pif/Harbinger, and hAT superfamilies. Three families, EFT-3m, EFT-27m, and EFT-28m were classified as unknown. These families most closely resemble CACTA elements although in the case of EFT-3m and EFT-27m, the TIR terminal sequences (CTCC) did not exactly match the consensus found in either fungi (CCC, DeMarco et al. 2006) or plants (CMCWR, Feschotte 2008). EFT-28m TIRs do terminate in CCC, but a 2-bp TSD was not always present.
Degenerate Autonomous Elements EFT-8, EFT-25, and EFT-26
Considering that MITEs require autonomous elements for transposition, we analyzed the E. festucae genome for the presence of putative parent elements. Analysis of linked instances of adjacent deleted MITE sequences containing single TIRs in the correct orientation (supplementary table S1, Supplementary Material online) revealed putative autonomous element relics that are the likely progenitor sequences of EFT-8m, EFT-25m, and EFT-26m. Analysis of linked EFT-8m TIRs revealed a 3076-bp autonomous element relic (EFT-8) with two full-length copies and another four sequences longer than 50% of full length in the genome assembly. BlastX analysis of the GenBank nonredundant databases showed the predicted translation of a 794-bp region of EFT-8 to have 29% identity (E = 2 × 10−13) with a hypothetical protein Os08g0459400 from Oryza sativa (rice) and similar percent identity but over a smaller region to numerous Pif-like transposases from various plants and fungi. This sequence contained 25 stop codons, and the high AT percentage of the full EFT-8 sequence (71%) suggested that the element had undergone repeat-induced point mutation (RIP; Cambareri et al. 1989). RIP results in C:G to T:A transitions in repeat sequences and was observed for other identified autonomous elements in epichloae (Young et al. 2005; Fleetwood et al. 2007). Alignment of full-length and deleted sequences showed a strong bias for C to T and G to A transitions, supporting RIP as the main cause of the degeneration.
A putative autonomous EFT-25 relic was identified on contig 1003 as two deleted sequences of EFT-25m separated by 1,313 bp but not containing other nested elements. All three reading frames of the sequence between the TIRs contained numerous stop codons, and BlastX analysis did not match any transposase sequences in the databases; however, the sequence was highly AT rich (78%) and likely to be the degenerate product of RIP. Only a single copy of this putative autonomous relic of EFT-25 was found in the E. festucae genome, but eight sequences corresponding to greater than 10% of the full-length sequence were present. In all but one instance, these were truncated at the end of a small contig. Alignment of the EFT-25 sequence with EFT-25m showed the MITE to be a direct deletion derivative of the full-length element.
A putative autonomous EFT-26 was identified on contig 754, which contained 2709 bp between TIRs. BlastX analysis of this sequence revealed a 1,386-bp region sharing 29% identity (E = 3 × 10−39) to a hypothetical protein in Chaetomium globosum (EAQ85500) and 32% identity over a smaller region to the Hop mutator transposase from F. oxysporum (AAP31248). This sequence contained far fewer stop codons (5) than the other two autonomous elements. This result and the lower AT percentage (55%) suggest this element has not been subjected to RIP to the same extent as the other two elements, perhaps suggesting a more recent origin. There was only a single full-length EFT-26 in the assembly with only one other non-MITE sequence aligning over 593 bp of the full-length element at the end of a small contig (contig 2450). Alignment of the full-length EFT-26 sequence with the EFT-26m consensus sequence showed that, as for the other two autonomous relics, the MITE is likely to be a deletion derivative of the full-length element.
Early MITE Invasion of Epichloid Genomes
The high sequence variation and large number of indels between copies of individual elements suggest that MITEs are ancient features of epichloid genomes. To determine the taxonomic extent of MITE colonization, we first searched the public databases for the presence of different families in published sequences from different Epichloë and Neotyphodium species not containing E. festucae parentage. We identified EFT-3m, EFT-11m, EFT-14m, EFT-24m, and EFT-26m in one or both of the two LOL gene clusters in N. uncinatum (E. typhina × E. bromicola) and EFT-11m in the LOL cluster of Neotyphodium sp. PauTG-1 (E. typhina × E. elymi) (supplementary table S2, Supplementary Material online).
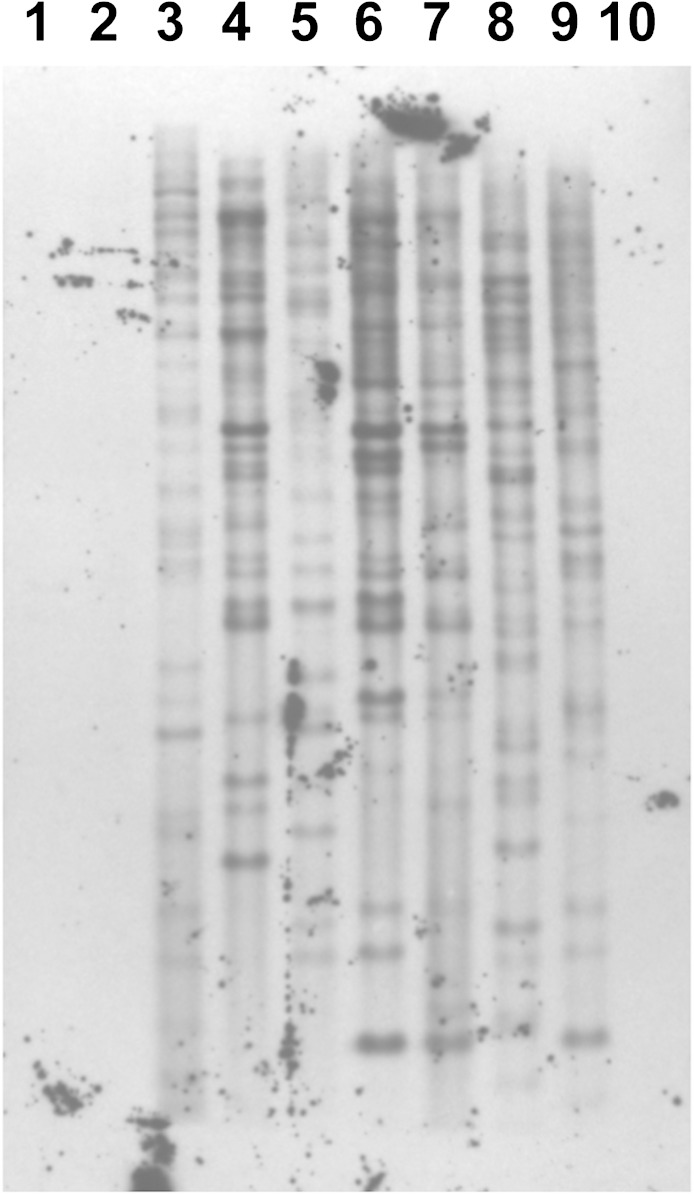
To further examine the distribution of MITEs, we performed Southern blot analysis of a range of epichloae with EFT-14m (fig. 1). This element was found in similar copy number across the range of Epichloë species tested, further supporting an early origin in this genus. Weak nonspecific hybridization was observed for A. fumigatus and N. crassa (data not shown), but no hybridization was seen for C. cynodontis, a member of a clavicipitaceous genus closely related to Epichloë/Neotyphodium in recent analyses (Sung et al. 2007). Due to the degeneracy of the MITE sequences and the subsequent low stringency hybridization conditions required, we were unable to obtain specific hybridization data for other MITEs tested, EFT-3m and EFT-9m.
FIG. 1.—
Taxonomic distribution of EFT-14m. Southern blot analysis was performed using genomic DNA extracted from various epichloae and closely related fungal species, transferred to a nylon membrane and hybridized with an AlkPhos direct labeled (GE Healthcare) EFT-14m probe amplified by PCR using primers EFT-14F and EFT-14R. 1, Fusarium graminearum; 2, Claviceps cynodontis; 3, Neotyphodium uncinatum E167; 4, Epichloe clarkii E426; 5, E. sylvatica E503, 6, E. festucae E2368; 7, E. festucae Fl1, 8, E. typhina E8; 9, N. coenophialum E19; 10, Phymatotrichosis omnivora.
MITEs Are Enriched Upstream of Genes and Near Secondary Metabolite Genes
MITEs are often found in genic regions of genomes. To determine the precise integration sites of E. festucae MITEs, MITE locations on supercontigs, as annotated by the MITE library genome-wide search, were compared with the locations of ORFs. MITE instances were classified as to whether they had inserted near (within 500 bp) to an ORF and near to 5′ or 3′ ends of ORFs (table 3). MITEs were frequently found within 500 bp of a predicted ORF (table 3), consistent with similar analyses for other organisms. To further analyze how frequently MITEs are found upstream of ORFs in putative regulatory regions, MITE instances were classified as to whether they had inserted near (within 500 bp) to 5′ or 3′ ends of ORFs (table 3). Binomial tests (using random insertion in the genome not including MITEs as the null distribution) on these data indicated MITE insertions were somewhat enriched 5′ of ORFs (x = 357, n = 1,077, P = 0.3038, P value = 0.051) and were strongly depleted at 3′ ends of ORFs (x = 207, n = 1,077, P = 0.2771, P value = 1.34 × 10−10).
Table 3.
Integration Site Data for Epichloë festucae MITEs
| Family | Total Copies | % Within 500 bp of ORF | % Within 500 bp 5′ of ORF | % Within 500 bp 3′ of ORF | % Near SM Gene |
| EFT-3mA/Toru | 126 | 44 | 33 | 14 | 5 |
| EFT-3mB | 87 | 44 | 21 | 28 | 3 |
| EFT-5m/Rima | 19 | 47 | 32 | 26 | 11 |
| EFT-8m | 110 | 32 | 18 | 17 | 4 |
| EFT-9m | 162 | 49 | 38 | 16 | 3 |
| EFT-11m | 18 | 28 | 17 | 11 | 0 |
| EFT-14m | 185 | 50 | 35 | 24 | 2 |
| EFT-24m | 124 | 44 | 35 | 15 | 2 |
| EFT-25m | 40 | 33 | 25 | 15 | 0 |
| EFT-26m | 96 | 47 | 32 | 18 | 1 |
| EFT-27mA | 34 | 68 | 44 | 35 | 0 |
| EFT-27mB | 31 | 42 | 39 | 13 | 0 |
| EFT-28m | 40 | 38 | 30 | 10 | 0 |
| EFT-29m | 30 | 47 | 33 | 20 | 3 |
| EFT-30m | 18 | 39 | 39 | 6 | 6 |
| All MITEs | 1,120 | 47*** | 33* | 19*** | 3** |
Note.—SM, secondary metabolism. % 5′ + % 3′ ≥% within 500 bp of ORF due to some elements being near adjacent ORFs.
*P ≤ 0.1 (enriched), **P ≤ 0.05 (enriched), ***P ≤ 0.005 (depleted).
As MITEs in epichloae were initially discovered within gene clusters for secondary metabolite production, we next looked at whether MITE integrations were significantly enriched near secondary metabolite genes within the E. festucae genome. We first examined previously annotated LOL, EAS, and LTM gene clusters (for loline, ergot alkaloid, and indole diterpene synthesis, respectively) for MITE insertions. (Note: Whereas initial analysis was performed on an early assembly of the E. festucae E2368 genome, contig numbers quoted in this section relate to the more complete 201006 assembly, which is publicly accessible.) Within the LOL cluster, which contains 11 genes on three contigs in the E. festucae E2368 assembly (contigs 1349, 1659, and 4990), we identified eight MITE insertions (3 × EFT-3m, 2 × EFT-11m, 2 × EFT-14m, and 1 × EFT-24). The EAS cluster consists of 11 genes and is found on contig 1654. Nine MITEs were found in this cluster, seven EFT-3m, one EFT-5m, and one EFT-9m, which was nested within one of the EFT-3m copies. Half of the 10-gene LTM cluster is absent from the genome assembly (E. festucae E2368 is a non–indole diterpene-producing strain); however, a truncated cluster of ltmP, ltmQ, ltmF, ltmC, and ltmB genes is found on contig 597. Four MITEs, two EFT-14m, one EFT-3m, and one EFT-8m are found in this cluster.
Given the number of MITE insertions in these gene clusters for previously characterized secondary metabolites, we wished to extend this analysis to include uncharacterized secondary metabolite gene loci. As secondary metabolite gene clusters other than those for previously known alkaloids were not yet annotated, we analyzed whether insertions were enriched within 10 kb of genes predicted to encode the secondary metabolite biosynthetic proteins NRPSs and PKSs, based on preliminary annotations derived using a combination of SMURF (Khaldi et al. 2010), Blast, and manual annotation (Epichloë Genome Consortium, unpublished data). This analysis indicated a strong preference for integration site bias near this class of gene (x = 31, n = 1,077, P = 0.0183, P value = 0.016) (table 3).
Rearrangements Mediated by EFT-3 Recombination
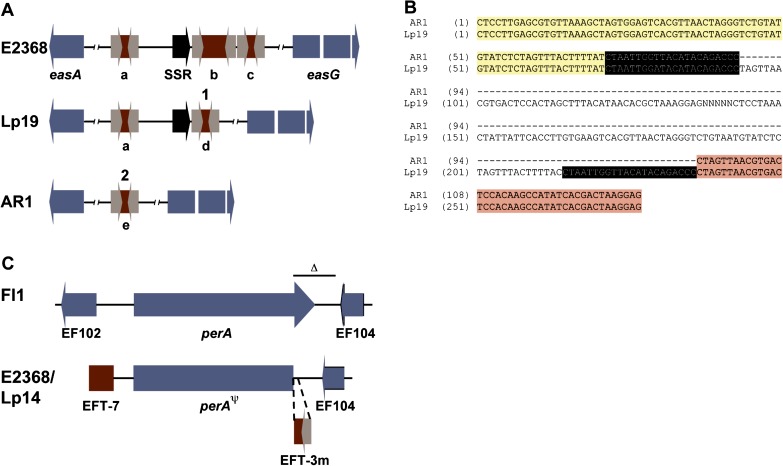
Recombination between repeat sequences provided by transposable elements can cause genomic rearrangements. Sequence analysis at the easA–easG and perA loci lead us to examine the effect of the EFT-3m element on local rearrangements in E. festucae. Comparison of the E. festucae E2368 easA–easG intergenic region with that of the previously sequenced N. lolii Lp19 (N. lolii = E. festucae anamorph) locus (accession EF125025) revealed a third EFT-3m integration in E2368 directly adjacent to one of the two found in this region in Lp19 (fig. 2). Comparison of the sequence of these two EFT-3m elements revealed that the “second” element in Lp19 was not 100% identical to either of the adjacent elements at the E2368 locus. Interestingly, the arbitrary left 44 bp were identical to the left side of the second E2368 element (b in fig. 2A), whereas the right 90 bp of the Lp19 element were identical to the right side of the “third” E2368 element (c in fig. 2A), with the intervening 18 bp identical in both E2368 elements. This indicated that the common ancestral locus likely contained the arrangement found in E2368 and that recombination between the adjacent elements led to a deletion event that recapitulated a single EFT-3m element as observed in Lp19.
FIG. 2.—
EFT-3–mediated rearrangements at secondary metabolite gene loci. (A) Rearrangements in easA–easG intergenic region in two Neotyphodium strains compared with the Epichloë festucae E2368 locus. 1. Recombination between adjacent EFT-3mB (b) and EFT-3mA (c) MITEs caused a deletion, resulting in the recapitulation of one EFT-3mA copy in Neotyphodium lolii Lp19 (d). 2. Recombination between EFT-3mA copies (a and d) deleted intervening sequence leaving a single copy of EFT-3mA (e—aligned with a and d in B) in N. lolii AR1. (B) Alignment of N. lolii AR1 easA–easG EFT-3mA sequence (e in A) with the two instances from N. lolii Lp19 (a and d in A). Regions of 100% sequence identity between the AR1 EFT-3mA sequence and the left and right Lp19 instances are highlighted in yellow and red, respectively. Sequence highlighted in black is identical between all three instances. Lp19 sequence between the two EFT-3mA instances is not shown in the alignment and replaced by 5 × N. (C) The perA locus in E. festucae E2368 and Fl1 and N. lolii Lp14. The overlined region marked Δ is deleted in E2368 and Lp14 compared with Fl1. The expanded region shows a deleted EFT-3 sequence at the deletion point at the 3′ end of perA. EFT-7 is an uncharacterized retrotransposon relic.
This observation led us to examine the easA–easG locus in two other strains with E. festucae lineages. PCR products amplified from the easA–easG region in the different strains revealed a different length polymorphism in each. Sequencing of these showed one of the strains Neotyphodium sp. Lp1 was identical to Lp19 but with an expansion of the simple sequence repeat (SSR) found between the two MITE insertions. In N. lolii AR1, a nonproducer of ergot alkaloids, the locus is deleted compared with other strains, with a single EFT-3m insertion and no SSR (fig. 2). Sequence analysis (fig. 2B) revealed that the left 71 bp of this element were identical to the left end of the “left” element found in Lp19 (a in fig. 2A), whereas the right 56 bp of the AR1 element were identical to the “right” element in Lp19 (d in fig. 2A), with 22 bp of intervening sequence identical in both Lp19 elements. This indicated that recombination between the left and the right elements is likely to have caused a deletion of the 405 bp between the two insertions.
EFT-3m also appears to have been involved in a deletion event in some epichloid strains at the perA locus, a single gene required for synthesis of the anti-insect secondary metabolite peramine. Examination of the sequence of the nonfunctional perA gene in the E. festucae E2368 genome revealed a deletion of 1,223 bp of the 3′ end of the gene, with a 50-bp deleted instance of an EFT-3m element located at the deletion point adjacent to the remaining perA sequence. Previous analysis had shown that the perA gene was present in N. lolii Lp14, although this strain does not produce peramine (Scott et al. 2009). To determine whether the Lp14 perA gene also contained a deletion, we sequenced a PCR product amplified from the perA-EF104 region and remarkably found an identical deleted perA as in E2368, indicative of shared ancestry of these two strains.
Discussion
In this study, we identified 13 different families of MITEs, the most diverse collection characterized in fungi to date. We used a computational pipeline, which utilizes several different algorithms and some manual input for the analysis. Previous analysis of two MITE families in epichloae showed that these elements were highly degenerate (Fleetwood et al. 2007). This was also the case for the 10 other families characterized in this study, with average pairwise identity ranging from 49% (EFT-24) to 90% (EFT-25) with most around 60–85% identical.
For three of the MITEs identified, we were able to identify relics of progenitor autonomous transposons. In each case, the MITEs seem to have been derived by deletion of internal sequences. This is one way in which MITEs can arise (Jiang et al. 2003), although more commonly they appear to arise by the chance occurrence of sequences related to autonomous element TIRs being found in the appropriate orientation in the genome (Jiang et al. 2004). Although we did not identify any MITEs that had been formed in this way, the presence of EFT-27 subfamilies that are dissimilar to each other outside of the TIR sequences suggests that this has occurred in the epichloae. Each of the three autonomous elements identified have been rendered nonfunctional by RIP and contain multiple stop codons and very high AT percentages. Therefore, the MITEs derived from these elements are also highly unlikely to be functional as they require a functioning transposase on a parent element. For most MITEs, we could not identify parent elements. This may mean that the corresponding autonomous elements have degenerated to the extent that we were no longer able to recognize them or may suggest that these MITE families use a transposase from autonomous elements sufficiently dissimilar to the MITEs that they were not able to be recognized by similarity to the MITE TIRs. Whether autonomous elements exist in the genome for these MITEs or not, they are unlikely to be currently mobilizable based on the degeneracy of these families.
At least half of the MITE families appear to be ancient in the Epichloë genus. All are highly degenerate, and this is unlikely to be due to RIP as almost all the families are smaller than the 400 bp identified in N. crassa as the minimum length for RIP to function (Cambareri et al. 1989). Additionally, alignments show no obvious bias for C:T or G:A transitions characteristic of RIP (Cambareri et al. 1989). Thus, this degeneracy is likely to be due to basal levels of mutation over a very long period. The age of the elements could not be estimated due to the lack of a molecular clock; however, the very high degeneracy suggests an ancient origin, and taxonomic distribution data place the invasion of many of the MITEs at least as being early in the evolution of Epichloë. The EFT-3m, EFT-11m, EFT-14m, EFT-24m, and EFT-26m MITEs were found in sequences from Neotyphodium species that do not have E. festucae parentage (asexual Neotyphodium species are derivatives of sexual Epichloë species and often hybrids), supporting their early invasion. These were all in a single secondary metabolite gene cluster, however, and few epichloae sequences outside of these clusters are present in public gene databases. Thus, the absence of the remaining MITE families in these sequences does not preclude their equally ancient origin. Further evidence was found for EFT-14, which was found in all epichloid species examined (fig. 1). A number of diverse Epichloë and Neotyphodium species are currently being sequenced, and analysis of these genomes will confirm whether all the MITEs are as old as EFT-14. If, as seems likely, each of the MITEs were ancient invaders of epichloae, this is in contrast to the other repeat elements thus far identified in this genus. The Tahi and Rua retrotransposons were shown by Southern analysis to have a much more limited taxonomic distribution (Young et al. 2009), suggesting they invaded epichloae just prior to the radiation of E. festucae and E. baconii.
In other organisms, including the mimp element in F. oxysporum (Bergemann et al. 2008), MITEs are frequently found in genic regions of genomes, sometimes including within introns or 5′ or 3′ UTRs (Bureau and Wessler 1992, 1994a; Santiago et al. 2002; Yang et al. 2005; Ohmori et al. 2008). Our analysis of the genomic location of MITEs in E. festucae indicates that E. festucae is no exception. Of all MITEs, 47% were found within 500 bp of a gene, some of which may be within 5′ or 3′ UTRs although we did not have sufficient EST data to determine this. Further analysis indicated that for most families, more instances were found within 500 bp of the 5′ end of a gene than the 3′ end (table 3). This is well within the distance from the ORF start codon that is likely to be cis regulatory sequence, and it seems likely that at least a proportion of these integrations have affected expression of downstream genes in some way. Transposons integrated into promoter regions in other organisms have been shown to affect expression by disruption of existing, or provision of new, cis regulatory sequences or through alteration of the chromatin environment (Kidwell and Lisch 1997; Feschotte 2008), although this has rarely been tested for MITEs. How many genes have altered expression and what kind of effects the MITEs have had on expression in epichloae cannot be determined from analysis of a single genome but with multiple diverse Epichloë and Neotyphodium genomes currently being sequenced; along with complementary transcriptome analyses, these questions may soon be addressed.
A second aspect of MITE integration site bias of interest in this study was the finding that MITEs are nonrandomly enriched near genes predicted to be involved in secondary metabolite biosynthesis. Secondary metabolite biosynthetic genes are well studied in epichloae due to the protective effects of these natural products on the host grass, and MITEs were initially identified within some of these gene clusters. Further analysis here of gene clusters for the known alkaloids, lolines, indole diterpenes, and ergot alkaloids, showed that these gene clusters contain a striking number of MITE instances from several families. Furthermore, MITEs are significantly more likely than chance to be found near (within 10 kb) a PKS or NRPS gene, which are usually secondary metabolite biosynthetic genes (table 3). To have so many transposable elements within such a small sequence, as we observe in the LOL, EAS, and LTM, clusters is remarkable. This coclustering of MITEs with secondary metabolite gene clusters is not a phenomenon that has been described in other species to our knowledge. Indeed, the epichloid biosynthetic gene clusters show a level of complexity somewhat higher than that of most characterized gene clusters in other fungi, with “mini-clusters” of genes and MITEs separated by nested autonomous retrotransposons and DNA transposons (Young et al. 2006; Fleetwood 2007).
Whether this coclustering has occurred due to an evolutionary benefit to gene clusters containing such a large number of MITEs, by affecting either gene regulation or evolution, or whether this arrangement simply arises passively through a tendency for both gene clusters and transposons to be found in certain genomic regions, particularly telomeres and centromeres, is not easily tested. However, recent work showing a role for transposon sequences in regulation of a secondary metabolite gene cluster in Aspergillus nidulans (Shaaban et al. 2010) supports a role in regulation. A further hint to the possible role that MITEs may have played in the evolution of gene clusters in epichloae was the finding that MITEs have mediated recombination events at a local level in one and likely two secondary metabolite gene loci. The EFT-3m element has led to deletion events in the EAS cluster, whereas the presence of an EFT-3m deleted instance at the deletion point at the 3′ end of the perA gene suggests an involvement in that deletion also. The deletion of the 3′ end of the perA gene and regulatory sequence of the easA and/or easG genes has likely led or contributed to the lack of peramine and ergot alkaloid production in the respective strains, a major impact on the grass endophyte symbioses in which these plant-protective natural products play a major role. It seems unlikely that such a restricted analysis would have identified the only instances in which MITEs have mediated local genome rearrangements, and with the large number of deleted instances in the genome, it seems likely that MITE repeat sequences have played a large role in genome evolution of epichloae.
In this study, we described a large number of MITEs in the E. festucae genome and provide evidence for a likely role in genome regulation and evolution in epichloae. Why have MITEs then been so rarely characterized in other fungi? A major reason is likely to be the small size of many of the elements because the cutoff for repeat sequences in repeat searches of fungal genomes is often larger than the size of many MITEs. The epichloid MITEs were also highly degenerate, possibly below the similarity thresholds used by researchers studying other fungal genomes. It seems likely to us that fungal genomes contain more MITEs than currently described, and indeed a preliminary analysis of the N. crassa, F. oxysporum, and M. grisea genomes revealed a number of new MITE families in each genome alongside the previously characterized Guest and mimp elements (Khan A and Fleetwood D, unpublished data). This class of nonautonomous elements is clearly deserving of more research in fungi, and studies of the impact of the elements on genome evolution and regulation will be of very considerable future interest.
Supplementary Material
Supplementary data and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Mr Alan McCulloch for methodological advice; Mr Craig Miskell for maintaining computational servers and software; Dr Shaun Lott for laboratory resources; Ms Jennifer Webb for help sequencing the E. festucae genome; Dr Jolanta Jaromczyk and Mr Charles T. Bullock for genome assemblies; and Ms Elissaveta Arnaoudova, Mr Neil Moore, and Mr Daniel Harris for genome annotations. This work was supported by the New Zealand Foundation for Research Science and Technology grants (C10X080 and C10X0815), the US National Research Foundation grant (NSF EF-0523661), and the US Department of Agriculture grants (2005-35319-16141 and 2009-34457-20125).
References
- Benjak A, Boue S, Forneck A, Casacuberta JM. Recent amplification and impact of MITEs on the genome of grapevine (Vitis vinifera L.) Genome Biol Evol. 2009;1:75–84. doi: 10.1093/gbe/evp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann M, Lespinet O, M’Barek SB, Daboussi M-J, Dufresne M. Genome-wide analysis of the Fusarium oxysporum mimp family of MITEs and mobilization of both native and de novo created mimps. J Mol Evol. 2008;67:631–642. doi: 10.1007/s00239-008-9164-7. [DOI] [PubMed] [Google Scholar]
- Blankenship JD, et al. Production of loline alkaloids by the grass endophyte, Neotyphodium uncinatum, in defined media. Phytochemistry. 2001;58:395–401. doi: 10.1016/s0031-9422(01)00272-2. [DOI] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994a;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci U S A. 1994b;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AD, Schardl CL, Songlin PJ, Mogen KL, Siegel MR. The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne) Curr Genet. 1990;18:347–354. doi: 10.1007/BF00318216. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, Selker EU. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Christensen MJ, Leuchtmann A, Rowan DD, Tapper BA. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis) and perennial ryegrass (Lolium perenne) Mycol Res. 1993;97:1083–1092. [Google Scholar]
- Chung K-R, Hollin W, Siegel MR, Schardl CL. Genetics of host specificity in Epichloe typhina. Phytopathology. 1997;87:599–605. doi: 10.1094/PHYTO.1997.87.6.599. [DOI] [PubMed] [Google Scholar]
- DeMarco R, Venancio TM, Verjovski-Almeida S. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol Biol. 2006;6:89. doi: 10.1186/1471-2148-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne M, et al. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics. 2007;175:441–452. doi: 10.1534/genetics.106.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler SR. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington (DC): American Society for Microbiology Press; 2002. pp. 1147–1158. [Google Scholar]
- Fierro F, Martin JF. Molecular mechanisms of chromosomal rearrangement in fungi. Crit Rev Microbiol. 1999;25:1–17. doi: 10.1080/10408419991299185. [DOI] [PubMed] [Google Scholar]
- Fleetwood DJ. Molecular characterisation of the EAS gene cluster for ergot alkaloid biosynthesis in epichloë endophytes of grasses [dissertation] [New Zealand]: Massey University; 2007. [Google Scholar]
- Fleetwood DJ, Scott B, Lane GA, Tanaka A, Johnson RD. A complex ergovaline gene cluster in epichloë endophytes of grasses. Appl Environ Microbiol. 2007;73:2571–2579. doi: 10.1128/AEM.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Daviere J-M, Kaper F, Langin T, Daboussi M-J. Genome organization in Fusarium oxysporum: clusters of class II transposons. Curr Genet. 2000;37:339. doi: 10.1007/s002940050537. [DOI] [PubMed] [Google Scholar]
- Jiang N, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Jiang N, Feschotte C, Zhang X, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr Opin Plant Biol. 2004;7:115–119. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Juretic N, Bureau TE, Bruskiewich RM. Transposable element annotation of the rice genome. Bioinformatics. 2004;20:155–160. doi: 10.1093/bioinformatics/bth019. [DOI] [PubMed] [Google Scholar]
- Khaldi N, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 1997;94:7704. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, et al. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek SM, Hansen K, Romanish M, Thorn RG. Molecular systematics of the cotton root rot pathogen, Phymatotrichopsis omnivora. Persoonia Mol Phylogeny Evol Fungi. 2009;22:63–74. doi: 10.3767/003158509X430930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek SM, Muller RA, Walker NR. First report of ergot of bermudagrass caused by Claviceps cynodontis in Oklahoma. Plant Dis. 2006;90:376. doi: 10.1094/PD-90-0376C. [DOI] [PubMed] [Google Scholar]
- Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Moon CD, Craven KD, Leuchtmann A, Clement SL, Schardl CL. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol Ecol. 2004;13:1455–1467. doi: 10.1111/j.1365-294X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Moon CD, Tapper BA, Scott B. Identification of epichloë endophytes in planta by a microsatellite-based PCR fingerprinting assay with automated analysis. Appl Environ Microbiol. 1999;65:1268–1279. doi: 10.1128/aem.65.3.1268-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Gertz EM, Schaffer AA, Agarwala R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ohmori Y, Abiko M, Horibata A, Hirano H-Y. A transposon, Ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant Cell Physiol. 2008;49:1176–1184. doi: 10.1093/pcp/pcn093. [DOI] [PubMed] [Google Scholar]
- Ramussen JP, et al. Guest, a transposable element belonging to the Tc1/mariner superfamily is an ancient invader of Neurospora genomes. Fungal Genet Biol. 2004;41:52–61. doi: 10.1016/j.fgb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Santiago N, Herraiz C, Goni JR, Messeguer X, Casacuberta JM. Genome-wide analysis of the emigrant family of MITEs of Arabidopsis thaliana. Mol Biol Evol. 2002;19:2285–2293. doi: 10.1093/oxfordjournals.molbev.a004052. [DOI] [PubMed] [Google Scholar]
- Scott B, et al. Regulation and functional analysis of bioprotective metabolite genes from the grass symbiont Epichloë festucae. In: Gullino ML, editor. Plant Pathology in the 21st Century. Dordrecht (The Netherlands): Springer; 2009. pp. 199–213. [Google Scholar]
- Shaaban M, et al. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet Biol. 2010;47:423–432. doi: 10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- Sung GH, et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-F, Siegel MR, Schardl CL. Transformation of Acremonium coenophialum, a protective fungal symbiont of the grass Festuca arundinacea. Curr Genet. 1992;22:399–406. doi: 10.1007/BF00352441. [DOI] [PubMed] [Google Scholar]
- Tu Z. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2001;98:1699–1704. doi: 10.1073/pnas.041593198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S. Graph clustering by flow simulation [dissertation] [The Netherlands]: University of Utrecht; 2000. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yang G, et al. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–1568. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- Yeadon PJ, Catcheside DEA. Guest: a 98 bp inverted repeat transposable element in Neurospora crassa. Mol Gen Genet. 1995;247:105–109. doi: 10.1007/BF00425826. [DOI] [PubMed] [Google Scholar]
- Young CA, et al. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol Genet Genomics. 2005;274:13–29. doi: 10.1007/s00438-005-1130-0. [DOI] [PubMed] [Google Scholar]
- Young CA, et al. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet Biol. 2006;43:679–693. doi: 10.1016/j.fgb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Young CA, et al. Indole-diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. Appl Environ Microbiol. 2009;75:2200–2211. doi: 10.1128/AEM.00953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]