Abstract
The expression of tumor suppressor Arf is tightly repressed during normal cell growth at a young age, and is activated by oncogenic insults and during aging, resulting in p53 activation and cell cycle arrest to prevent hyperproliferation. The mechanisms of both transcriptional repression and activation of Arf are not understood. We show that p53 binds to and represses Arf expression and that this repression requires the function of both histone deacetylase (HDAC) and polycomb group (PcG) proteins. Inactivation of p53 leads to increased Arf transcription in both mouse embryonic fibroblasts (MEFs) cultured in vitro and in tissues and organs of p53 null mice. Activation of endogenous p53 enhances Arf repression, and reintroduction of p53 back into p53 null MEFs restores Arf repression. Both DNA binding and trans-activation activities of p53 are required for Arf repression. We show that p53 is required for both HDAC and PcG to repress Arf expression. Bindings of both HDAC and PcG to Arf are disrupted by inactivation of p53 and can be restored in p53 null MEFs by the reintroduction of wild-type, but not mutant, p53. These results indicate that p53 recruits both HDAC and PcG to Arf locus to repress its expression, and this repression constitutes a second feedback loop in p53 regulation.
Keywords: Arf, p53, Ring1B, transcriptional regulation, feedback inhibition
Introduction
Transcription factor p53 mediates cellular response to a wide range of genotoxic and growth stresses, including DNA damage and oncogenic insults (1). Activated p53 increases the expression of numerous genes and elicits three distinct types of cellular outcomes—temporary cell cycle arrest, permanent cell senescence and apoptotic cell death—to prevent damaged or stressed cells from continuing proliferation (2). It is generally believed that escape from the p53-mediated checkpoint pathway is a necessary step for the development of most, if not all, types of tumors (3, 4).
Much has been learned on the function of p53 in both senescence and apoptosis (5). Equally important is the regulation of p53 in causing temporary cell cycle arrest. Unlike senescence or apoptosis, both of which are irreversible, a mechanism to inhibit activated p53 is critically important in releasing temporarily arrested cells to resume the cell cycle once the damage is repaired or stress is relieved. One such mechanism to reversibly regulate p53 is the feedback inhibition loop in which p53 activates the expression of its principle inhibitor (6-8), MDM2, which binds to and inhibits the function of p53 by both repressing the transactivating activity of p53 (9, 10), as well as targeting p53 for ubiquitin-mediated degradation (11-13).
The ARF-p16 locus, which is altered in an estimated 30 – 40% of human tumors, encodes two distinct tumor suppressor gene products, p16INK4a and p14ARF (p19 in mouse), via the use of separate promoters and alternative reading frames (14-16). While p16INK4a binds to and inhibits cyclin Ds-dependent CDK4 and CDK6 to retain the growth suppressive activity of Rb family proteins (17), ARF binds to and antagonizes the activity of MDM2, thereby stabilizing and activating p53 (18-20). The ARF gene is expressed at a low level in normally growing young cells. Previous studies have linked two histone modifying complexes, histone deacetylases [HDAC, (21-23)] and polycomb proteins [PcG, (24-30)], to the repression of Arf expression. How these two histone modifying complexes, which do not recognize a specific DNA sequence, are recruited to Arf locus is not known.
ARF is activated by various oncogenic insults or during cell aging, leading to the notion that ARF mediates an oncogene-checkpoint pathway to prevent oncogenic-stimulated cells from hyperproliferating (15, 31). The potent activity of ARF in binding to and inhibiting the function of MDM2 raises the question of how p53 is reversibly regulated in oncogenically insulted cells where ARF, if continuously expressed, would prevent MDM2 from inhibiting p53 and thus disrupt the MDM2-p53 feedback regulatory loop. It was noted early on that in both human (19, 32) and mouse cells (33, 34), ARF expression exhibits a strong inverse correlation with the functional status of p53, suggesting a possible feedback repression of ARF expression by p53. The molecular mechanism underlying this feedback regulation is unknown and is the focus of this study.
Materials and Methods
Cell culture, western analysis and antibodies
Mouse embryonic fibroblasts (MEFs) and 293T cells were cultured in DMEM with 10% FBS. Cells were lysed with RIPA buffer. Antibodies to Bmi1 (F6, Upstate), Ring1B (ab-3832, Abcam), Suz12 (ab-12073, Abcam), Ezh2 (ab-3748, Abcam), HDAC1 (ab-7028, Abcam), Acetyl-Histone H4 (06-598, Upstate), p53 (FL-393X, Santa Cruz), p19Arf (ab-80, Abcam), mouse p16 (M-156; Santa Cruz), 3m-H3K27 (ab-6002, Abcam), E2F3 (sc-878X, Santa Cruz), Tubulin (Ab-2 DM1A, Neomarkers), Normal Mouse IgG (Neomarkers), Normal Rabbit IgG (Neomarkers) and Actin (C-11; Santa Cruz) were purchased commercially.
Retroviral procedures
The retroviral vector expressing mouse Bmi1 and microRNA targeting 3′-UTR of mouse p53 was provided by Dr. Ned Sharpless and Yizhou He, respectively. Human p53 cDNA was cloned into pBabe-puro retrovirus vector, and point mutations were made by site-directed mutagenesis and verified by DNA sequencing. Retroviruses encoding shRNAs silencing mBmi1, mE2f3, mHdac1, and mHdac2 were constructed by ligating respective oligonucleotides (see details in Supplementary Materials and Methods) into a PMKO-puro vector. Detailed experimental procedures for retroviral production and infection are described in (30).
Quantitative-RT-PCR
Detailed protocol has been described in (30). Sequences of PCR primers are described in Supplementary Materials and Methods. Mean values and standard deviations were calculated from triplicates of 3 independent repeats.
ChIP assay
ChIP analysis was performed as described in (30). PCR was performed using Platinum Taq polymerase (Invitrogen) and primers on mouse Arf locus (see more details in Supplementary Materials and Method). For ChIP-Q-PCR, purified DNA was added to a Q-RT–PCR mixture that contained 1× SYBR Green PCR master mix and 150 nM gene-specific primers. Assays were performed in triplicate on a 7900 HT sequence detection system. Mean values and standard deviations were calculated from triplicates of 3 different repeats.
Results
p53 represses Arf expression in vivo
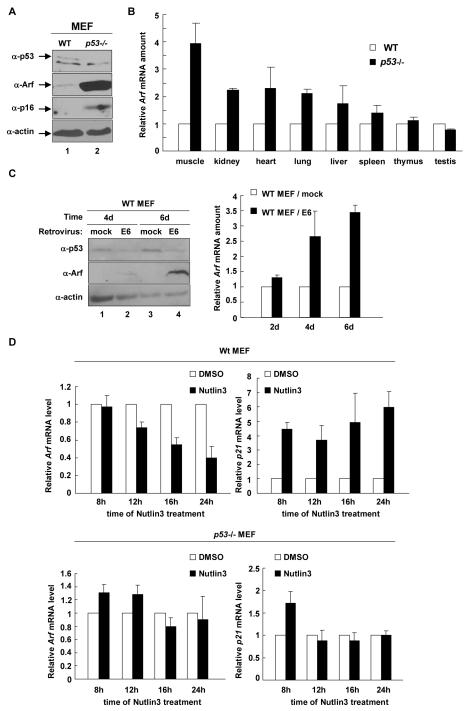
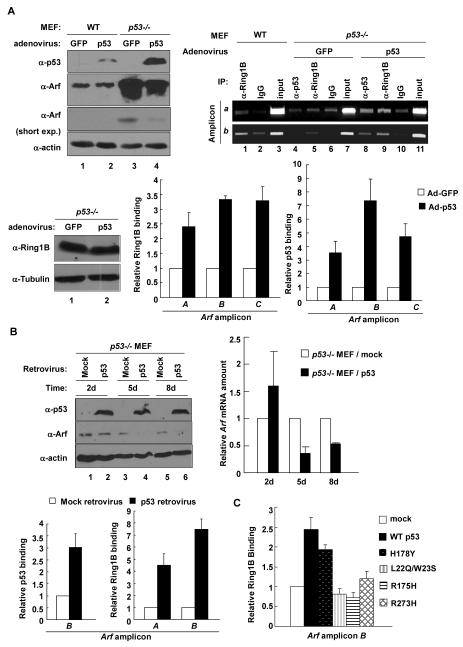
Confirming previous observations (33, 35), we found that the steady state level of Arf protein is significantly increased in p53-deficient MEFs (Fig. 1A). We also observed a clear increase of p16 protein level in p53-/- MEFs, which could be caused by the decrease of p21 expression and then a reduction in function of the Rb pathway, which collaborates with Polycomb Repressive Complex (PRC) to repress p16 gene transcription in a feedback loop (30). A demonstration of Arf repression by p53 in vivo, however, has been lacking. Therefore, we dissected three pairs of age-matched (one pair at 5-weeks of age and two at 8-weeks of age) wild-type (WT) and p53 null mice and determined the Arf expression in 8 different organs/tissues. This study demonstrated that Arf expression was significantly increased by p53 loss in 4 organs (muscle, kidney, heart and lung), moderately in two (liver and spleen) and unchanged in two (thymus and testis) (Fig. 1B). This result provides the first evidence demonstrating p53-dependent repression of Arf in vivo.
Figure 1. p53 represses Arf expression.
(A) The steady state levels of Arf and p16 proteins were determined in WT and p53-/- MEFs at passage 5 (p5) by immunoblotting. (B) 3 pairs of age-matched WT and p53-/- mice were dissected and total RNA was extracted from 8 different organ/tissues. The level of Arf mRNA was determined by Q-RT-PCR. (C) WT MEFs (p2) were infected with mock or E6-expessing retrovirus and selected by G418 treatment. The level of p53 and Arf proteins or mRNA was determined by immunoblotting or Q-RT-PCR. (D) WT (p2) and p53-/- MEFs were treated with 1% DMSO or 10 μM Nutlin3 and the levels of Arf and p21 mRNA were determined by Q-RT-PCR.
To exclude the possibility that Arf accumulation in p53-null MEFs was caused indirectly by other potential mutations and/or adaptive changes accumulated during multiple rounds of cell division, we transduced WT MEFs with a retrovirus expressing the type 16 papilloma virus-encoded E6 oncoprotein that binds to and targets the degradation of p53, or a microRNA retrovirus targeting 3′-UTR of mouse p53. Ectopic expression of E6 or mip53resulted in a detectable increase of Arf protein and mRNA as early as 2 days after viral transducton and a continual increase of Arf (Fig. 1C and Supplementary Figure A and B). This result supports a direct role of p53 in the repression of Arf transcription and also suggests a continuous need of p53 to maintain the repression.
Nutlins are a group of small compounds that can bind MDM2 in the p53-binding pocket and disrupt the p53-MDM2 interaction, leading to p53 activation (36). We treated WT MEFs at passage 2 with Nutlin-3 (10 μM) or DMSO over 24 hours and then examined Arf expression. Confirming the activation of p53, Nutlin-3 increased p21 mRNA in WT MEFs, but had no effect on p21 level in p53-/- MEFs. Nutlin-3 treatment enhanced Arf repression in WT MEFs within 24hs, resulting in a time-dependent decrease of Arf expression by 60%, but no effect on Arf level in p53-/- MEFs despite the much higher Arf transcription (Fig. 1D). We thus conclude that p53 represses Arf at a level of transcriptional regulation through a mechanism that requires a direct and continuous role of p53
p53 binds to Arf locus
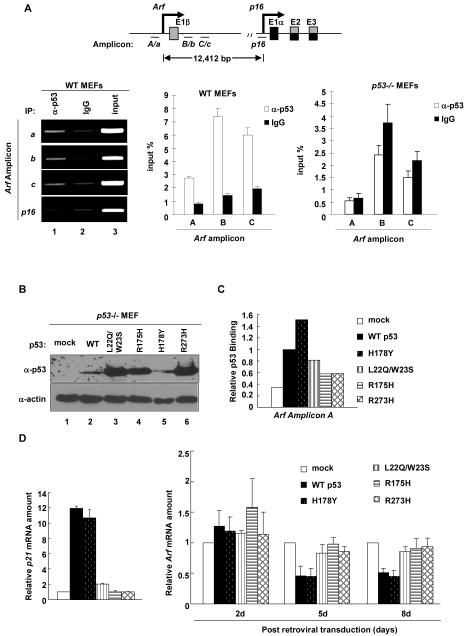
That p53 is directly involved in Arf repression led us to determine whether p53 binds to the Arf locus. We performed ChIP analysis using a panel of 35 pairs of oligonucleotide primers that span 4kb upstream and 4kb downstream of the transcription start site of mouse Arf. We detected direct p53 binding to a region immediately upstream and downstream of exon1β (amplicons a, b, c for regular PCR; corresponding amplicons A, B, C for Q-PCR) of Arf (Fig. 2A). In contrast, there was very little binding of p53 to the p16 promoter, nor any p53 binding detected by using p53-/- MEF as negative control in ChIP analysis. These results demonstrate that p53 affects the expression of Arf and p16 differently and indicate a direct role of p53 in the repression of Arf expression.
Figure 2. Both transactivation and DNA binding activity of p53 are required for p53 to bind and repress Arf.
(A) WT p2 and p53-/- p3 MEFs were analyzed for p53 binding on Arf and p16 locus. The amount of DNA immunoprecipitated by p53 or rabbit IgG were expressed relative to the percentage of input DNA. (B) p53-/- MEFs were infected with mock or WT p53 and mutant p53 retrovirus. Cells were selected by puromycin treatment, collected at indicated time after infection and the expression levels of p53 were determined by immunoblotting. (C) Cells in (B) collected 8 days after infection were analyzed for binding of p53 on Arf locus by ChIP-Q-PCR. (D) p21 mRNA levels of cells in (B) were determined 2 days after infection by Q-RT-PCR, and cells collected at different times after infection were analyzed for Arf mRNA levels by Q-RT-PCR.
p53 mediated repression of Arf needs both transactivation and DNA binding activity
To provide further evidence supporting a direct role of p53 in Arf repression, we examined four p53 mutants in Arf repression, including two well-characterized p53 hot-spot mutants—R175H, which grossly disrupts p53’s protein conformation and R273H, which retains p53’s native conformation but loses contact with DNA [see a recent review on the genetic and biochemical properties of different p53 mutants (37)]. In addition, we also examined a double mutant—L22Q/W23S—which disrupts p53’s transcriptional activity, as well as the binding with Mdm2 (38) and a hyperactive p53 mutant—H178Y—that could rescue the inactivated function of a common mutation, G245S (39). p53-/- MEFs were transduced with a retrovirus expressing the WT and individual mutants of p53. As expected, the three functional inactivation mutants—p53L22Q/W23S, p53R175H and p53R273H—were expressed at high levels, whereas cells could only tolerate a much lower level expression of both WT p53 and hyperactive p53H178Y mutant (Fig. 2B). The activity of the WT and individual p53 mutants was functionally verified by examining the expression of p21 (Fig. 2D).
ChIP and qRT-PCR assay showed p53H178Y bound to Arf stronger than the WT p53 and p53L22Q/W23S exhibited decreased Arf binding. Both p53R175H and p53R273H, however, bound very weakly to Arf (Fig. 2C). We then determined if these p53 mutants could restore Arf repression in p53 null MEFs. Starting 5 days after viral transduction, both WT and the hyperactive p53H178Y mutant reduced the Arf mRNA level by 50% (Fig. 2D). In contrast, ectopic expression of p53L22Q/W23S, p53R175H and p53R273H at very high levels had little effect in restoring the repression of Arf. Together, these results demonstrate that p53 represses Arf by directly binding to the Arf locus and that both transactivity and DNA binding of p53 are essential to repress Arf expression.
HDAC binds to and represses Arf and is recruited to mArf in a p53-dependent manner
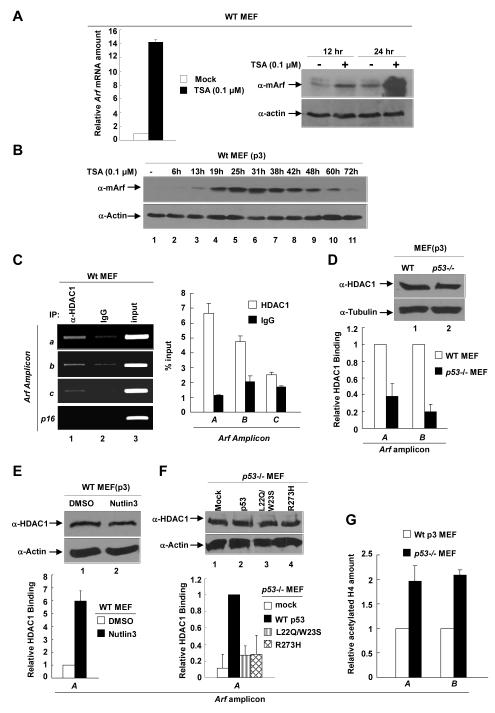
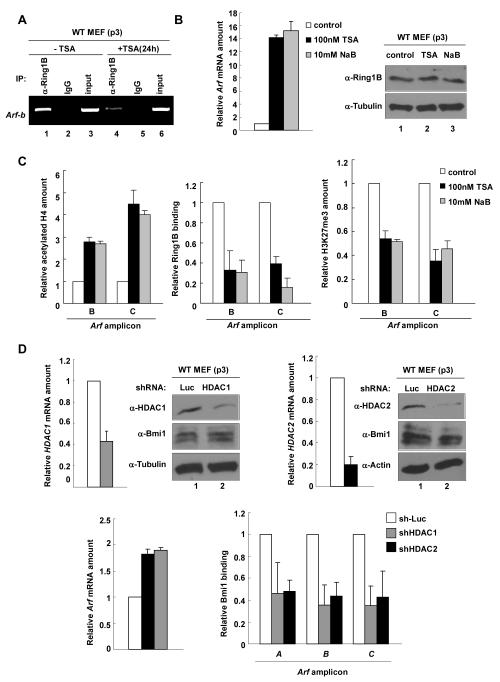
Histone deacetylases (HDACs) associate with many transcriptional repressive complexes and have also been reported to participate in the transcriptional repression of both human and mouse ARF genes (22, 23). To determine whether HDACs are involved in p53-mediated repression of Arf transcription, we treated WT MEFs with TSA, an inhibitor of class I/II HDACs. A low concentration of TSA (0.1 μM) increased mouse Arf mRNA level by >15-fold within 24hs (Fig. 3A). TSA treatment resulted in a detectable Arf protein increase as early as 6 hours, peaking around 25hs and lasting to as long as 60hs (Fig. 3B). These resultsdemonstrate a potent role of HDACs in the repression of mouse Arf transcription.
Figure 3. HDAC1 binds to and represses Arf in a p53-dependent manner.
(A) WT p3 MEFs were treated with 0.1μM TSA for 12hs or 24hs. The levels of Arf protein or mRNA were determined by immunoblotting or Q-RT-PCR. (B) WT p3 MEFs were treated with 0.1μM TSA for 0~72hs. Cell pellets were collected at indicated time and steady state level of protein was determined by immunoblotting. (C) Binding of HDAC1 to Arf locus was examined in WT p2 MEFs by ChIP analysis. The amount of DNA immunoprecipitated by HDAC1 or rabbit IgG were expressed relative to the percentage of input DNA. (D, G) Binding of HDAC1 on Arf, and amount of acetylated histone H4 on Arf were quantified in WT and p53-/- MEFs by ChIP-Q-PCR. (E) WT p3 MEFs were treated with 1% DMSO or 10μM Nutlin-3 and cells were collected after 16hs. The binding of HDAC1 on Arf was determined by ChIP-Q-PCR with indicated primers. (F) p53-/- MEFs were infected with mock or WT and mutant p53 retrovirus. Cells were selected by puromycin treatment, collected at 5 days post-infection, and analyzed for HDAC1 binding to Arf by ChIP-Q-PCR.
We next tested the binding of HDAC1 on Arf in WT MEFs. ChIP showed that HDAC1 directly binds to a region immediately upstream of the transcription starting site of Arf, whereas no binding of HDAC1 was detected on the nearby p16 locus (Fig. 3C). Importantly, the binding of HDAC1 to mouse Arf was greatly decreased, by more than 60% (for amplicon A) to 80% (amplicon B), in p53-/- MEFs (Fig. 3D) or in WT MEFs with p53 knocked down by microRNA (Supplementary Figure C), providing the first evidence linking HDAC-mediated repression of Arf transcription to the function of p53. Supporting a functional dependency of deacetylation of Arf promoter on p53 function, activation of p53 by Nutlin-3 in WT MEFs resulted in a 6-fold increase of HDAC1 binding to Arf (Fig. 3E) and reintroduction of WT p53, but not p53L22Q/W23S and p53R273H, back into p53-/- MEFs restored the binding of HDAC1 to the Arf locus (Fig. 3F). Finally, we showed that H4 acetylation of the Arf promoter in p53-/- MEFs was 2-fold higher than that in WT MEFs (Fig. 3G). Together, these results demonstrate that HDAC1 directly binds to and deacetylates Arf in a p53-dependent manner.
p53 is required for polycomb repressive complex to bind and repress Arf
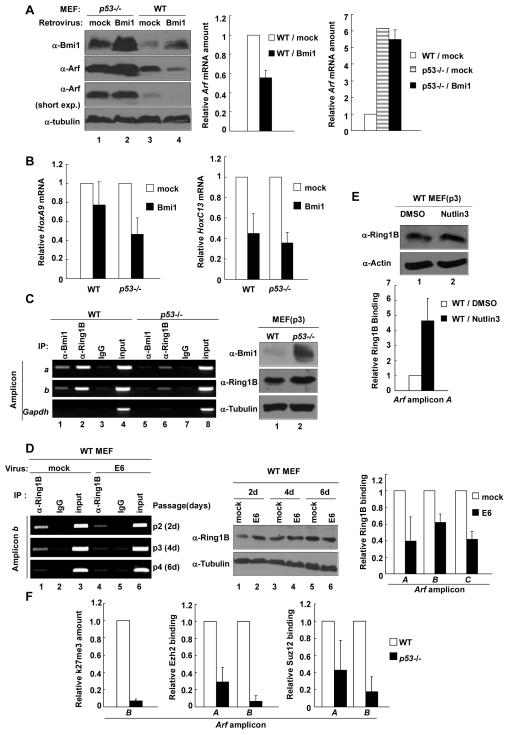
Previous studies have shown that oncogene Bmi1 promotes cell proliferation and extends the life span of fibroblasts, in part, through repressing both p16Ink4a and p19Arf expression (24, 25). More recently, polycomb group proteins have been shown to directly bind and repress both p16 and ARF in mouse cells (29, 30). To determine the functional interplay between p53 and PcG in repressing Arf expression, we transduced both WT and p53-/- MEFs with a retrovirus expressing the Bmi1 gene and determined Arf expression. Nine days after retrovirus transduction (passage 5), both the steady state level of the Arf protein and mRNA were decreased substantially in Bmi1-overexpressing WT MEFs (Fig. 4A). In contrast, although Arf is expressed at a much higher level in p53-/- MEFs, overexpression of Bmi1 did not significantly affect either Arf protein or mRNA level. To determine the specificity of the functional dependency of Bmi1-mediated repression of Arf on p53, we examined the expression of two classical Bmi1 repressive targets—HoxA9 and HoxC13 (40). Ectopic expression of Bmi1 was capable of repressing both HoxA9 and HoxC13 genes regardless of the p53 status (Fig. 4B). Together, these results demonstrate a specific functional dependency of Bmi1-mediated repression of Arf transcription on p53.
Figure 4. p53 is required for PRC to bind and repress Arf.
(A, B) p2 WT and p53-/- MEFs were infected with mock or Bmi1-expressing retrovirus. 9 days after infection, MEFs were collected and the protein levels of Bmi1 and Arf were determined by immunoblotting; the mRNA levels of Arf (A), HoxA9, and HoxC13(B) were determined by Q-RT-PCR. (C) Binding of Bmi1 and Ring1B to Arf locus was tested in WT p2 and p53-/- MEFs by ChIP. (D) WT p2 MEFs were infected with mock or E6 retrovirus and G418 selected. Cells were collected at indicated time after infection and analyzed for Ring1B binding on Arf. (E) WT p3 MEFs were treated with 1% DMSO or 10μM Nutlin3, and cells were collected after 16hs. The binding of Ring1B on Arf was determined by ChIP-Q-PCR. (F) ChIP-Q-PCR was used to determine the trimethylation of H3K27 and binding of Ezh2 and Suz12 on Arf locus in WT p3 and p53-/- MEFs.
Bracken et al previously found that the Polycomb Repressive Complex (PRC) can bind directly to the mouse Arf locus to repress its expression (29). To search for the mechanism of the p53-dependency of Arf repression by Bmi1, we examined by ChIP and confirmed the binding of Bmi1 and Ring1B, another component of PRC, to the Arf locus in WT and p53-/- MEFs. Notably, the bindings of both Bmi1 and Ring1B to the Arf locus were greatly decreased in p53-/- MEFs (Fig. 4C). Confirming the p53-dependency, binding of Ring1B to the Arf locus in WT MEFs was also greatly reduced after transduction with an E6-expressing or p53-targeting microRNA retrovirus (Fig. 4D and Supplementary Figure C) and conversely increased in WT MEFs when p53 was activated by Nutlin-3 (Fig. 4E). Together, these results demonstrate a requirement of p53 for the binding of PRC1 to Arf locus.
Previous studies on Hox gene silencing by PRC suggest a sequential model whereby PRC2-mediated H3K27 trimethylation facilitates the recruitment of PRC1, which causes H2A-K119 ubiquitylation to repress Hox gene expression (41, 42). We further analyzed the binding of PRC2 and H3K27 trimethylation on Arf, and quantification showed a dramatic decrease of this repressive marker in p53-/- MEFs to less than 7% (Fig. 4F). Likewise, the binding of two components of PRC2, Ezh2 and Suz12, to the Arf locus were also substantially reduced in p53-/- MEFs to 29% and 6.7% for Ezh2 and to 43% and 18% for Suz12 of WT MEFs at sites A and C, respectively (Fig. 4F). Hence, the p53 function is required for both PRC1 and PRC2 to bind to the Arf locus.
Both transactivity and DNA binding activities of p53 are required for PRC1 binding to Arf
To further demonstrate the requirement of p53 in facilitating the binding of PRC1 to the Arf locus, we re-introduced p53 back into p53-/- MEFs which partially restored the repression of both Arf protein and mRNA (Fig. 5A and 5B). The binding of Ring1B to Arf was also restored when p53 was ectopically expressed, resulting in a 2- to 3-fold and 4- to 7-fold increase of Ring1B binding to Arf in p53-/- MEFs after adenovirus- and retrovirus-mediated p53 expression, respectively (Fig. 5A and 5B). We then determined if mutant p53 restored PcG binding to Arf. Both WT p53 and the hyperactive p53H178Y mutant, but not three inactivation mutants, restored Ring1B binding to Arf, leading to a 2.4-fold and 1.9-fold increase, respectively (Fig. 5C). Together these results demonstrate that both the trans-activating and DNA binding activities of p53 are required for the binding of PcG to Arf.
Figure 5. Both transactivity and DNA binding activity of p53 are required for PRC1 binding to Arf.
(A) WT (p3) and p53-/- MEFs were infected with either GFP- or p53-expressing adenovirus. Two days after infection, cell lysates were analysed by WB for expression of p53 and Arf, and crosslinked cell pellets were collected and analyzed for binding of p53 and Ring1B to Arf locus. (B) p53-/- MEFs were infected with either mock or p53-expressing retrovirus and selected by puromycin. The levels of p53 and Arf proteins or mRNA were determined by immunoblotting or Q-RT-PCR. Cells collected at 5 day after infection were analyzed for p53 and Ring1B binding on Arf by ChIP-Q-PCR. (C) p53-/- MEFs were infected with either mock WT or mutant p53 retrovirus. Cells were selected by puromycin and analyzed for bindings of Ring1B to Arf by ChIP-Q-PCR using Arf amplicon B.
HDAC function is required for PcG to bind mouse Arf
Given that p53 is required for the binding of both HDAC and PcG complexes to the Arf locus and that HDAC has been reported to be in the same repressive complex with and facilitates the repressive function of PcG (43, 44), we next determined the requirement of HDAC function for PcG binding on Arf by two different approaches, pharmacological inhibition of HDAC activity and shRNA-mediated depletion of HDACs. Treatment of WT MEFs with TSA and another pharmacological inhibitor of HDAC, sodium butyrate (NaB), both significantly increase Arf expression, but neither affected Ring1B level (Fig. 6B). Inhibition of HDAC by either TSA or NaB increased H4 acetylation, yet decreased Ring1B binding and H3K27 tri-methylation on Arf promoter (Fig. 6A and C).
Figure 6. HDAC function is required for PRC to bind Arf.
(A) WT p3 MEFs were treated with 0.1 μM TSA for 24hs. Cell pellets were collected and analyzed for binding of Ring1B on Arf by ChIP. (B) WT p3 MEFs were treated with 0.1μM TSA or 10mM NaB for 30hs, and cells were collected and analyzed for Arf mRNA amount by Q-RT-PCR. (C) Cells in (B) were collected and analyzed for histone H4 acetylation, histone H3K27 trimethylation and Ring1B binding on Arf locus by ChIP-Q-PCR. (D) WT MEFs were infected with either an sh-Luciferase or shHDAC-expressing retovirus, selected by puromycin for 2 days and collected 4 days after infection. The mRNA levels of HDAC1, HDAC2 and Arf were determined by Q-RT-PCR, and the binding of Bmi1 to Arf locus were determined by ChIP-Q-PCR.
We then used shRNA to knock down HDAC1 and HDAC2 in WT MEFs. Although only a partial depletion was achieved, we observed a decrease of Bmi1 binding to Arf and am increase of Arf expression (Fig. 6D). These results demonstrate that the activity of HDAC is required for PcG to bind to and repress the expression of Arf, and both HDAC1 and HDAC2 contribute to this inhibition.
HDAC and PcG function are required for p53 to repress Arf expression
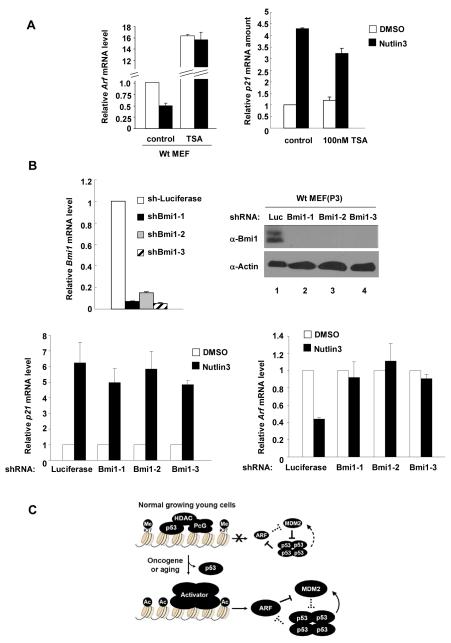
The functional dependency of both HDAC and PcG on p53 in the repression of Arf expression led us to determine whether these two histone modifying enzymes are required for p53 to repress Arf expression. To this end, we inhibited endogenous HDAC with TSA in WT MEFs and then determined whether activation of endogenous p53 by Nutlin can still repress Arf. While activation of p53 by Nutlin-3 resulted in a > 50% decrease of Arf expression in control MEFs, it had no detectable effect on Arf expression when the HDACs were inhibited by TSA, even though the Arf mRNA level was elevated by as much as more than 16-fold in the TSA-treated cells (Fig. 7A). As a control for p53 activation, inhibition of HDACs by TSA did not affect the increase of p21 expression by Nutlin-activated p53.
Figure 7. HDAC and PcG function are required for p53 to repress Arf.
(A) WT p2 MEFs were treated with TSA and Nutlin-3 either alone or in combination for 24hs. Cells were collected and analyzed for mRNA levels of p21 and Arf by Q-RT-PCR. (B) WT MEFs (p2) were infected with either an sh-Luciferase or shBmi1-expressing retrovirus, selected by 2 μM puromycin for 2 days, continuosly cultured for 5 days after infection and then treated with Nutlin-3 for 24hs. Cells were collected and analyzed for Bmi1, p21 and Arf mRNA level by Q-RT-PCR. (C) A schematic model illustrating p53-mediated Arf repression by HDAC and PcG. See text for more details.
To determine whether the function of PcG is required for p53 to repress Arf expression, we knocked down the expression of Bmi1 in WT MEFs and examined the effect of Bmi1 silencing on Arf repression by Nutlin-activated p53. All three shBmi1 viruses efficiently reduced the expression of Bmi1 (Fig. 7B), and none of them affected the activation of p21 expression by Nutlin-activated p53. While activation of p53 by Nutlin-3 effectively reduced Arf expression by more than 50% in WT MEFs infected with shRNA control virus targeting Luciferase, it had no significant effect to reduce Arf expression in MEFs transduced with any of the three shRNA virus targeting Bmi1. Hence, the function of p53 in the repression of Arf expression is also dependent on PcG.
Discussion
The results demonstrate a direct role of p53 in the repression of mouse Arf transcription by showing the direct binding of p53 to the mouse Arf locus and that p53 is required for recruiting HDAC and PcG proteins to the Arf locus. Our results are consistent with a model where p53 binds specifically to the Arf locus and then recruits histone deacetylases to deacetylate the Arf locus. Deacetylation of the Arf locus then facilitates the recruitment of PRC to the Arf locus, leading to H3K27 trimethylation and silencing of Arf expression (Fig. 7C). This model is supported by three lines of evidence. First, Arf expression is elevated in vivo in multiple p53-deficient tissues or organs, is rapidly elevated upon functional inactivation of p53 and is further repressed upon the activation of endogenous p53 in early passage of WT MEFs. Second, p53 directly binds to Arf locus, and that both transactivation and DNA binding activities of p53 are required for the repression of Arf and, importantly, for the binding to and repression of Arf by both HDAC and PRC. Third, we have also shown that both HDAC and PcG are conversely required for p53 to repress Arf expression.
The findings presented here shed mechanistic insights on the p53-mediated oncogenic checkpoint pathway: one on the mechanism of Arf activation and the other on the feedback regulation of p53. Although it has been observed for more than a decade that many hyperproliferative oncogenes can activate Arf expression (31), the molecular mechanism underlying oncogenic activation of Arf is unknown. Our demonstration that Arf is bound and repressed by p53 during normal cell growth suggests a critical step—dissociating p53 from the Arf locus—for an oncogene to activate Arf expression. It will be interesting to determine how an oncogene causes p53 dissociation in the presence of increased level of p53 since Arf activation would lead to p53 stabilization.
Feedback inhibition is a regulatory strategy commonly used in biochemical reactions such as the inhibition of threonine dehydrase by isoleucine (46). The accumulation of an end-product inhibits the enzyme involved in its synthesis to avoid excessive accumulation and waste of resources. A similar strategy is also widely employed in cell regulations, especially those involved in cell growth and proliferation, to ensure a balanced homeostasis and cell physiology. Feedback inhibition is particularly needed for the control of the function of a gene such as p53 whose activity, if not feedback inhibited, could lead to an irreversible consequence to the cell such as permanent cell cycle arrest or cell death. At low, non-lethal levels of DNA damage, cell cycle progression is delayed by the activation of p53 and then p21 to give cells time to repair the DNA and then resumed when the repair is completed. The resumption of cell cycle progression is achieved through the p53-MDM2 feedback loop in which p53 activates the transcription of its primary inhibitor (6-8). ARF gene expression exhibits a strong inverse correlation with the functional status of p53 in both human (19, 32) and mouse cells (33, 34), suggesting a possible feedback repression of ARF expression by p53. Ours results provide a molecular basis supporting this feedback regulation. The significance of evolving this second feedback inhibition loop is that the first p53-MDM2 negative feedback loop would not be effective to inhibit p53 to resume the cell cycle if ARF expression is not repressed: the continuously synthesized ARF would bind to and prevent MDM2 from degrading p53 (Fig. 7C).
Our study also adds to the understanding of p53-mediated transcriptional repression, an area that is much less understood than p53-mediated transcriptional activation although an estimated 15% of genes containing a p53 response element can be repressed by p53 [see recent review by (2, 50)]. p53 has been reported to directly bind with mSin3A, a transcriptional co-repressor and a member of class I HDAC complexes and recruit HDACs to a specific promoter such as Map4 or Nanog (51, 52). We also confirmed the association between p53 and HDAC by detecting p53-HDAC1 binding in WT MEFs (Supplementary Figure D). Our study provides two separate lines of evidence supporting a role of histone deacetylation in p53-mediated repression of Arf. First, we showed that treatment of MEFs with a low concentration of TSA drastically increased Arf mRNA (>15-fold) and this effect is seen as early as 6 hours (Fig. 3). Second, we demonstrate that HDAC1 directly binds to and deacetylates Arf in a p53 dependent manner. We further identify a new mechanism—recruiting PRC—for p53-mediated transcriptional repression. To the best of our knowledge, this represents the first evidence that p53-mediated repression involves polycomb repressive complex which contains histone modifying activities known to function in silencing gene expression. Conversely, identification of a sequence-specific binding factor—p53—in the recruitment of PRC to a specific locus also helps to better understand how PcG is recrutied to their targets, a puzzling issue associated with the repression of many PRC-regulated genes in mammalian cells.
Supplementary Material
Acknowledgments
We thank Yi Zhang, Ned Sharpless and members of Xiong lab for the discussions throughout this study, Yizhou He for providing the retroviral construct targeting p53, and Dr. Zhi Liu for p53-/- mice. This study was supported by NIH grant CA68377 to Y.X.
References
- 1.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13(6):951–61. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 2.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–12. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 4.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 6.Barak Y, Juven T, Haffner R, Oren M. Mdm-2 expression is induced by wild-type p53 activity. EMBO J. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto A, Deppert W. Upregulation of mdm-2 expression in Meth A tumor cells tolerating wild-type p53. Oncogene. 1993;8(9):2591–603. [PubMed] [Google Scholar]
- 8.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 9.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 10.Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2, a dual mechanism. Genes & Dev. 1997;11:1974–86. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 12.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 13.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letter. 1997;420:25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 14.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667–77. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 15.Kim WY, Sharpless NE. The Regulation of INK4/ARF in Cancer and Aging. Cell. 2006;127(2):265–75. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6(9):663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 17.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell cycle control causing specific inhibition of cyclin D CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The INK4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 19.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17(17):5001–14. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez S, Klatt P, Delgado S, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440(7084):702–6. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 22.Matheu A, Klatt P, Serrano M. Regulation of the INK4a/ARF locus by histone deacetylase inhibitors. J Biol Chem. 2005;280(51):42433–41. doi: 10.1074/jbc.M508270200. [DOI] [PubMed] [Google Scholar]
- 23.Yarosh W, Barrientos T, Esmailpour T, et al. TBX3 is overexpressed in breast cancer and represses p14 ARF by interacting with histone deacetylases. Cancer Res. 2008;68(3):693–9. doi: 10.1158/0008-5472.CAN-07-5012. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–8. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes & Dev. 1999;13:2678–90. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes & Dev. 2005;19(12):1438–43. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Dev. 2005;19(12):1432–7. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 29.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21(5):525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes & Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12(19):2984–91. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 32.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18(11):6457–73. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12(15):2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Stanchina E, McCurrach ME, Zindy F, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12(15):2434–42. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95(14):8292–7. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 37.Lozano G. The oncogenic roles of p53 mutants in mouse models. Curr Opin Genet Dev. 2007;17(1):66–70. doi: 10.1016/j.gde.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Chen J, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to MDM-2 and the adenovirus 5 E1B 55-kD protein. Genes & Dev. 1994;8:1235–46. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 39.Otsuka K, Kato S, Kakudo Y, Mashiko S, Shibata H, Ishioka C. The screening of the second-site suppressor mutations of the common p53 mutants. Int J Cancer. 2007;121(3):559–66. doi: 10.1002/ijc.22724. [DOI] [PubMed] [Google Scholar]
- 40.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(6):845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14(5):637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(6):845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 43.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23(4):474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 44.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128(2):275–86. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 45.Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18(12):1413–22. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umbarger HE. Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science. 1956;123(3202):848. doi: 10.1126/science.123.3202.848. [DOI] [PubMed] [Google Scholar]
- 47.Kia S Kheradmand, Kartalaei P Solaimani, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2(1):16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agger K, Cloos PA, Rudkjaer L, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barradas M, Anderton E, Acosta JC, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23(10):1177–82. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9(10):724–37. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 51.Murphy M, Ahn J, Walker KK, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13(19):2490–501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165–71. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.