Abstract
RNase P is a highly conserved ribonucleoprotein enzyme that represents a model complex for understanding macromolecular RNA-protein interactions. Archaeal RNase P consists of one RNA and up to five proteins (Pop5, RPP30, RPP21, RPP29, and RPP38/L7Ae). Four of these proteins function in pairs (Pop5-RPP30 and RPP21–RPP29). We have used nuclear magnetic resonance (NMR) spectroscopy and isothermal titration calorimetry (ITC) to characterize the interaction between Pop5 and RPP30 from the hyperthermophilic archaeon Pyrococcus furiosus (Pfu). NMR backbone resonance assignments of free RPP30 (25 kDa) indicate that the protein is well structured in solution, with a secondary structure matching that observed in a closely related crystal structure. Chemical shift perturbations upon the addition of Pop5 (14 kDa) reveal its binding surface on RPP30. ITC experiments confirm a net 1 : 1 stoichiometry for this tight protein-protein interaction and exhibit complex isotherms, indicative of higher-order binding. Indeed, light scattering and size exclusion chromatography data reveal the complex to exist as a 78 kDa heterotetramer with two copies each of Pop5 and RPP30. These results will inform future efforts to elucidate the functional role of the Pop5-RPP30 complex in RNase P assembly and catalysis.
1. Introduction
Ribonuclease P (RNase P) is a ribonucleoprotein (RNP) complex primarily responsible for cleaving the 5′ leader sequence of precursor-tRNA (pre-tRNA) molecules in all domains of life [1–3]. The RNase P RNA subunit (RPR) constitutes the Mg2+-dependent catalytic moiety and supports pre-tRNA cleavage on its own in vitro [4–6]. The bacterial RNase P holoenzyme contains one large RPR and one conserved protein (RNase P protein, RPP) that is essential for function in vivo [7]. The bacterial RPP aids RPR catalysis by increasing the affinity of the holoenzyme for the substrate and for the Mg2+ cofactor [8–11]. Eukaryal and archaeal genomes do not encode sequence homologs of the bacterial RPP [12]. Instead, eukaryal and archaeal RNase P holoenzymes comprise multiple RPPs (up to 10 in eukarya and 5 in archaea) together with an RPR [12, 13].
The known archaeal RPPs are homologous to eukaryal proteins Pop5, RPP21, RPP29, RPP30, and RPP38 [21–23]. Recombinantly expressed RPPs have been assembled with the in vitro transcribed cognate RPR to reconstitute the holoenzyme from several archaea, including Methanothermobacter thermoautotrophicus (Mth) [24, 25] Pyrococcus horikoshii (Pho) [23], Pyrococcus furiosus (Pfu) [26], Methanocaldococcus jannaschii (Mja) [25, 27], and Methanococcus maripaludis [22]. Reconstitution studies with Mja, Mth, and Pfu RNase P indicate that four RPPs work in two distinct pairs: Pop5 with RPP30 and RPP21 with RPP29 (designated Pop5-RPP30 and RPP21-RPP29, resp.). This finding is consistent with results from yeast two-hybrid assays on RPPs from other archaea (Mth [28], Pho [29]) and eukarya [Saccharomyces cerevisiae (Sce)] [30], which indicate binding interactions between these pairs of proteins. Kinetic studies continue to provide insights into the functional roles of these binary RPP complexes; for example, single-turnover experiments with Mth RNase P revealed that Pop5-RPP30 increases by 60-fold the rate of pre-tRNA cleavage by the RPR and that RPP21-RPP29 enhances substrate affinity by 15-fold [25]. Although various biochemical studies demonstrate functional cooperation between the RNA and protein subunits of archaeal RNase P [18, 22, 25–27, 31], high-resolution structures are critical to fully understand both the assembly of the holoenzyme and the role of RPPs in assisting RPR catalysis.
X-ray crystallography and nuclear magnetic resonance spectroscopy (NMR) have been employed to determine the structure of several archaeal RPPs, including RPP29 from Mth [24], Archaeoglobus fulgidus [32], Pho [33], and Pfu [34], Pop5 from Pfu [35] and Pho [36], Pho RPP30 [37], RPP21 from Pho [38] and Pfu [39] and RPP38/L7Ae [40, 41]. Structures for the RPP21-RPP29 binary complex were obtained by crystallography for Pho [36] and by NMR for Pfu [34], and of the Pho Pop5-RPP30 pair by crystallography [14].
We have characterized the Pfu Pop5-RPP30 interaction by using NMR spectroscopy and isothermal titration calorimetry (ITC). Backbone resonance assignments have been obtained for the 214-residue Pfu RPP30 (Figure 1) in its free state. Size exclusion chromatography (SEC) and dynamic light scattering (DLS) data indicate that the free protein is monomeric, and secondary chemical shifts are consistent with the secondary structure previously observed for Pho RPP30 [37]. The addition of Pop5 to uniformly 13C/15N-labeled RPP30 induces many site-specific changes in the heteronuclear NMR spectra and leads to significant broadening of most resonances. Chemical shift perturbations reveal the Pop5 binding surface on RPP30, and complement previous chemical shift perturbation maps on Pop5 [35]. NMR and ITC data indicate that these two proteins form a tight 1 : 1 complex, while SEC and DLS data indicate the Pfu Pop5-RPP30 complex is heterotetrameric under a range of experimental conditions. We discuss possible functional implications of these findings.
Figure 1.
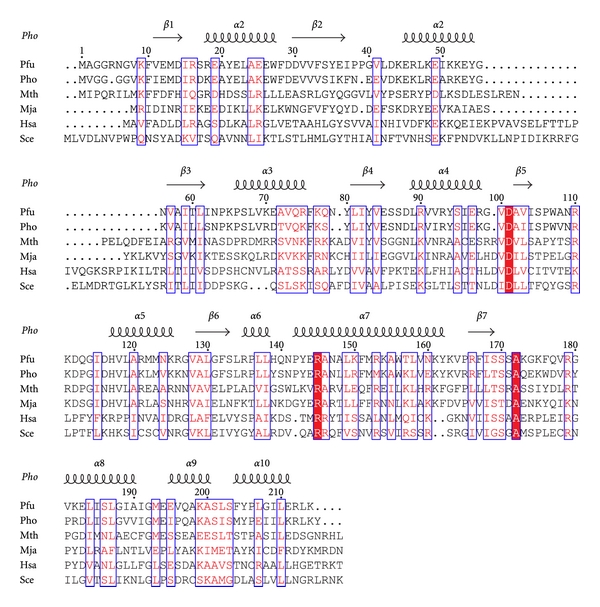
Sequence alignment of RPP30 homologs from four archaea, Pyrococcus furiosus (Pfu; NP_579643), Pyrococcus horikoshii (Pho; NP_143706), Methanothermobacter thermautotrophicus (Mth; NP_275831), and Methanocaldococcus jannaschii (Mja; NP_248131) and two eukaryotes Homo sapiens (Hsa; NP_006404) and Saccharomyces cerevisiae (Sce; NP_011929). Block letters indicate invariant residues and boxes indicate residues with a global similarity score >0.7. The secondary structure observed in the homologous Pho crystal structure [14] is depicted above the alignment. Alignment was performed using CLUSTALW [15] and colored according to similarity with a Risler scoring matrix [16] using the program ESPript [17].
2. Results
2.1. Oligomeric State
Size exclusion chromatography and dynamic light scattering were used to determine the oligomeric state of the Pop5-RPP30 complex (Figure 2). Free Pfu RPP30 eluted from the gel filtration column with an apparent molecular weight of 24 kDa, matching its monomer mass (24,495 Da). The mixture of RPP30 with an excess of Pop5 yielded two peaks, one corresponding to monomeric Pop5 (13,839 Da) and the other eluting with an apparent mass in excess of that of a heterodimer. Dynamic light scattering measurements showed that Pfu RPP30 and Pop5 are each monodisperse and monomeric when alone in solution (data not shown), while the 1 : 1 Pop5-RPP30 complex had a hydrodynamic radius of ~3.65 nm, corresponding to that calculated for the heterotetramer [(Pop5-RPP30)2] using HYDROPRO [18] (Figure 2). Although the complex seemed to aggregate at pH values above 6, it was monodisperse at lower pH values (and under a wide range of experimental conditions). These findings are in agreement with the heterotetrameric state observed in crystals of the Pho Pop5-RPP30 complex [14].
Figure 2.
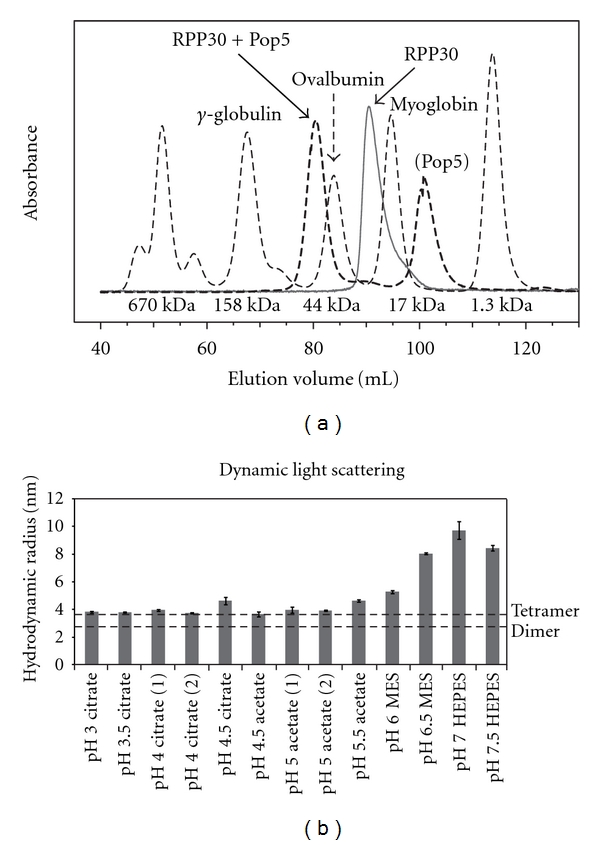
Size exclusion chromatography and dynamic light scattering of the Pop5-RPP30 complex. (a) Gel filtration chromatogram of RPP30 alone (solid), RPP30 in the presence of excess Pop5 (dashed), and molecular weight standards (dotted; thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.3 kDa)). The mixture of RPP30 with excess Pop5 yielded one peak corresponding to free Pop5, and a second peak with an apparent mass in excess of that expected for a Pop5-RPP30 heterodimer (39 kDa). (b) Measured hydrodynamic radii of the Pop5-RPP30 complex from dynamic light scattering under varying buffer conditions. The dotted lines indicate the calculated hydrodynamic radii from HYDROPRO [18] for both a heterotetramer or a heterodimer. The smallest hydrodynamic radius is consistent with formation of a heterotetramer [(Pop5-RPP30)2, 3.65 nm].
2.2. Backbone Resonance Assignments of Free RPP30
Pfu RPP30 samples were highly soluble in a range of buffer conditions at 55°C, yielding well-dispersed NMR spectra indicative of a well-structured protein (Figures 3(a) and 3(b)). However, spectral quality degraded significantly at lower temperatures (e.g., 25°C), perhaps reflecting nonspecific aggregation (data not shown). Because 55°C was found to be the optimum temperature for assembly and activity of the Pfu RNase P holoenzyme [26] and high-quality spectra could be obtained at that temperature, all NMR experiments were performed at 55°C.
Figure 3.
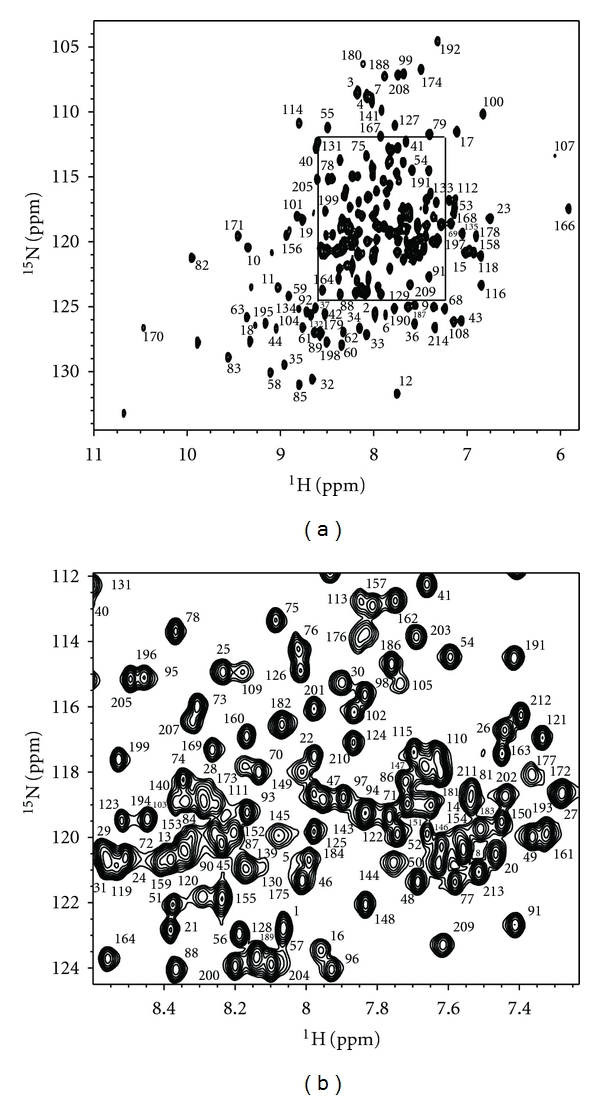
Two-dimensional 1H-15N correlation spectrum of Pfu RPP30. Residue numbers indicate assigned backbone amide resonances. The boxed region of the full spectrum (a) is expanded (b) for clarity.
Of the protein's 214 native residues (Figure 1), 193 backbone amide resonances could be assigned (Figure 3). Excluding the nine proline residues, this represents approximately 94% of the assignable backbone resonances. Most unassigned residues are adjacent to prolines. Residues 137–142 comprising helix α6 and the α6-α7 loop are flanked by prolines 136 and 143. Intense signals were observed for the first eight N-terminal residues, suggesting that these are highly flexible in solution. Secondary structural features were predicted from analysis of backbone chemical shifts using TALOS+ [19] (Figure 4). The secondary structural features match well with the TIM barrel fold observed in the crystal structure of Pho RPP30, consistent with their high degree of sequence homology (68% identity, 82% similarity [15]).
Figure 4.
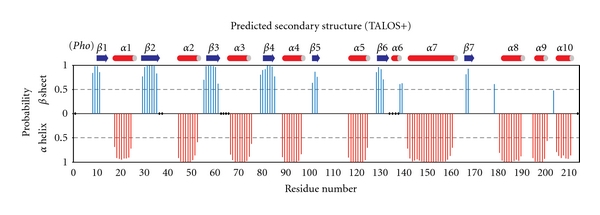
Chemical-shift-based secondary structure prediction of free RPP30 is consistent with its structural homology to Pho RPP30. Graph shows probability for each residue assigned to be either β sheet (blue above axis) or α helix (red below axis), as determined with the TALOS+ software [19]. Assigned secondary structure is given to each residue based on the highest probability for the residue to be either α Helix, β Sheet, or Loop. Loop residues are not indicated, while dots on the x-axis represent residues for which TALOS+ gave no prediction. For comparison, the secondary structure observed in crystals of the homologous Pho RPP30 is shown above the graph.
2.3. Backbone Resonance Assignments of RPP30 in Complex with Pop5
Compared to the free proteins, the solubility of the Pfu Pop5-RPP30 complex was much more sensitive to buffer conditions and mode of preparation. Data collection at lower temperatures proved intractable as spectral quality degraded significantly, while interpretable spectra could be recorded at 55°C (Figure 5). Spectra of [U-15N,13C]-RPP30 in complex with Pop5 at pH 5, 55°C remained well dispersed but highly overlapped due to increased linewidth resulting from the slower rotational correlation time of the heterotetramer. Backbone assignments of RPP30 in complex with Pop5 were obtained by tracing Cα connectivity in HNCA and HN(CO)CA spectra, using the free RPP30 assignments as a starting point, and the HNCO spectra to resolve some ambiguities. Due to increased linewidths and signal overlap in the heterotetrameric complex, only 180 backbone RPP30 amide resonances (~88%) could be assigned.
Figure 5.
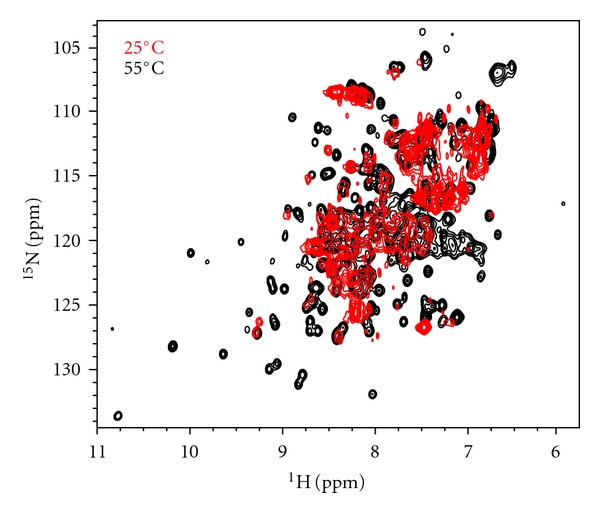
Elevated temperatures yield interpretable NMR spectra for the 78 kDa heterotetrameric Pfu Pop5-RPP30 complex. Overlay of two-dimensional 1H-15N correlation spectra of [U-15N]-RPP30 in complex with unlabeled Pop5 at 55°C (black) and 25°C (red) reveal excessive broadening at 25°C that is partly overcome at 55°C.
2.4. Pop5-Induced Chemical Shift Perturbations
The addition of unlabelled Pop5 to RPP30 induced changes to many RPP30 amide chemical shifts (Figure 6). The chemical shift perturbation pattern reflects the asymmetric interaction between each RPP30 and the two copies of Pop5 in the heterotetramer (Figure 7). In the crystal structure of the Pho Pop5-RPP30 heterotetrameric complex [14], the RPP30 protomers do not contact each other, but make contacts to both protomers of Pop5, burying almost 3800 Å2. The Pop5-RPP30 interface is dominated by α6, the loop between β7 and α8, as well as α8 of RPP30, which contact helices α1–α3 of both copies of Pop5. In the Pho crystal structure, additional contacts are observed between the unstructured N-terminus of Pop5 and helices α4 and α5 of RPP30 (in blue, Figure 7). While the Pfu NMR data are generally consistent with the Pho crystal structure, they indicate that only the first interface mentioned above is present in solution.
Figure 6.
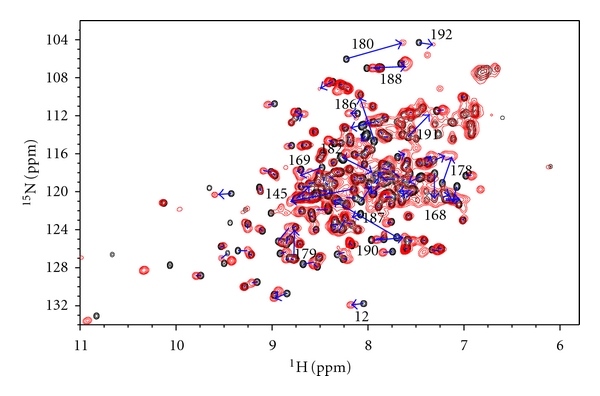
Overlay of 1H-15N correlation spectra of [U-15N]-RPP30 free (black) and bound to unlabeled Pop5 (red). Arrows highlight chemical shift perturbations induced by the addition of Pop5. Select perturbed amides are labeled with the residue assignment.
Figure 7.
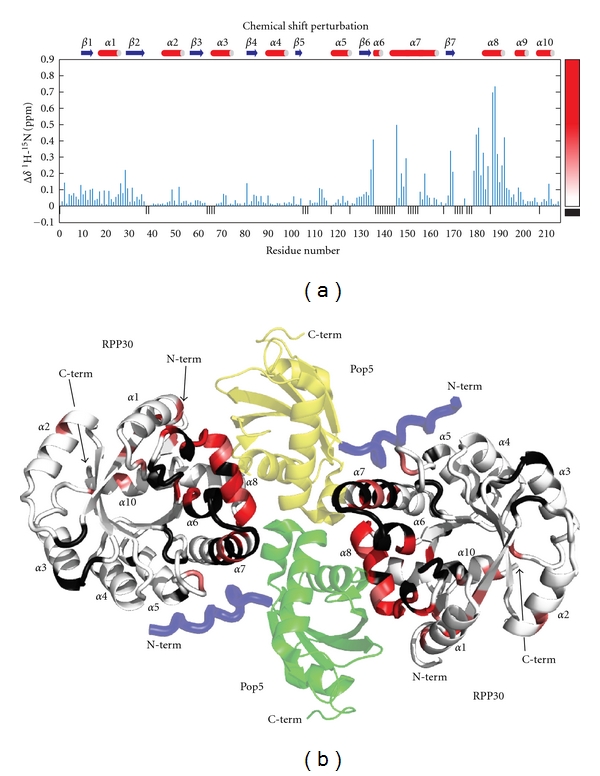
Chemical shift perturbations on RPP30 induced by Pop5 binding. (a) Secondary structure and per residue weighted average amide 1H and 15N chemical shift perturbations [20]. The color gradient used for mapping the data to the structure is shown on the right. Small black negative bars indicate residues for which the free and Pop5-bound RPP30 shifts could not be compared due to incomplete assignments or proline residues. (b) Cartoon diagram of a homology model of the Pfu Pop5-RPP30 complex based on the crystal structure of the complex from Pho [14]. Protomers of RPP30 are colored from white to red according to increasing shift perturbation. Black indicates no CSP data. The two protomers of Pop5 in the heterotetramer are green and yellow. An N-terminal segment observed in the Pho Pop5-RPP30 crystal structure, but not the Pfu Pop5 structure, is shown in blue.
Large chemical shift perturbations could be mapped to the major Pop5-RPP30 interface, comprising α6 and α8 of Pfu RPP30 (Figures 6 and 7). Amides of residues Gly180, Lys182, Leu187, Gly188, and Ala190 experience particularly large 1H shift perturbations (Δδ 1H > 0.3 ppm; Figures 6 and 7); these large perturbations can be attributed to strong ring current effects due to packing against aromatic residues Tyr78 and Phe82 in α3 of Pop5, or changes in position with regard to aromatic residues Trp27 and Phe28 of RPP30 (Figure 8). Residues Arg179, Ser186, Leu187, and Ile191 each experience large perturbations of the 15N chemical shifts (Δδ 15N > 2.5 ppm). These 15N perturbations, which are less sensitive to aromatic ring current effects [42], are likely to be caused by either changes to sidechain orientation or local backbone conformation.
Figure 8.
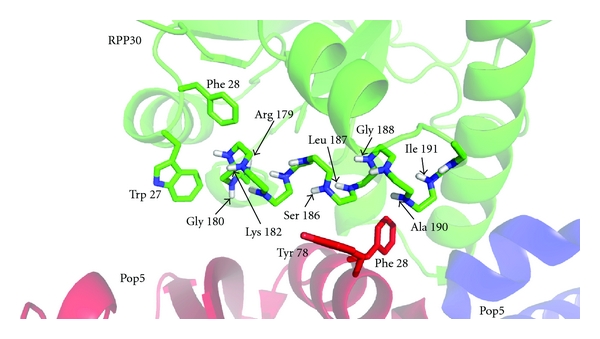
Close-up view of the Pop5-RPP30 binding interface. RPP30 is green and the two Pop5 subunits are red and blue. The backbone of α8 in RPP30 is shown as sticks with the amide nitrogen and proton atoms shown in blue and white, respectively. The sidechains of aromatic residues from Pop5 and RPP30 near α8 in RPP30 are shown as sticks. Electronic ring currents from such aromatic residues can result in large chemical shift perturbations for amides in the protein-protein interface.
In addition to this interface, the crystal structure of the Pho complex reveals a smaller Pop5-RPP30 interface in which the N-terminal portion of α7 of RPP30 contacts α2 of Pop5, and the N-terminal residues of Pop5 contact helices α4 and α5 of RPP30 [14]. Although the Pfu RPP30 backbone assignments are incomplete for the N-terminal portion of α7, the residues assigned in this region do show significant chemical shift perturbations, consistent with the heterotetrameric model. Unlike α7, all the nonproline residue of α4 and α5 are assigned but exhibit no significant chemical shift perturbations. This observation suggests that the N-terminal region of Pfu Pop5 does not make intimate contact with Pfu RPP30, which while not supportive of the heterotetramer model, is consistent with (i) the high crystallographic temperature factors observed for the N-terminus of Pop5 in the Pho complex [14], and (ii) the findings of complementary solution studies of labeled Pfu Pop5 bound to unlabeled RPP30, in which this region was observed to be largely unstructured in the absence or presence of RPP30 [35]. Thus, while the chemical shift perturbations are consistent with formation of a Pfu Pop5-RPP30 heterotetramer, they provide additional insight into the relative intimacy of the two sets of intermolecular contacts observed crystallographically for the Pho complex. The integrity of the heterotetrameric complex is further supported by hydrodynamic (see above) and thermodynamic data (see below).
2.5. Thermodynamics of the Pop5-RPP30 Interaction
The thermodynamics of the Pop5-RPP30 interaction were analyzed by isothermal titration calorimetry (ITC). The first two titrations demonstrate that the proteins form a high-affinity complex with an overall stoichiometry of 1 : 1 in the absence of other RNase P components (Figures 9(a) and 9(b)). The curves obtained under these conditions fit well to a one-site model but do not reveal a thermodynamic signature for formation of the expected 2 : 2 complex. An additional set of titrations were performed with a higher Wiseman “c” value [43] with smaller injections more carefully discern the presence of more complex binding modes. These titrations show clearly multiple binding modes with a combined stoichiometry of ~1, consistent with the expected 2 : 2 binding interaction. Although the data could be fit to a curve corresponding to a two-site binding model, given the three intermolecular interfaces observed in the complex (one Pop5-Pop5 interface, and two Pop5-RPP30 interfaces, Figure 7(b)), a more complex binding mechanism may be required to explain the thermodynamic data; exploring the details of such models is beyond the scope of the present study. Titrations performed under low ionic strength (10 mM sodium acetate) exhibited weaker binding (K d of 124 nM versus 3.5 nM) and less favorable binding enthalpy than those at higher ionic strength (140 mM, 160 mM, acetate plus salt). These observations are consistent with a significant contribution to binding from the hydrophobic effect as predicted from the nature of the observed intermolecular interfaces [14, 35].
Figure 9.
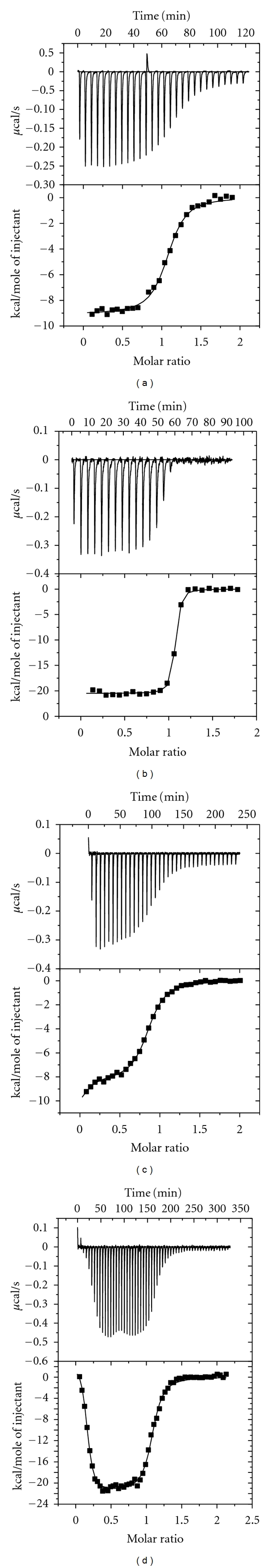
Titration calorimetry of Pop5-RPP30 binding reveals net 1 : 1 stoichiometry but higher-order binding. Top panels: Representative ITC thermograms obtained upon titration of Pfu Pop5 into Pfu RPP30 at 55°C. Bottom panels: Time-integration for each peak in the thermogram after normalizing per mol of injectant (squares). Titrations, (a, no salt) and (b, 130 mM KCl), were performed with 9.89 and 4.95 μM RPP30, respectively, in the cell and can be fit by a single site binding model, with apparent dissociation constants of 124 ± 13 and 3.5 ± 0.6 nM, respectively. Experiments performed with 14.8 μM RPP30 in the cell (c: no salt, d: 150 mM NaCl) yielded more complex thermograms that could only be fit by invoking two-site, or more complex binding models, consistent with higher-order assembly. Comparison of data at lower and higher ionic strength indicates binding is strongly favored by inclusion of moderate salt (e.g., 130 mM KCl) in the solutions.
3. Discussion
3.1. Stoichiometry and Folding of RPP Binary Complexes
These biophysical investigations of the interactions between Pfu RNase P proteins Pop5 and RPP30 have provided definitive evidence for the formation of a tight 2 : 2 Pop5-RPP30 complex in solution in the absence of other Pfu RNase P components. NMR chemical shift perturbations map the binding interface of Pop5 on RPP30. Together with previously reported chemical shift perturbations of Pop5 [35], these results confirm that the crystallographic observations from Pho Pop5-RPP30 [14] generally hold true in solution for the closely related Pfu Pop5-RPP30, except that the NMR data do not reveal evidence for interaction between RPP30 and the highly conserved, relatively unstructured N-terminus of Pop5. The finding of a heterotetrameric Pop5-RPP30 pair contrasts with the observation that the other archaeal RPP pair, RPP21-RPP29, forms only a heterodimer, both in crystals and in solution [34, 36].
Another notable difference between these binary complexes is in the effect of binding on the structures of the individual proteins. NMR studies of Pfu RPP21 and RPP29, both individually and as a complex, revealed that disordered regions present in these proteins fold upon heterodimerization [34, 39]. These structural changes, which are essential for generating the protein-protein interface, are especially pronounced in RPP29 where 52 new (from a total of 124) backbone amide resonances, associated with residues in α-helices, are observed in the 15N-edited spectra of the RPP21–RPP29 complex. Unlike Pfu RPP21 and RPP29, the structures of Pfu Pop5 and RPP30 appear to be well formed in the absence of the other. Interestingly, despite a tight interface mediated by structured cores of the interacting partners in Pfu Pop5-RPP30 and RPP21–RPP29 [14, 34, 36], both complexes display one or more unstructured termini, rich in basic residues, which could potentially play a critical role in RNA binding.
3.2. Necessity for Formation of Binary RPP Complexes
Assembly of multicomponent complexes represents a combinatorial challenge to the cell. In this regard, preassembly of some RNase P components might confer several advantages. First, with fewer individual RNA-protein binding steps, progression towards the final, native holoenzyme is facilitated. Second, if formation of heterodimers (e.g., RPP21–RPP29) is obligatory for generating the RNA-binding platform, then the affinity of the individual subunits for the RNA could be increased significantly by employing binary complexes; this strategy may help eliminate misfolded RNAs that offer binding sites for only one protein in the dimer, or limit binding of individual proteins to unintended targets. These ideas are well illustrated by the recent studies of the RPP20–RPP25 complex, which is associated with human RNases P and MRP [44, 45]. ITC experiments revealed that the binding affinity of human RPP20–RPP25 for the P3 helix of the cognate MRP RNA increases 1000-fold relative to those observed with either RPP20 or RPP25 [44]. Consistent with this finding, a crystal structure of the Saccharomyces cerevisiae Pop6-Pop7 dimer (homologous to human RPP20–RPP25) bound to the RNase MRP RNA's P3 domain shows how juxtaposing POP6 and POP7 in a binary complex positions their respective RNA-binding surfaces for cooperative and specific recognition of the P3 helix [45].
3.3. Possible Roles of the Pop5-RPP30 Heterotetramer
While it is easy to appreciate the utility of pairwise interactions between RPPs, the functional significance of the Pfu Pop5-RPP30 heterotetramer is presently unclear. To examine this question in Pho RNase P, a six-residue internal deletion mutant of Pho Pop5 was examined for its ability to for heterotetramers and reconstitute activity [14]. The deleted residues correspond to a loop between Pop5 helices α1 and α2 that constitutes an interface between the two copies of Pop5 in the Pho Pop5-RPP30 heterotetramer crystal structure. Although this Pho Pop5 mutant assembles into a heterodimer with RPP30, it was reported to not support RPR-mediated pre-tRNA cleavage (data are not shown in [14]). However, since the mutation might affect other unintended aspects of RNase P assembly or function, it is difficult to conclude whether the heterotetrameric form of Pop5-RPP30 is indeed required for RNase P activity.
If Pop5-RPP30 were heterotetrameric in fully assembled archaeal RNase P, symmetry considerations would argue that the holoenzyme (RNA + proteins) is also dimeric. Currently, neither the stoichiometry of individual subunits nor the molecular weight of the archaeal RNase P holoenzyme is well determined. In the absence of such data, insights might be gleaned from previous findings on the eukaryal and eubacterial RNase P variants.
Various experimental observations hint towards higher-order structure in eukaryotic RNase P. Yeast two-hybrid studies have established self-association of different human/yeast RPPs, leading to expectations that multiple copies of some of these subunits might be present in their respective native holoenzymes [30, 46]. Protein quantitation by SYPRO Ruby fluorescence staining in RNase MRP, an RNP complex involved in rRNA and mRNA processing and shares several protein subunits with RNase P [47], suggests that multiple copies of nine of the ten protein subunits are associated with the MRP RNA [48]. Additional experimental evidence is needed to corroborate and extend these stoichiometric estimates.
Hints at functional dimerization have also emerged from biochemical and biophysical studies of eubacterial RNase P. Small angle X-ray scattering (SAXS) studies revealed that although the Bacillus subtilis RNase P RNA exists as a monomer in solution, addition of its cognate protein cofactor results in formation of a holoenzyme dimer [49, 50]. The oligomerization state is sensitive to the ionic conditions, with dimers and monomers favored at low and high ionic strength, respectively. Moreover, at low ionic strength, the holoenzyme dimer was observed to dissociate upon addition of a monomeric pre-tRNA substrate, while a mixture of monomeric and dimeric ES complexes were observed upon addition of a dimeric substrate.
While there is no direct experimental evidence for presence of the RNase P dimer in vivo, the K D value of 50 nM (at 0.1 M NH4Cl) reported for the dimerization of B. subtilis RNase P would permit formation of the dimer at the estimated intracellular concentrations of bacterial RNase P (up to 42 nM in B. subtilis [50] and 680 nM in E. coli [51]). Among possible payoffs from a holoenzyme dimer, Pan and coworkers considered both cooperative substrate binding and increased processivity during maturation of polycistronic transcripts containing multiple tRNAs (e.g., the largest tRNA operon in B. subtilis which codes for 21 tRNAs). Whether polycistronic substrates accumulate at steady state is unclear, given the concerted actions of various nucleases (e.g., RNase E in some bacteria [52, 53]) that simplify the task of RNase P by generating monomeric intermediates from polycistronic precursors.
Oligomerization of RNase P might confer gains in addition to processivity. The half-life of the RNA moiety might be enhanced in a higher-order RNP complex. Or, certain oligomeric states, even if comprised of only the RPR and a partial suite of RPPs, might represent either inactive forms that prevent nonspecific cleavage of unintended cellular RNAs or intermediates primed to assemble into the holoenzyme. Further studies are clearly needed to gain a full understanding of the functional relevance of oligomeric forms of RNase P.
4. Materials and Methods
4.1. Protein Preparation
Unless otherwise noted, Pop5 and RPP30 refer to the Pfu proteins throughout. Unlabelled Pop5 (NP_579107) and RPP30 (NP_579643) samples were prepared as described previously [35]; the Cys72Ser variant of Pop5 was used in this study due to its decreased tendency to aggregate. Uniformly doubly labeled RPP30 ([U-13C, 15N]-RPP30) was purified from cells grown in M9 minimal medium supplemented with 1% (v/v) Eagle Basal Vitamin Mix (Life Technologies, Gaithersburg, MD) [54], containing 1 g/L 15N ammonium chloride as the sole nitrogen source and 3 g/L 13C glucose as the sole carbon source. The final RPP30 protein used includes three additional residues (Gly-Glu-Phe) at the N-terminus, which remain after TEV protease cleavage of a (His)6-maltose binding protein tag used for affinity purification. Labeled RPP30 was dialyzed into NMR buffer [10 mM sodium acetate, pH 5, 0.02% (w/v) sodium azide] and concentrated to 1 mM. D2O was added to 5% (v/v). Unlabeled proteins were dialyzed at room temperature for 3 days into NMR buffer to ensure complete equilibration in preparation for ITC.
4.2. Gel Filtration
Size exclusion chromatography was performed with each protein alone and with RPP30 mixed with an excess of Pop5. The proteins were dialyzed into NMR buffer with 500 mM NaCl and run on a Hiload 16/60 Superdex-200 gel filtration column (GE healthcare).
4.3. Dynamic Light Scattering
Dynamic light scattering (DLS) was used as a preliminary screen to identify conditions that produced favorable NMR spectra. Samples for DLS were prepared by dialyzing 100 μL of each protein (90 μM) into each buffer tested (Figure 2). Stoichiometric amounts of Pop5 were then slowly pipetted into RPP30 resulting in a 45 μM sample of the complex. Each sample was filtered through a 0.22 μm filter (Costar Spin-X Centrifuge Tube Filter, Corning Inc.) and analyzed with a DynaPro-801 Molecular Sizing Instrument (Protein Solutions, Inc.). The behavior of each sample was examined in terms of its estimated hydrodynamic radius, as well as a low polydispersity over radius (polyD/r). Polydispersity is representative of the particle size distribution width, thus a low polyD/r value indicates a homogeneous mixture. The buffer conditions sampled ranged from pH 3 to 8 in increments of 0.5; reported values are the mean and standard deviation from 18 measurements (Figure 2). DLS measurements in salt concentrations from 0 to 1 M NaCl showed that the proteins had a tendency to aggregate above 250 mM NaCl (data not shown). Below 250 mM NaCl, DLS measurements over a range of protein concentrations from 50 to 500 μM were consistent with a single, heterotetrameric species.
4.4. Preparation of the Pop5-RPP30 Complex
The NMR sample of labeled RPP30 in complex with unlabeled Pop5 was prepared by dialyzing each protein separately into a buffer containing 10 mM sodium acetate (pH 5) with 0.02% (w/v) sodium azide. RPP30 and Pop5 were concentrated to 575 and 1,575 μM, respectively. Equimolar amounts of each protein were mixed slowly by use of Spectra/Por CE dialysis tubing 100,000 MWCO (Spectrum Laboratories, Inc). The resulting solutions yielded a Pop5-RPP30 complex at a concentration of 425 μM in buffer containing 10 mM sodium acetate (pH 4), 0.02% (w/v) sodium azide. For NMR experiments, D2O was added to 5% (v/v). Free U-[13C,15N]-RPP30 (0.5 mM) was prepared in the same buffer for direct comparison of NMR spectra of RPP30, free and bound to Pop5.
4.5. NMR Spectroscopy
Triple resonance TROSY [55] spectra (HNCO, HNCA, HNCACB, and CBCA(CO)NH) [56] were recorded on U-[13C,15N]-RPP30 at pH 5 for backbone resonance assignments. Triple resonance spectra without the TROSY effect (HNCO, HNCA, HN(CO)CA, CBCA(CO)NH) [56] were recorded on U-[13C, 15N]-RPP30 in complex with unlabeled Pop5 at pH 4 for backbone assignments of the complex. All NMR experiments were performed at 55°C on a Bruker DRX-600 spectrometer equipped with a cryogenically cooled triple resonance single-axis gradient probe. Data were processed with NMRPipe [57] and analyzed using NMRViewJ [58]. Backbone assignments of free RPP30 were obtained by manual inspection of the data, with the assistance of the Probabilistic Interaction Network of Evidence (PINE) algorithm [59]. Pop5-induced chemical shift perturbations on RPP30 were quantified using the chemical shift differences of amide 1H and 15N resonance frequencies as [20]. Assignments for free and Pop5-bound RPP30 have been deposited in the BioMagResBank (BMRB: http://www.bmrb.wisc.edu/) with accession numbers 17189 and 17190, respectively.
Isothermal Titration Calorimetry, (ITC) —
Protein concentrations were determined by absorbance at 280 nm in 20 mM sodium phosphate (pH 6.5), 6 M guanidium hydrochloride using molar extinction coefficients predicted from their amino acid sequences [60]: RPP30 ε 280 = 28,590 M−1 cm−1, Pop5 ε 280 = 19,750 M−1 cm−1.
The first set of titrations performed was Pop5 (87.2 μM) titrated into RPP30 (9.89 μM and 4.95 μM) (Figures 9(a) and 9(b), resp.). All titrations were performed at 55°C on a MicroCal VP-ITC microcalorimeter, with the first injection being 5 μL and all subsequent injections being 10 μL with 240 seconds between injections; the first 5 μL injection was discarded from analysis. The heat of dilution was estimated by averaging the last 3 to 5 nonreacting injections in which excess Pop5 was injected into saturated RPP30. The titrations were carried out in the absence (Figure 9(a)) and presence of 130 mM KCl (Figure 9(b)). Data were fit to a one-site model with the Origin software v. 7 (MicroCal, Inc.) to obtain the binding stoichiometry n, association constant K A, and binding enthalpy ΔH. Although these data revealed a net 1 : 1 stoichiometry, they could not distinguish a 1 : 1 heterodimer from the 2 : 2 stoichiometry of the heterotetramer.
To reveal the thermodynamic signature of forming a heterotetrameric complex, a second set of titrations was performed while aiming to vary the Wiseman “c” value [43]. When Pop5 (148 μM) was titrated into RPP30 (14.8 μM) in the absence of salt (Figure 9(c)), the first injection was 4 μL and all subsequent injections were 7.5 μL, with 400 s between injections. The titration with 150 mM sodium chloride (Figure 9(d)) had a first injection of 2.5 μL and all subsequent injections of 5 μL with 350 s between injections. The injection volume was decreased in order to increase the likelihood of observing multiple binding sites. Data were fit to a two-site model by Origin software v. 7 (MicroCal, Inc.).
4.6. Homology Modeling
A homology model of Pfu Rpp30 was obtained by threading its amino acid sequence into the coordinates of Pho RPP30 (PDB entry 2CZV, chain b) [14] using SWISS-MODEL [61]. A model of the Pfu heterotetramer was assembled by aligning the coordinates of Pfu Pop5 [35] with the corresponding coordinates of Pho Pop5 in the same coordinate set (2CZV chains c, d). Superposition and surface calculations were performed with Pymol (http://www.pymol.org/).
Acknowledgments
The authors thank members of the Foster and Gopalan laboratories for spirited discussions and technical assistance; M. Adams (University of Georgia, Athens), J. Williamson (The Scripps Research Institute, La Jolla, CA), and D. Waugh (National Cancer Institute, Bethesda) for valuable reagents; and the Ohio State University (OSU) Campus Chemical Instrument Center staff for assistance with NMR instrumentation. This work was supported by Grant GM067807 from the NIH to V. Gopalan and M. P. Foster, and a grant MCB 0843543 from the NSF to V. Gopalan; B. L. Crowe and R. C. Wilson were supported in part by an NIH Chemistry-Biology Interface Training Grant (T32 GM008512); R. C. Wilson received additional support from an American Heart Association predoctoral fellowship.
References
- 1.Altman S. A view of RNase P. Molecular BioSystems. 2007;3(9):604–607. doi: 10.1039/b707850c. [DOI] [PubMed] [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends in Biochemical Sciences. 2006;31(6):333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Letters. 2010;584(2):287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Gardiner K, Marsh T. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35(3):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 5.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(14):7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(7):2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schedl P, Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNA(Asp) Biochemistry. 1998;37(8):2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 10.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P·Pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41(30):9545–9558. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13(9):1505–1515. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblad MA, López MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Research. 2006;34(18):5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarrous N, Gopalan V. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Research. 2010;38(22):7885–7894. doi: 10.1093/nar/gkq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. Journal of Molecular Biology. 2006;357(2):583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risler JL, Delorme MO, Delacroix H, Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. Journal of Molecular Biology. 1988;204(4):1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 17.Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 18.García De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophysical Journal. 2000;78(2):719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM. Identification by NMR of the binding surface for the histidine- containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry. 1997;36(15):4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 21.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8(3):296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14573–14578. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzuma Y, Mizoguchi M, Takagi H, et al. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochemical and Biophysical Research Communications. 2003;306(3):666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 24.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen WY, Pulukkunat DK, Cho IM, Tsai HY, Gopalan V. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Research. 2010;38(22):8316–8327. doi: 10.1093/nar/gkq668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(44):16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468(7325):784–791. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall TA, Brown JW. Interactions between RNase P protein subunits in Archaea. Archaea. 2004;1(4):247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kifusa M, Fukuhara H, Hayashi T, Kimura M. Protein-protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Bioscience, Biotechnology and Biochemistry. 2005;69(6):1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- 30.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazantsev AV, Krivenko AA, Harrington DJ, et al. High-resolution structure of RNase P protein from Thermotoga maritima. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7497–7502. doi: 10.1073/pnas.0932597100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43(44):14128–14138. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- 33.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10(9):1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. Journal of Molecular Biology. 2009;393(5):1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archael RNase P protein. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. Journal of Molecular Biology. 2008;384(3):652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 37.Takagi H, Watanabe M, Kakuta Y, et al. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochemical and Biophysical Research Communications. 2004;319(3):787–794. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 38.Kakuta Y, Ishimatsu I, Numata T, et al. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: an archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44(36):12086–12093. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- 39.Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry. 2008;47(45):11704–11710. doi: 10.1021/bi8015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443(7109):302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 41.Fukuhara H, Kifusa M, Watanabe M, et al. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochemical and Biophysical Research Communications. 2006;343(3):956–964. doi: 10.1016/j.bbrc.2006.02.192. [DOI] [PubMed] [Google Scholar]
- 42.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Nuclear Magnetic Resonance of Biological Macromolecules A. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 43.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry. 1989;179(1):131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 44.Hands-Taylor KLD, Martino L, Tata R, et al. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Research. 2010;38(12):4052–4066. doi: 10.1093/nar/gkq141. Article ID gkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO Journal. 2010;29(4):761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear RNase P. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(3):920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes and Development. 1998;12(11):1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. Journal of Biological Chemistry. 2005;280(12):11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- 49.Fang XW, Yang XJ, Littrell K, et al. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA. 2001;7(2):233–241. doi: 10.1017/s1355838201001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrera A, Fang X, Jacob J, Casey E, Thiyagarajan P, Pan T. Dimeric and monomeric Bacillus subtilis RNase P holoenzyme in the absence and presence of Pre-tRNA substrates. Biochemistry. 2002;41(43):12986–12994. doi: 10.1021/bi020416k. [DOI] [PubMed] [Google Scholar]
- 51.Dong H, Kirsebom LA, Nilsson L. Growth rate regulation of 4.5 S RNA and M1 RNA the catalytic subunit of Escherichia coli RNase P. Journal of Molecular Biology. 1996;261(3):303–308. doi: 10.1006/jmbi.1996.0461. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8(1):97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes and Development. 2002;16(9):1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Plainview, NY, USA: Cold Spring Harbor Lab; 1989. [Google Scholar]
- 55.Rance M, Loria JP, Palmer AG. Sensitivity improvement of transverse relaxation-optimized spectroscopy. Journal of Magnetic Resonance. 1999;136(1):92–101. doi: 10.1006/jmre.1998.1626. [DOI] [PubMed] [Google Scholar]
- 56.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34(2):93–158. [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in Molecular Biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 59.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Computational Biology. 2009;5(3) doi: 10.1371/journal.pcbi.1000307. Article ID e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, NJ, USA: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 61.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]


