Abstract
Metabolic adaptation to environmental changes is crucial for the long-term survival of an organism. Signaling mechanisms that govern this adaptation thus influence lifespan. One such mechanism is the insulin/insulin-like growth factor signaling (IIS) pathway, a central regulator of metabolism in metazoans. Recent studies have identified the stress-responsive Jun-N-terminal kinase (JNK) pathway as a regulator of IIS signaling, providing a link between environmental challenges and metabolic regulation. JNK inhibits IIS activity and, thus, promotes lifespan extension and stress tolerance. Interestingly, this interaction is also at the center of age-related metabolic diseases. Here, we review recent advances illuminating the mechanisms of the JNK--IIS interaction and its implications for metabolic diseases and lifespan in metazoans.
Insulin/IGF signaling and the control of lifespan
Invertebrates exhibit dramatic metabolic adaptation to environmental challenges, responding to oxidative stress, starvation, low or high temperatures and other stressors by curtailing reproduction, anabolism and growth. This so-called diapause is commonly believed to enable the transfer of energy resources from growth and reproduction to somatic maintenance under deleterious environmental conditions [1,2]. Although metabolic responses to environmental changes are less pronounced in vertebrates, extensive work over the past decade has established considerable similarities in the regulation of metabolic adaptation in invertebrates and the control of metabolic homeostasis in vertebrates. In particular, the impact of these regulatory mechanisms on lifespan was found to be conserved.
Insulin/IGF signaling and lifespan in flies and worms
Genetic studies in the nematode Caenorhabditis elegans and the fruitfly Drosophila melanogaster have established that genes involved in the endocrine regulation of diapause influence lifespan and stress tolerance of the adult organism under optimal conditions. A major regulator of both diapause and longevity identified in these studies is the insulin/IGF signaling (IIS) pathway. Worms carrying strong loss-of-function alleles for the insulin-receptor homolog dauer formation-2 (daf-2), for example, enter larval diapause (‘dauer’ state) even under favorable environmental conditions, whereas animals carrying weaker loss-of-function alleles for daf-2 are temperature-sensitive for dauer formation and have dramatically longer adult lifespans than wild-type counterparts at all temperatures [3–7]. Similarly, inactivation of daf-2 in adulthood using RNAi results in lifespan extension [3–6].
The two physiological consequences of daf-2 repression, dauer induction and extended lifespan, are mediated by the transcription factor DAF-16, the homolog of mammalian and Drosophila Forkhead Box O (FoxO) proteins [5,8,9]. DAF-16 is retained in the cytoplasm and inactivated in response to IIS. Signal transduction through the canonical IIS pathway results in Akt-mediated phosphorylation of DAF-16 and subsequent interaction with 14–3-3 proteins, which mediate its cytoplasmic retention [10–16] (Figure 1). In IIS mutants, elevated DAF-16 activity induces the expression of DAF-16 target genes that encode cell-cycle inhibitors, growth regulators, anti-oxidant proteins and DNA-repair molecules [4,17–20].
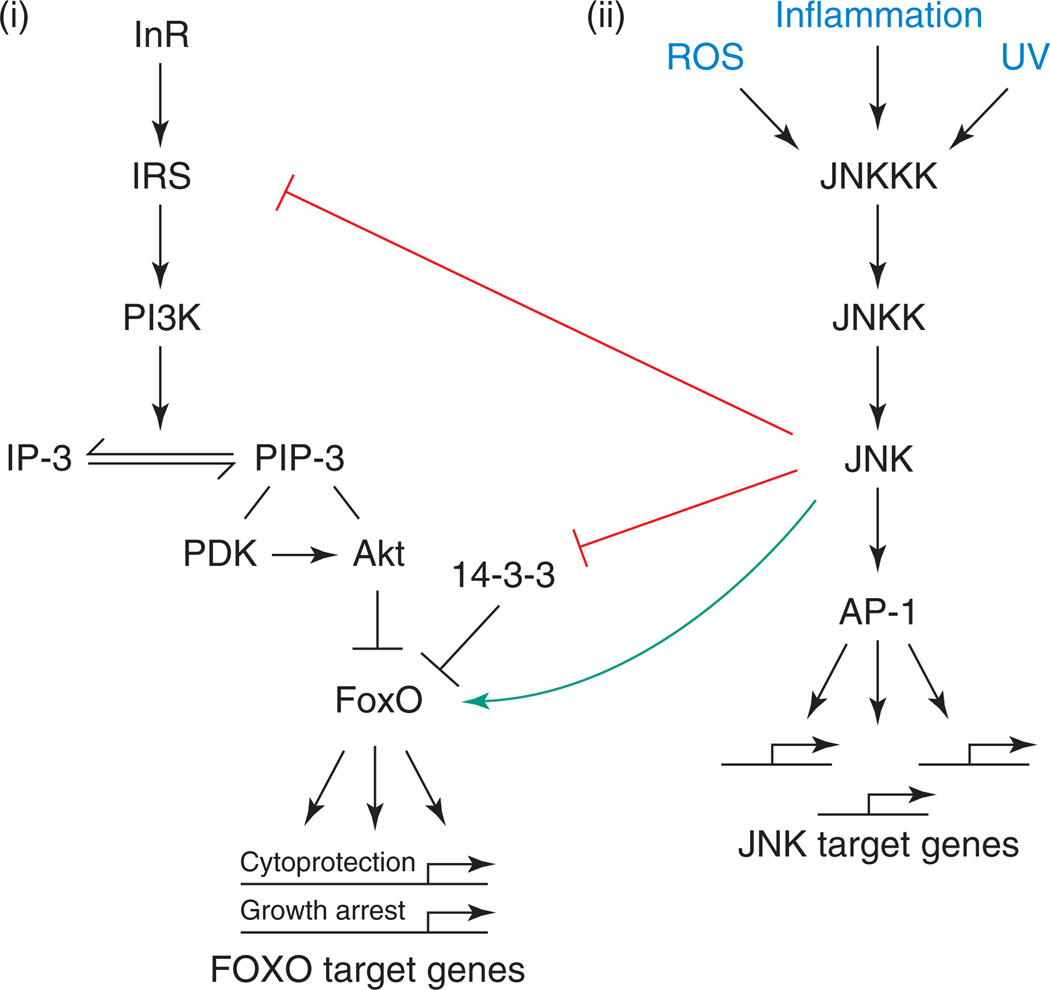
Figure 1.
Signaling interactions between IIS- and JNK-signaling pathways. Depicted are the generic, evolutionarily conserved JNK and IIS pathways. (i) Insulin activates the insulin receptor (InR), initiating a signaling cascade that results in activation of the protein kinase Akt. Akt phosphorylates the forkhead transcription factor FoxO, causing its cytoplasmic retention. IIS activation thus results in repression of FoxO-target genes. (ii) Upon activation by various stressors, specific JNKKKs (Jun-N-terminal kinase kinase kinases) phosphorylate JNK kinase (JNKK), which, in turn, phosphorylates JNK. Induction of gene transcription by JNK occurs primarily through the transcription-factor activator protein-1 (AP-1), a heterodimeric protein composed of the proteins Jun and Fos. Note that IRS-1 proteins are not conserved in C. elegans. JNK has been described as phosphorylating IRS-1, 14–3-3 proteins and FoxO proteins in various systems.
The regulation of metabolic adaptation by IIS is evolutionarily conserved, highlighting its importance for the fitness of the organism [2,21–23]. Flies and mosquitoes, for example, undergo reproductive diapause under unfavorable conditions (such as short day length and low temperature), and this transition correlates with reduced IIS activity [1,24–26]. Insulin signal transduction in Drosophila proceeds through the canonical pathway of conserved molecules (the insulin receptor, InR; insulin-receptor substrate, IRS (Chico in Drosphila); phosphatidylinositol-dependent kinase, PI3Kinase; protein kinase B (also called Akt); 14–3-3; and the Drosophila Forkhead Box O transcription factor, dFoxO) and regulates lifespan, stress tolerance, metabolic homeostasis and growth [27–29] (Figure 1). Lifespan and oxidative stress tolerance are increased significantly in fly mutants for InR [30] and chico [31] and in flies overexpressing dFoxO [32,33]. These beneficial effects of reduced IIS activity come at a cost, however, because reduced IIS signaling results in impaired growth and metabolic homeostasis [27,30–35].
IIS and lifespan in vertebrates
Considerable similarities also exist between the regulation of diapause in lower organisms and mechanisms that curtail anabolic functions in response to environmental and intrinsic stress signals in vertebrates [22]. The IIS pathway in vertebrates diverged into the insulin-like growth factor (IGF)- and insulin-signaling pathways, indicating potentially specific functions for each of these pathways in regulating metabolic adaptation, longevity and stress tolerance. Although this divergence is well documented for IGF or insulin effects on growth and metabolic control [36,37], current findings indicate that both pathways can also influence longevity in mice [38,39].
Mammalian FoxO homologs can induce metabolically dormant states in mammalian cells while increasing cytoprotective gene expression [40,41]. They are also known to promote DNA repair and prevent oxidative damage accumulation in quiescent cells [42,43]. Recent work in mouse hematopoietic stem cells further indicates that FoxO proteins are crucial for maintaining regenerative potential of stem cell populations in aging vertebrates [44–46]. The impact of reduced IIS activity and, thus, increased FoxO-mediated cytoprotection on lifespan has been tested in mice; inactivation of the insulin receptor in adipose tissue, as well as systemic reduction of IGF-1 signaling, results in increased lifespan [38,39,47]. Furthermore, reducing insulin-receptor substrate-2 (IRS-2) throughout the body or reducing IRS-2 specifically in the brain extends lifespan of mice [48]. Along the same lines, lower IGF activity has been found in human centenarians, indicating a similar role for IIS activity in regulating lifespan in humans [49].
Regulation of lifespan by JNK--IIS interactions
Although reduced IIS activity is well established as beneficial for overall stress tolerance and longevity in vertebrates and invertebrates, the mechanism(s) by which IIS activity is regulated in response to stress is only beginning to be understood. In mammalian cell culture, oxidative stress causes nuclear translocation of FoxO [50,51]. Similarly, C. elegans and Drosophila FoxO homologs are translocated to the nucleus in response to a variety of environmental insults [11,35,52]. In the nucleus, FoxO not only induces expression of cytoprotective genes but also induces expression of cell-cycle inhibitors and translational repressors [18,35,42,51,52]. FoxO thus emerges as a central, evolutionarily conserved hub in the cellular stress response that is necessary to balance anabolic processes with repair and/or maintenance mechanisms. One signal that promotes FoxO activation in response to environmental stress has been identified as the Jun-N-terminal kinase (JNK)-signaling pathway [53–55]. In the following section, we discuss the genetic and molecular interactions between IIS- and JNK-signaling pathways identified to date, in addition to the significance of these interactions in regulating longevity and metabolic homeostasis.
Regulation of stress tolerance and lifespan by JNK signaling
JNK is an evolutionarily conserved stress-activated protein kinase (SAPK) that is induced by a range of intrinsic and environmental insults (e.g. UV irradiation, reactive oxygen species, DNA damage, heat and inflammatory cytokines) (Figure 1). It is activated by a mitogen-activated protein (MAP) kinase cascade and ultimately regulates transcription factors that lead to changes in gene expression. JNK activation has highly context-dependent consequences for the cell, ranging from apoptosis over morphogenetic changes to increased survival [56–59].
Interestingly, moderate activation of JNK signaling results in increased stress tolerance and extended lifespan in flies and worms. Flies heterozygous for the JNK phosphatase Puckered (puc), or in which the JNK kinase Hemipterous (JNKK/Hep) is overexpressed in neuronal tissue, are resistant to the oxidative-stress-inducing compound Paraquat [60]. Similarly, JNKK/Hep mutant flies exhibit increased stress sensitivity and are deficient in their ability to induce a transcriptional stress response [60]. Supporting the protective role for JNK signaling in flies, puc heterozygotes or Hep-overexpressing animals are long-lived under normal conditions [60,61].
Similar consequences of JNK activation have been described in C. elegans. Worms with increased JNK activity caused by inhibition of the JNK phosphatase VH1 dual-specificity phosphatase are protected against heavy metal toxicity, and overexpression of JNK increases lifespan under normal conditions [54,62].
Crosstalk between IIS and JNK signaling
The protective effects of JNK signaling described above are mediated by DAF-16/FoxO, indicating an antagonism between IIS and JNK signaling in worms and flies [54,55] (Figure 1). Findings that support a role for DAF-16/FoxO downstream of JNK signaling include that expression of JNK-responsive genes is reduced in dfoxO mutant flies and that reducing the dfoxO gene dose is sufficient to repress JNK-induced growth inhibition and apoptosis [55,63]. Activation of JNK can, furthermore, dominantly suppress IIS-induced cell growth and induce nuclear translocation of FoxO. In addition, the long lifespan of puc heterozygotes is reverted to wild-type levels when dfoxO loss-of-function alleles are introduced [55,63]. In C. elegans, JNK activation also results in nuclear translocation of DAF-16, and functional DAF-16 is required for JNK-mediated lifespan extension [54,64,65]. Activation of FoxO proteins by JNK has further been described in mammalian cells, indicating that the interaction between IIS and JNK signaling is conserved [53,66]. Similar to IIS loss-of-function conditions, FoxO thus has a crucial role in promoting lifespan extension and stress tolerance when JNK is activated.
Interestingly, the antagonism between IIS and JNK signaling seems to be mediated by molecular interactions at multiple levels within the two pathways, as well as by endocrine mechanisms (Figures 1 and 2 and Table 1). In vertebrates, JNK phosphorylates the insulin-receptor substrate (IRS), inhibiting insulin signal transduction [67,68]; JNK can also activate FoxO4 directly by phosphorylation [53]. Recent studies further indicate that phosphorylation of 14–3-3 proteins by JNK results in release of their binding partners, including FoxO, which then translocates to the nucleus [69–71] (Figure 1). The exact mechanisms of interaction between JNK and IIS in invertebrates have not been extensively addressed. Although C. elegans JNK was shown to phosphorylate DAF-16 [54], no homolog for IRS proteins has been identified in worms. Furthermore, sequence alignment shows that the JNK phosphorylation site in mammalian IRS proteins (Ser307 of human IRS-1 [63]) is not conserved in Drosophila Chico. The same divergence occurs in FoxO; JNK phosphorylation sites identified in mammalian FoxO4 (Thr447 and Thr451 of human FoxO4 [53]) are not conserved in either DAF-16 or Drosophila FoxO. The JNK phosphorylation sites on mammalian 14–3-3 proteins (Ser184 in 14–3-3ζ and Ser186 in 14–3-3σ [69–71]), however, are conserved in Drosophila 14–3-3ζ and 14–3-3ε, as well as in the C. elegans 14–3-3 homolog PAR-5 (but not in the other 14–3-3 homolog, Ftt-2), indicating that similar interactions might exist in flies and worms.
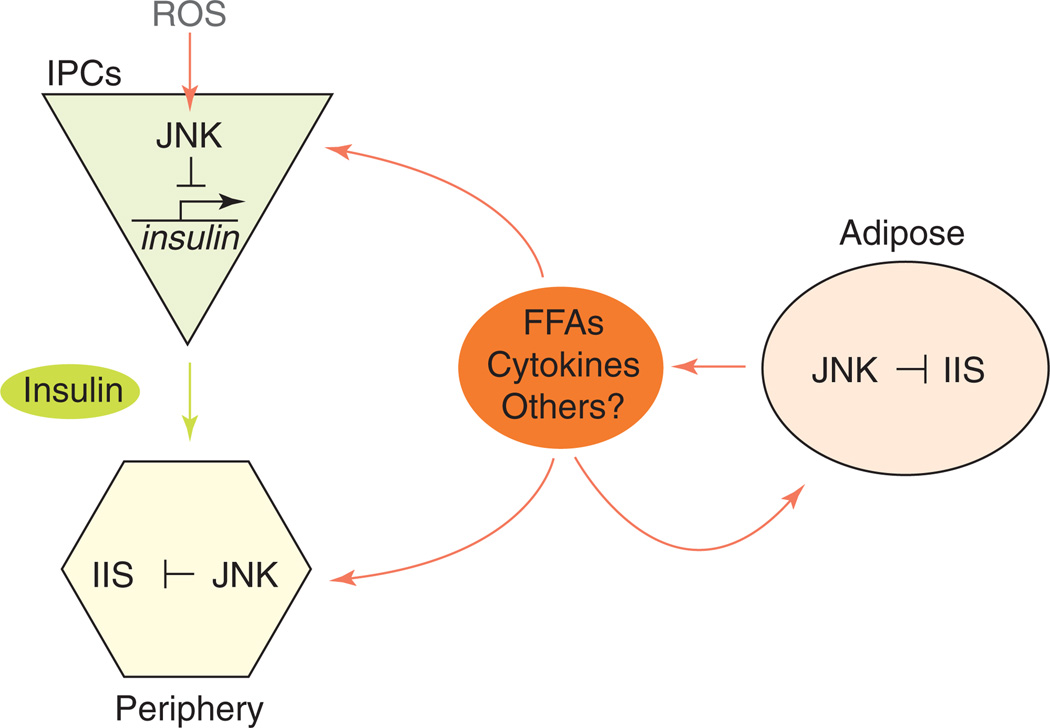
Figure 2.
Endocrine interactions regulating metabolic homeostasis and lifespan. Free fatty acids (FFAs) and inflammatory cytokines are secreted by adipose tissue. These factors activate JNK signaling in insulin-target tissues and promote insulin resistance. Activation of JNK in insulin-producing cells (IPCs) results in transcriptional repression of insulin or insulin-like peptides, systemically depressing IIS activity.
Table 1.
Summary of interactions between JNK and IIS characterized in flies, worms and vertebrates
| Regulation of lifespan by JNK and interactions between JNK and IIS | Refs |
|---|---|
| C. elegans | |
| JNK phosphorylates DAF-16, promoting its nuclear localization | [54] |
| Mutants for the JNK phosphatase VHP-1 are protected against heavy metal toxicity | [62] |
| D. melanogaster | |
| JNK extends lifespan and increases stress tolerance | [60] |
| JNK signaling promotes Foxo nuclear localization in adipose tissue and represses insulin-like peptide transcription in insulin-producing cells | [55] |
| Vertebrates | |
| JNK phosphorylates the insulin-receptor substrate | [67,68,78] |
| JNK phosphorylates and activates FoxO4 | [53] |
| JNK phosphorylates 14–3-3 proteins, releasing Foxo | [69–71] |
| JNK represses insulin transcription in β cells | [72,93] |
Although the overall antagonism between JNK and IIS seems to be evolutionarily ancient, the exact mechanism by which this antagonism is achieved seems to have diverged in vertebrates. Detailed analysis of signal crosstalk between JNK signaling and IIS in both vertebrates and invertebrates thus seems crucial to establish the relative importance of the various mechanisms for the JNK–IIS interaction in humans and, ultimately, to identify promising targets for rational therapies of metabolic diseases.
Endocrine control of IIS activity by JNK signaling
An additional mechanism by which JNK signaling antagonizes IIS in vertebrates and flies is through its ability to repress insulin and insulin-like peptide transcription in insulin-producing cells (β cells in mammals and neurosecretory cells in flies) [55,72]. JNK signaling thus systemically represses IIS activity, enabling coordinated metabolic adaptation of insulin-target tissues to environmental changes and extending lifespan of flies [55]. This mechanism is part of a complex system of endocrine interactions between insulin-producing tissues, fat-storing tissues and peripheral insulin-target tissues that regulate lifespan [22].
Endocrine control of metabolic adaptation and lifespan was initially characterized in worms: ablation of ASI neurons (two chemosensory neurons located in the head of C. elegans) promotes dauer formation [73]. These neurons express the insulin-like peptide DAF-28, which is repressed in response to starvation and dauer pheromone [74]. Of note, a role for JNK signaling in this repression has not yet been tested. These effects of DAF-28 repression on dauer formation are mediated in part through the IIS pathway and DAF-16 [74]. Systemic control of IIS-pathway activity by varying levels of DAF-28 expression thus influences DAF-16 function in peripheral tissues, regulating metabolic homeostasis and lifespan. Confirming a role for peripheral DAF-16 activity in regulating lifespan, it was found that DAF-16 expression in the intestine is sufficient to extend lifespan [75]. The intestine is also the major fat-storing tissue in the worm, indicating that DAF-16 function in adipose tissue is crucial for lifespan regulation [75].
Similar effects have been described in Drosophila, in which ablation of neurosecretory insulin-producing cells (IPCs) is sufficient to extend lifespan [76,77]. As in worms, overexpression of FoxO in the peripheral fat body also leads to lifespan extension accompanied by apparent feedback repression of insulin-like peptide gene expression in IPCs [32,33].
Highlighting the similarity between endocrine mechanisms ensuring systemic metabolic adaptation in invertebrates and in vertebrates, it was found that deletion of the insulin receptor (and thus activation of FoxO) specifically in adipose tissue of mice extends lifespan [38]. The consequences of JNK-mediated repression of insulin production in pancreatic β cells for lifespan of mammals have not yet been addressed.
Taken together, these findings support the notion that complex, evolutionarily conserved, endocrine interactions between insulin-producing and insulin-target tissues influence metabolic homeostasis and lifespan. Stress signaling through JNK seems to be an integral part of this regulatory network. The exact nature of the endocrine factors mediating these interactions (in particular the feedback regulation of insulin-like peptide transcription by adipose tissue) remains to be resolved, but studies focusing on the etiology of diabetes and the metabolic syndrome point to a variety of potential cytokines and lipokines as potential mediators (see below) (Figure 2).
The other side of the coin: JNK--IIS interactions in pathology
The studies reviewed here illustrate an emerging model for the importance of metabolic adaptation to environmental challenges in the control of lifespan of evolutionarily diverse organisms. The antagonism between IIS and JNK signaling has an important role in this metabolic adaptation, positively influencing lifespan. Interestingly, however, inhibition of IIS activity by JNK has also been identified as a major factor in the etiology of type II diabetes, implying that a tight balance between JNK-mediated adaptive responses and deleterious effects has to be maintained to achieve optimal metabolic homeostasis and lifespan (Figure 3). JNK activity was found to be abnormally elevated in peripheral tissues (liver, muscle and adipose tissue) in genetic and dietary-induced models of obesity and insulin resistance [78]. Accordingly, the absence of JNK1 results in improved insulin sensitivity and enhanced insulin-receptor-signaling capacity in mouse models of obesity [78]. Further genetic evidence indicates that increased JNK activity, caused by loss-of-function mutations in the JNK scaffold protein JIP1, causes type 2 diabetes in humans [79]. Liver-specific knockdown of JNK1 indeed lowers circulating glucose and insulin levels [80], and deletion of JNK1 from the hematopoietic lineage protects mice from diet-induced inflammation and insulin resistance [81]. Similarly, MAP kinase phosphatase 4 protects against the development of insulin resistance by dephosphorylating and inactivating JNK [82], further implicating JNK overactivation in the development of diabetes.
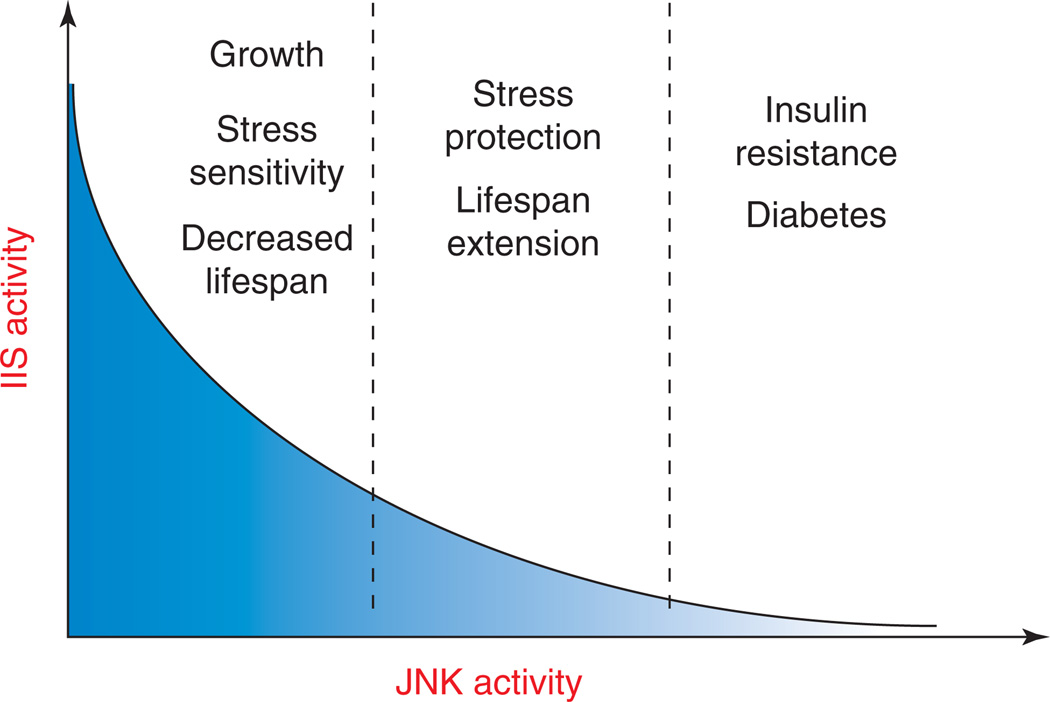
Figure 3.
Relationship between JNK and IIS activity, lifespan and metabolic homeostasis. Moderately increased JNK signaling increases lifespan by reducing IIS activity. In conditions of chronic or excessive JNK activation, however, strong repression of IIS activity results in systemic insulin resistance, promoting diabetes and metabolic syndrome.
Excessive activation of JNK in obese conditions seems to be due primarily to increases in adipose-derived inflammatory signals such as free fatty acids (FFAs) and TNF-α and the concurrent activation of Toll-like receptors [83]. Activation of JNK by these receptors then leads to inhibitory phosphorylation of IRS-1, resulting in attenuated insulin signaling in cells exposed to TNF-α or elevated FFAs and in the development of systemic insulin resistance in obese animals and humans [67,78,84–87].
Another trigger that causes excessive JNK activation and insulin resistance is endoplasmic reticulum (ER) stress [85,88,89]. The presence of dysfunctional proteins in the ER (‘ER stress’) initiates the unfolded-protein response, a mechanism designed to restore homeostasis in the ER [90]. ER stress is induced by changes in nutrient and energy availability, as well as by exposure to pathogens [90,91]. In obese individuals, ER stress causes cellular insulin resistance by activating JNK, which suppresses insulin signaling through the phosphorylation of IRS-1 [92].
In pancreatic β cells, oxidative stress that is increased under diabetic conditions also results in excessive JNK activation [72,93]. Such excessive JNK activation leads to suppression of insulin-gene expression, promoting systemic insulin deficiency and β-cell dysfunction [72,93]. The JNK pathway, therefore, emerges as a crucial mediator of insulin resistance and the metabolic syndrome in obese individuals (Figure 3).
It seems clear that optimizing the interaction between insulin signaling and JNK is a potential therapeutic strategy for age-related metabolic diseases. Before successfully designing JNK-targeted rational therapies for such diseases, however, numerous questions will have to be answered.
Why have various points of interaction between JNK signaling and IIS evolved, and what is the significance of local versus systemic regulation?
As described above, JNK represses IIS activity by interacting with various IIS signal transducers (Figure 1). It remains to be established, however, what the significance of these various points of interaction are and how this signaling network is regulated in different contexts and tissues. Answering this question is important because it will enable a better understanding of pleiotropic consequences of the interaction between stress-signaling pathways and growth-factor-signaling pathways (such as IIS). Considerable progress towards addressing this question has been achieved using systems biology approaches in mammalian tissue culture [94,95], but in vivo studies are needed to further address the complexities of these interactions [63].
How is the interaction between JNK signaling and IIS integrated into other signaling mechanisms that control cellular responses to stress?
JNK signaling is only one of several stress response pathways in metazoans. It remains to be established how other stress-response mechanisms – such as signaling through the related kinase p38, through DNA damage-response pathways or through the NF-E2-related transcription factor (Nrf-2) – impact metabolism and whether and/or how they affect IIS activity. Interestingly, recent studies have found that the p38 SAPK activates the C. elegans homolog of the Nrf-2 transcription factor, SKN-1, which affects cellular responses to IIS repression by co-regulating subsets of target genes with DAF16/FoxO [96–98]. In this system, SKN-1 is also phosphorylated and turned off by Akt, enabling the inactivation of this response mechanism under high levels of IIS activity [98]. At the same time, SKN-1 has been shown to systemically influence metabolism, lifespan and dauer formation by acting in ASI neurons [99]. The mechanisms mediating this endocrine interaction, however, are not well understood.
How do intrinsic processes of degeneration, cellular senescence and genotoxic stress influence IIS activity and lifespan?
Work on premature aging (progeria) models in mice has established that defects in DNA repair mechanisms result in depression of the growth-hormone–IGF axis, indicating an adaptive response that promotes cell survival [100–103]. The signaling mechanisms involved in this interaction, however, are not understood. It is intriguing to speculate that DNA-damage-induced JNK activity might play a part in this interaction.
How can beneficial versus detrimental effects of IIS repression by JNK be balanced for optimal lifespan?
Reduced IIS activity has beneficial effects on lifespan and stress tolerance in invertebrates (although they are accompanied by metabolic imbalance) but can adversely affect overall health in mammals. These differences indicate a fundamental divergence in the metabolic regulation of invertebrates and vertebrates, which is further supported by the paradoxic increase in whole-body lipids in IIS mutant worms and flies [27]. Current understanding indicates, however, that these differences are probably because of the context in which and the degree to which IIS activity is impaired. Importantly, complete inhibition of IIS in flies (e.g. in flies carrying null alleles for InR) is lethal, and strong ablation of IIS activity (e.g. in flies carrying non-lethal but strong loss-of-function alleles of InR or in which IPCs were ablated) results in developmental delay, growth defects and sterility, as well as in metabolic phenotypes that are similar to vertebrate diabetes, such as hyperglycemia [27,104,105].
Furthermore, it has been established that the tissue-specific effects of IIS repression play a major part in determining the ultimate overall metabolic phenotypes of IIS mutations, and the increased complexity of vertebrates warrants analyzing in more detail the effects of IIS repression in various tissues on lifespan. Interestingly, reduction of IRS-2 in the brain of mice increases lifespan, while resulting in obesity, hyperinsulinemia and glucose intolerance [48]. Deletion of the insulin receptor in adipose tissue, on the other hand, results in increased lifespan associated with resistance to obesity [38]. At the same time, repression of IIS signal transduction by JNK in adipose tissue and liver of mice contributes to diabetes and increased mortality [89,106]. The effects of IIS repression on lifespan are, thus, tissue specific and can be uncoupled from consequences on metabolic diseases. Addressing the cause of these divergent functions will be important to fully understand the control of metabolic homeostasis and lifespan by JNK–IIS interactions. The development of rational therapies for age-related metabolic diseases will depend on this understanding.
Update
Recently, Sabio et al. generated mice with JNK1 deficiency in adipose tissue and found reduced insulin resistance in the liver of mice reared on a high-fat diet. Furthermore, this work shows that JNK1 in adipose tissue of obese animals can regulate the circulating levels of the cytokine IL-6, which is known to be involved in the development of insulin resistance, specifically in the liver. This study provides crucial evidence that stress signaling, through JNK, in peripheral tissues such as fat can have profound effects on IIS and metabolic homeostasis through a network of endocrine interactions. Sabio, G. et al. (2008) A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science 322, 1539–1543.
Acknowledgements
We would like to thank Dae-Sung Hwangbo and Dirk Bohmann for comments on the manuscript. This work was supported by the National Institute on Aging (RO1 AG028127).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Denlinger DL. Regulation of diapause. Annu. Rev. Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- 2.Tatar M, Yin C. Slow aging during insect reproductive diapause: why butterflies, grasshoppers and flies are like worms. Exp. Gerontol. 2001;36:723–738. doi: 10.1016/s0531-5565(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 3.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 6.Kimura KD, et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 7.Gems D, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin K, et al. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 9.Ogg S, et al. The Fork head transcription factor DAF-16-transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 10.Berdichevsky A, et al. C. elegans SIR-2.1 interacts with 14–3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 12.Lee RY, et al. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 13.Morris JZ, et al. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 14.Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, et al. The 14–3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev. Biol. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, et al. C. elegans 14–3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Hsu AL, et al. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 18.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 19.McElwee J, et al. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 20.Oh SW, et al. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 21.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 22.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat. Rev. Mol. Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 23.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 2008;70:191–24. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 24.Saunders DS, et al. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci U. S. A. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc. Natl. Acad. Sci U. S. A. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams KD, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garofalo RS. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metabol. 2002;13:156–162. doi: 10.1016/s1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- 28.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 2007;32:180–188. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen MD, et al. 14–3-3ε antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell. 2008;7:688–699. doi: 10.1111/j.1474-9726.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 31.Clancy DJ, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 32.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 33.Hwangbo DS, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 34.Bohni R, et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 35.Junger MA, et al. The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner H, et al. Similarities and differences between insulin and IGF-I: structures, receptors, and signalling pathways. Arch. Physiol. Biochem. 2008;114:17–22. doi: 10.1080/13813450801900694. [DOI] [PubMed] [Google Scholar]
- 37.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 38.Bluher M, et al. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 39.Holzenberger M, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 40.Buteau J, et al. Metabolic diapause in pancreatic beta-cells expressing a gain-of-function mutant of the forkhead protein Foxo1. J. Biol. Chem. 2007;282:287–293. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]
- 41.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 42.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 43.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 44.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Kurosu H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taguchi A, et al. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 49.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 51.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 52.Puig O, et al. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Essers MA, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh SW, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang MC, et al. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 56.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 57.Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- 58.Manning AM, Davis RJ. Targeting JNK for therapeutic benefit: from junk to gold? Nat. Rev. Drug Discov. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- 59.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 2002;12:14–21. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 60.Wang MC, et al. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 61.Libert S, et al. Realized immune response is enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol. Immunol. 2008;45:810–817. doi: 10.1016/j.molimm.2007.06.353. [DOI] [PubMed] [Google Scholar]
- 62.Mizuno T, et al. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004;23:2226–2234. doi: 10.1038/sj.emboj.7600226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo X, et al. Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. EMBO J. 2007;26:380–390. doi: 10.1038/sj.emboj.7601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumoto M, Accili D. All roads lead to FoxO. Cell Metab. 2005;1:215–216. doi: 10.1016/j.cmet.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Wolf M, et al. The MAP kinase JNK-1 of Caenorhabditis elegans : location, activation, and influences over temperature-dependent insulin-like signaling, stress responses, and fitness. J. Cell. Physiol. 2008;214:721–729. doi: 10.1002/jcp.21269. [DOI] [PubMed] [Google Scholar]
- 66.Jaeschke A, Davis RJ. Metabolic stress signaling mediated by mixed-lineage kinases. Mol. Cell. 2007;27:498–508. doi: 10.1016/j.molcel.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aguirre V, et al. The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J. Biol. Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 68.Mamay CL, et al. An inhibitory function for JNK in the regulation of IGF-I signaling in breast cancer. Oncogene. 2003;22:602–614. doi: 10.1038/sj.onc.1206186. [DOI] [PubMed] [Google Scholar]
- 69.Sunayama J, et al. JNK antagonizes Akt-mediated survival signals by phosphorylating 14–3-3. J. Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuruta F, et al. JNK promotes Bax translocation to mitochondria through phosphorylation of 14–3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida K, et al. JNK phosphorylation of 14–3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- 72.Kaneto H, et al. Involvement of c-Jun N-terminal kinase in oxidative stress-mediated suppression of insulin gene expression. J. Biol. Chem. 2002;277:30010–30018. doi: 10.1074/jbc.M202066200. [DOI] [PubMed] [Google Scholar]
- 73.Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 74.Li W, et al. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Libina N, et al. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 76.Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wessells RJ, et al. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 78.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 79.Waeber G, et al. The gene MAPK8IP1, encoding isletbrain-1, is a candidate for type diabetes. Nat. Genet. 2000;24:291–295. doi: 10.1038/73523. [DOI] [PubMed] [Google Scholar]
- 80.Yang R, et al. Liver-specific knockdown of JNK1 up-regulates proliferator-activated receptor gamma coactivator-beta and increases plasma triglyceride despite reduced glucose and insulin levels in diet-induced obese mice. J. Biol. Chem. 2007;282:22765–22774. doi: 10.1074/jbc.M700790200. [DOI] [PubMed] [Google Scholar]
- 81.Solinas G, et al. JNK1 in hematopoietically derived cells contributes to diet-induced inflammation and insulin resistance without affecting obesity. Cell Metab. 2007;6:386–397. doi: 10.1016/j.cmet.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 82.Emanuelli B, et al. Overexpression of the dual-specificity phosphatase MKP-4/DUSP-9 protects against stress-induced insulin resistance. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3545–3550. doi: 10.1073/pnas.0712275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsukumo DM, et al. Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes. 2007;56:1986–1998. doi: 10.2337/db06-1595. [DOI] [PubMed] [Google Scholar]
- 84.Hotamisligil GS. Mechanisms of TNF-α-induced insulin resistance. Exp. Clin. Endocrinol. Diabetes. 1999;107:119–125. doi: 10.1055/s-0029-1212086. [DOI] [PubMed] [Google Scholar]
- 85.Hotamisligil GS. Role of endoplasmic reticulum stress and c-Jun NH2-terminal kinase pathways in inflammation and origin of obesity and diabetes. Diabetes. 2005;54 Suppl. 2:S73–S78. doi: 10.2337/diabetes.54.suppl_2.s73. [DOI] [PubMed] [Google Scholar]
- 86.Hotamisligil GS, et al. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-α- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 87.Yin MJ, et al. The anti-inflammatory agents aspirin and salicylate inhibit the activity of IκB kinase-β. Nature. 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 88.Bouzakri K, Zierath JR. MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J. Biol. Chem. 2007;282:7783–7789. doi: 10.1074/jbc.M608602200. [DOI] [PubMed] [Google Scholar]
- 89.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 90.Kaufman RJ, et al. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 2002;3:411–421. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- 91.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51 Suppl. 3:S455–S461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 92.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type-2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 93.Kajimoto Y, Kaneto H. Role of oxidative stress in pancreatic β-cell dysfunction. Ann. N. Y. Acad. Sci. 2004;1011:168–176. doi: 10.1007/978-3-662-41088-2_17. [DOI] [PubMed] [Google Scholar]
- 94.Janes KA, et al. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 95.Janes KA, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 96.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue H, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 100.Hinkal G, Donehower LA. How does suppression of IGF-1 signaling by DNA damage affect aging and longevity? Mech. Ageing Dev. 2008;129:243–253. doi: 10.1016/j.mad.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoeijmakers JH. Genome maintenance mechanisms are critical for preventing cancer as well as other aging-associated diseases. Mech. Ageing Dev. 2007;128:460–462. doi: 10.1016/j.mad.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 102.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 103.van der Pluijm I, et al. Impaired genome maintenance suppresses the growth hormone–insulin-like growth factor-1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rulifson EJ, et al. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 105.Chen C, et al. The Drosophila insulin receptor required for normal growth. Endocrinology. 1996;137:846–856. doi: 10.1210/endo.137.3.8603594. [DOI] [PubMed] [Google Scholar]
- 106.Hotamisligil GS. Inflammatory pathways and insulin action. Int. J. Obes. Relat. Metab. Disord. 2003;27 Suppl. 3:S53–S55. doi: 10.1038/sj.ijo.0802502. [DOI] [PubMed] [Google Scholar]