Abstract
S100A4, a member of the Ca2+-activated S100 protein family, regulates the motility and invasiveness of cancer cells. Moreover, high S100A4 expression levels correlate with poor patient survival in several cancers. Although biochemical, biophysical, and structural data indicate that S100A4 is a noncovalent dimer, it is unknown if two functional S100A4 monomers are required for the productive recognition of protein targets and the promotion of cell invasion. To address this question, we created covalently linked S100A4 dimers using a glycine rich flexible linker. The single-chain S100A4 (sc-S100A4) proteins exhibited wild-type affinities for calcium and nonmuscle myosin-IIA, retained the ability to regulate nonmuscle myosin-IIA assembly, and promoted tumor cell invasion when expressed in S100A4-deficient colon carcinoma cells. Mutation of the two calcium-binding EF-hands in one monomer, while leaving the other monomer intact, caused a 30–60-fold reduction in binding affinity for nonmuscle myosin-IIA concomitant with a weakened ability to regulate the monomer–polymer equilibrium of nonmuscle myosin-IIA. Moreover, sc-S100A4 proteins with one monomer deficient in calcium responsiveness did not support S100A4-mediated colon carcinoma cell invasion. Cross-linking and titration data indicate that the S100A4 dimer binds a single myosin-IIA target peptide. These data are consistent with a model in which a single peptide forms interactions in the vicinity of the canonical target binding cleft of each monomer in such a manner that both target binding sites are required for the efficient interaction with myosin-IIA.
S100A4 is a member of the S100 Ca2+-binding family of proteins. The S100 designation is derived from the observation that these proteins are soluble in 100% saturated ammonium sulfate at neutral pH.1 At present, there are 21 identified human S100 family members,2,3 which make up a subclass of the larger EF-hand-containing family of proteins represented by calmodulin and troponin C.4,5 The S100 proteins are 25–65% identical with one another in terms of amino acid sequence and are expressed solely in vertebrates in a cell and tissue specific manner.2,6,7 This family of proteins is involved in diverse cellular processes and contributes to various disease states such as neurodegeneration, inflammation, and cancer.2,8,9
The S100 proteins are primarily α-helical, and most family members form noncovalent antiparallel dimers.10–15 The dimerization interface is composed of an X-type four-helix bundle that is comprised of helices 1 and 4 from each monomer.10–15 Each S100 monomer contains two helix–loop–helix (EF-hand) Ca2+-binding motifs connected by a hinge region: a 14-residue N-terminal pseudo-EF-hand and a 12-residue C-terminal canonical EF-hand.10–15 The canonical EF-hand (EF2) uses several side chain oxygen atoms, an oxygen from a water molecule, and two oxygen atoms from a glutamate side chain to coordinate the Ca2+ ion, whereas the N-terminal EF-hand (EF1) coordinates Ca2+ via several backbone carbonyl oxygens, an oxygen from water, and two oxygen atoms from a glutamate side chain.16 Consistent with these structural data, the reported Ca2+ equilibrium dissociation constants (KD) for S100A4 indicate that EF1 has a weak affinity for calcium (KD > 500 μM), whereas EF2 has a relatively high affinity for Ca2+ (KD = 2.6 ± 1.0 μM).17 Most S100 proteins, including S100A4, undergo a significant conformational rearrangement following binding of Ca2+ to the C-terminal EF-hand. This conformational rearrangement increases the interhelical angle between helices 3 and 4 and exposes a hydrophobic target binding cleft comprised of residues from helix 3, helix 4, and the hinge region.
The biological function of S100A4 has been characterized most extensively in the context of cancer. S100A4 overexpression in benign rat mammary tumor cells confers a metastatic phenotype with no apparent effect on tumor proliferation.18 In addition, overexpression of S100A4 in the mammary epithelium of MMTV-neu mice, which normally have a low incidence of metastasis, increased the incidence of metastasis by 72%.19 Conversely, the loss of S100A4 expression in highly metastatic MMTV-PyMT mice resulted in a 60% decrease in the number of metastatic lesions in the lungs.20 Consistent with these animal studies, S100A4 expression in breast cancer patients correlates with patient death.21
Intracellular S100A4 targets include the p53 homologues p63 and p73, nonmuscle tropomyosin, liprin β1, and nonmuscle myosin-IIA.22–25 For the S100A4–myosin-IIA interaction, S100A4 binding disrupts the monomer–polymer equilibrium of myosin-IIA by promoting the disassembly of preexisting filaments or by inhibiting the assembly of monomers into filaments.26–28 Moreover, S100A4 modulates the polarization and motility of mammary tumor cells through a direct interaction with myosin-IIA.29 Given the importance of myosin-II in modulating cell migration, the regulation of myosin-IIA assembly by S100A4 likely contributes to the ability of S100A4 to promote tumor invasion and metastasis.30
Although there has been some examination of the molecular determinants involved in S100A4 dimerization,31–33 it is not known whether both monomers of the S100A4 homodimer are required for the recognition of its protein targets and the promotion of cell invasion. Using covalently linked S100A4 monomers or single-chain S100A4 proteins (sc-S100A4), we demonstrate that two functional S100A4 monomers are necessary for the efficient interaction of S100A4 with myosin-IIA and for the promotion of tumor cell invasion.
EXPERIMENTAL PROCEDURES
Creation of sc-S100A4 Constructs
To create the sc-S100A4 constructs, the S100A4 cDNA was amplified using a primer to the 3′ end that contained the DNA sequence for the linker and a BamHI restriction site (monomer 1). The S100A4 polymerase chain reaction (PCR) product containing the linker sequence was cloned into pET23a using the NdeI and BamHI sites, and monomer 2 was cloned into the BamHI and HindIII sites. The resulting sc-S100A4 constructs were subcloned into pET26b using the NdeI and HindIII sites. The final linker sequence between the two S100A4 monomers was (GGGGS)nGS.
The EF-hand substitutions were created in the wild-type S100A4–ET23a construct using the QuikChange site-directed mutagenesis kit (Stratagene). Reverse primers contained either the E33A or E74A substitution. Single-EF-hand substitutions were introduced first, and the E33A/E74A double-EF-hand substitutions were introduced by subcloning the E33A mutant into the E74A mutant S100A4 construct using the NdeI and XmaI sites. To incorporate the EF-hand substitutions into the sc-S100A4 constructs, we amplified E33A/E74A S100A4 with primer sets containing the appropriate linker addition and subcloned it as described above. The sc-S100A4-GFP constructs were created by ligation-independent cloning.34,35 The pEGFP-N3 vector and S100A4 primers were as follows: vector forward primer, 5′TCTGTGCTGTCGTCGGTACCGCGGGCCC3′; vector reverse primer, 5′TGGTGCTGCTTCCGTCCGGTAGCGC-TAGCGGAT3′; S100A4 forward primer, 5′ACCGGACGGAAG-CAGCACCATGGCGTGCCCTCTG3′; and S100A4 reverse primer, 5′ACCGACGACAGCAACAGATTTCTTCCTGGG3′. pEGFP-N3 was amplified using KOD polymerase (Novagen), and S100A4 was amplified using a standard PCR protocol. The resulting PCR products were treated with T4 polymerase in separate reaction mixtures containing either 2.5 mM dATP (pEGFP-N3) or 2.5 mM dTTP (S100A4). For ligation-independent cloning, the reaction mixture contained equimolar amounts of T4-digested S100A4 and pEGFP-N3 and the reaction was performed at 22 °C for 25 min. The reaction was quenched by the addition of 10 mM EDTA (pH 8.0) and incubation at 22 °C for 5 min.
Cloning of the S100A4–GFP Fusion Protein
GFP was fused to the C-terminus of S100A4 using primer pairs that removed the S100A4 stop codon and added a 5′ HindIII site and a 3′ SacII site. The resulting PCR product was subcloned into the pEGFP-N3 (Clontech) vector using these sites.
Peptides
Myosin-IIA peptides [FITC-Ahx-DAMNR-EVSSLKNKLRR-CONH2 (FITC-MIIA1908–1923) and FITC-Ahx-TETADAMNREVSSLKNKLRRGDLP-CONH2 (FITC-MIIA1904–1927)] were synthesized by Biosynthesis Inc. (Lewisville, TX). The molecular weight and purity (>95%) of all peptides were verified using mass spectrometry and high-performance liquid chromatography (HPLC), respectively. The concentration of all peptide solutions was determined using the FITC moiety.
Protein Purification
MIIA1851–1960 and MIIA1338–1960 were expressed and purified as described previously.16,27 The wild-type sc-S100A4 constructs and S100A4–GFP fusion protein were expressed and purified as described for the native S100A4 dimer.14 The masses of the sc-S100A4 proteins and S100A4–GFP fusion protein were confirmed by mass spectrometry.
For purification of the EF-hand substitutions, BL21(DE3)* cells were transformed and grown in autoinduction TB medium for 20 h at 37 °C.36 The cells were pelleted and resuspended in 10 mL of lysis buffer [50 mM Tris (pH 7.5), 10% glycerol, 300 mM KCl, 2 mM DTT, 1 mM EDTA, 1 mM PMSF, and chymostatin, leupeptin, and pepstatin (5 μg/mL each)] per gram of cell pellet. Resuspended cell pellets were sonicated and then centrifuged at 16000g for 30 min. Ammonium sulfate was added to the supernatant to 30% saturation, and the sample was centrifuged at 16000g for 10 min. CaCl2 (2 mM) was added to the supernatant, and the sample was applied to a Phenyl-Sepharose (GE Healthcare) column equilibrated with buffer 1 [20 mM Tris (pH 7.5), 2 mM CaCl2, 300 mM KCl, 1 mM DTT, 1 mM EDTA, 16.4 g/100 mL ammonium sulfate, and 0.02% NaN3]. The column was washed with 3 column volumes of buffer 1 and washed with 3 column volumes of buffer 2 (buffer 1 without ammonium sulfate). sc-S100A4 EF-hand mutant proteins were eluted using buffer 3 [20 mM Tris (pH 7.5), 5 mM EGTA, 1 mM DTT, 1 mM EDTA, and 0.02% NaN3]. Collected protein fractions were dialyzed against 20 mM PIPES (pH 6.8), 20 mM KCl, 0.5 mM DTT, and 0.02% NaN3, centrifuged at 16000g for 15 min, and applied to a Mono S column (GE Healthcare) equilibrated with dialysis buffer. The column was washed with 10 column volumes of dialysis buffer, and sc-S100A4 EF-hand mutant proteins were eluted using a 0 to 0.5 M KCl gradient. Pooled protein fractions were dialyzed against 20 mM Tris (pH 7.5), 20 mM KCl, 1.5 mM DTT, and 0.02% NaN3. The dialyzed protein was centrifuged for 15 min at 16000g and concentrated using a Vivaspin 3 kDa cutoff concentrator (Sartorius). Concentrated sc-S100A4 EF-hand mutant proteins were applied to a Superdex 75 gel filtration column (GE Healthcare) equilibrated with dialysis buffer. On the basis of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), pure protein fractions were pooled and concentrated as described above. All protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad) and a S100A4 standard at a known concentration.
Analytical Size Exclusion Chromatography
Wild-type S100A4 (0.7–15 μM), 15 μM sc-S100A4 proteins, or 1 mg/mL protein standards [BSA (67 kDa), ovalbumin (43 kDa), chymotrypsin (27 kDa), and ribonuclease A (13 kDa)] were injected onto a Superdex 75HR 10/30 column (GE Health-care) equilibrated with 20 mM Tris (pH 7.5), 150 mM KCl, 2 mM CaCl2, 1 mM DTT, and 0.02% NaN3. For studies examining the elution of apo-S100A4, the calcium was replaced with 2 mM EGTA. Absorbance at 280 nm was used to monitor protein elution.
Chemical Denaturation
Guanidine hydrochloride (GdnHCl) stocks (7 M) were made using EGTA buffer [20 mM Tris (pH 7.5), 20 mM KCl, 2 mM EGTA, and 0.02% NaN3] or calcium buffer in which the EGTA was replaced with 2 mM CaCl2. S100A4 proteins were dialyzed into either EGTA or calcium buffer, and 5 μM S100A4 dimer was used per 200 μL reaction mixture with fresh DTT (1 mM) added the day of the experiment. Reaction mixtures contained GdnHCl concentrations ranging from 0 to 6.5 M. Individual reaction mixtures were incubated for 22 h at 22 °C. The intrinsic fluorescence of the two S100A4 tyrosine residues was monitored using a Fluoromax-3 spectrofluorometer (Jobin Yvon Inc.). The data were fit to a sigmoidal dose–response curve using GraphPad Prism version 5.0a (GraphPad Software Inc.), and the fraction unfolded and D1/2 were calculated using formulas described previously.37
Circular Dichroism
S100A4 proteins were dialyzed into 20 mM Tris (pH 7.5), 20 mM KC1, 1 mM DTT, and 0.02% NaN3. Either 0.3 mM CaCl2 or 2 mM EGTA was added to each reaction mixture containing 6.8 μM S100A4 dimer and the mixture incubated for 1 h prior to the acquisition of spectra. Samples were placed into a 0.1 cm cuvette, and spectra from 190 to 250 nm were obtained at 22 °C using a Jasco (Easton, MD) J-18 spectrometer. Molar ellipticity values were calculated using the formula θmolar = (θ× MW × 100)/(lc), where θis the measured ellipticity in degrees, MW is the molecular weight of the S100A4 dimer, 100 is the conversion of molar concentration to decimoles per cubic centimeter, l is the path length of the cuvette in centimeters, and c is the protein concentration in milligrams per milliliter.38
Calcium Binding
Calcium affinities were determined as described previously.17,39 Briefly, S100A4 and MIIA1851–1960 were dialyzed into calcium free buffer [20 mM Tris (pH 7.5), 150 mM KCl, 1 mM DTT, and 0.02% NaN3] that had been pretreated with Chelex 100 resin. Calcium was titrated into reaction mixtures containing 12.5 μM S100A4 dimer and 25 μM 5,5′Br2-BAPTA alone or in the presence 100 μM MIIA1851–1960 monomer, and the absorbance was monitored at 263 nm. Using Caligator, macroscopic binding affinities were obtained by fitting the resulting titration curves to two Ca2+-binding sites using a stepwise macroscopic binding equation in the presence of chelator.40
Myosin-IIA Binding Assay
Binding affinities of S100A4 for myosin-IIA were determined using a fluorescence anisotropy assay as described previously.17 FITC-MIIA1908–1923, which comprises the minimal S100A4 binding site, was used to monitor the interaction of the wild-type and sc-S100A4 proteins and myosin-IIA. Reaction mixtures (200 μL) contained 100 nM FITC-MIIA1908–1923 and 0–20 μM wild-type or sc-S100A4 dimer, or 0–500 μM EF-hand mutant sc-S100A4 in 20 mM Tris (pH 7.5), 150 mM KCl, 1 mM DTT, 0.02% NaN3, and 0.5 mM CaCl2. Anisotropy was measured with excitation at 494 nm and emission at 516 nm. Dissociation constants were obtained by fitting to a single-site saturation binding equation with a floating Ymin value using GraphPad Prism.
For fluorescence anisotropy titrations, 1 μM FITC-MIIA1904–1927 was titrated with 0–8 μM S100A4 dimer in 20 mM Tris (pH 7.5), 150 mM KCl, 1 mM DTT, 0.02% NaN3, and 0.5 mM CaCl2. Linear regression was used to determine the intersecting points of the titration curve.
Promotion of Disassembly
The ability of wild-type S100A4, sc-S100A4–5, sc-S100A4–10, and sc-S100A4 EF-hand mutant proteins to regulate myosin-IIA disassembly was assessed as described previously.27 Briefly, reaction mixtures containing 1.5 μM MIIA1338–1960 dimer and 0–15 μM S100A4 dimer in 20 mM Tris (pH 7.5), 300 μM CaCl2, 1.5 mM MgCl2, 150 mM NaCl, 1 mM DTT, and 0.02% NaN3 were incubated for 1 h at 22 °C and centrifuged at 80000 rpm for 15 min using an Optima TLX ultracentrifuge (Beckman). A sample of the mixture was obtained by removing a 10 μL aliquot after incubation for 10 min. The 5× sample buffer was added to aliquots of the mix and the supernatant, and they were loaded onto a 12% Tris-Tricine gel. The amount of myosin-IIA in the supernatant was quantified by densitometric analysis of the Coomassie-stained protein bands.
Cross-Linking Experiments
Wild-type S100A4 and MIIA1851–1960 were dialyzed into 20 mM HEPES (pH 7.5), 150 mM KCl, 1 mM DTT, and 0.02% NaN3. Reaction mixtures containing equimolar concentrations of S100A4 monomer and MIIA1851–1960 monomer (either 25 or 50 μM of each) were incubated with either 0.5 mM CaCl2 or 4 mM EGTA for 60 min at 22 °C. Disuccinimidyl tartrate (DST) was added to a final concentration of 1.2 mM, and the reaction mixture was incubated for an additional 60 min. Cross-linking reactions with S100A4 alone or MIIA1851–1960 alone were performed in the presence of 0.5 mM CaCl2. Reactions were quenched by the addition of 5× Laemmli gel loading dye or 50 mM Tris-HCl (pH 7.5). Samples were run on a 12% Tris-Tricine gel and stained with Coomassie brilliant blue. For immunoblot analysis, the 12% Tris-Tricine gel was transferred to a polyvinyl difluoride (PVDF) membrane (Millipore) and reacted with antibodies against S100A4 or the myosin-IIA C-terminus.41,42
Cross-linked S100A4–myosin-IIA complexes were evaluated by LC-ESI MS. Seventeen micrograms of the protein complexes was separated on a 1.0 mm × 50 mm C3 column (MicroTech Scientific) attached to an LC Packings Ultimate Plus HPLC system. The column was equilibrated in buffer A (2% acetonitrile and 0.1% formic acid). After a 5 min desalt, protein complexes were separated using a 15 min gradient from 15 to 60% buffer B (90% acetonitrile and 0.1% formic acid) with a flow rate of 75 μL/min. The effluent was directly delivered into an LTQ Linear Ion Trap (Thermo) for mass analysis, which has an error of ±5 Da for samples in this mass range.
Cell Culture
HCT116 parental and S100A4μ/μ cells, which will be described elsewhere, were maintained in McCoys 5A medium (Cellgro) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. BAC1.2F5 macrophages were maintained in αMEM medium supplemented with L-asparagine, L-glutamine, 10% FBS, and 3000 units/mL human recombinant CSF-1.
Transfection
HCT116 cells were plated in a 10 cm dish 24 h prior to transfection at a cell density that yielded approximately 40% cell confluence the next day. Cells were transfected with S100A4 plasmid DNA using the HeLaMON-STER reagent (Mirus). Maximal expression was observed 48 h post-transfection.
Immunoblots
Whole cell lysates were prepared from HCT116 cell lines at a cell density of 70–80% confluence. Cells were lysed in RIPA buffer [50 mM Tris, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, and leupeptin, chymostatin, and pepstatin (5 μg/mL each)], and total protein was quantified using the Dc Bio-Rad protein assay. Whole cell lysates (20 μg) were separated on a 12% Tris-Tricine gel, transferred to a PVDF membrane, and immunoblotted for β-actin and S100A4. Immunoreactive proteins were imaged using the West Pico chemiluminescence substrate (Pierce).
3D Invasion Assays
Invasion assays were conducted as described previously.43 Briefly, 80000 HCT116 cells labeled with Cell Tracker Green CMFDA and 200000 BAC1.2F5 cells labeled with CellTracker Red CMPTX were plated on 35 mm glass-bottom dishes in macrophage medium. After 18–24 h, the cells were overlaid with a ~1100 μm thick layer of 5.8 mg/mL collagen I (BD Biosciences). After 24 h, the cells were fixed with 4% formaldehyde for 30 min and analyzed by confocal microscopy on a Bio-Rad Radiance 2000 Laser Scanning Confocal Microscope. Optical z-sections were taken every 5 μm, beginning at the base of the dish to 70 μm into the collagen gel. The percent cell invasion was calculated by dividing the fluorescence from z-sections 20–70 μm by the sum of the fluorescence from all sections. Experiments were performed in duplicate, and three different fields from each dish were imaged. For S100A4–GFP or sc-S100A4–GFP protein rescue experiments, transfected HCT116 cells were used 48 h post-transfection in the 3D invasion assay. The GFP fluorescence was used to monitor the invasion of the collagen gel by tumor cells.
RESULTS
Development of the Single-Chain S100A4 Proteins
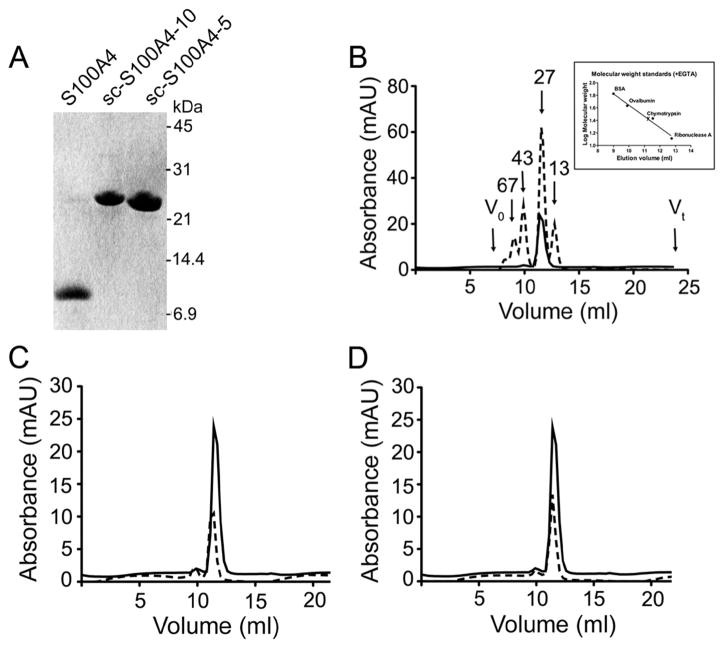
With the exception of S100G (also known as calbindin D9k), the S100 proteins form noncovalent dimers.44 Because each monomer is comprised of two EF-hands and a target binding site, the presumed functional unit of these proteins consists of four EF-hands and two target binding sites.2,10,14–16 To investigate whether both monomers in the S100A4 dimer are necessary for biochemical and biological function, we created an obligate S100A4 dimer. The two S100A4 chains were connected with a flexible linker comprised of four glycines and one serine, thus allowing S100A4 to fold properly even though it is encoded by a single polypeptide chain. Glycine residues were used to promote a flexible linker between the two S100A4 chains, and a serine residue was added to aid in the solubility and stability of the resulting single-chain S100A4 protein (sc-S100A4) (Figure 1).45,46 The linker sequence (GGGGS) was repeated one, two, and three times, and the proteins were designated sc-S100A4–5, sc-S100A4–10, and sc-S100A4–15, respectively. Both sc-S100A4–5 and sc-S100A4–10 expressed well in bacteria, whereas expression of sc-S100A4–15 was not detected in whole cell lysates (data not shown). On SDS–PAGE, the purified native S100A4 (wt-S100A4) migrated at approximately 11 kDa, consistent with a noncovalent dimer. Consistent with the mass spectrometry data, purified sc-S100A4–5 and sc-S100A4–10 migrated above 21 kDa as expected for two native S100A4 chains covalently attached via a linker sequence (Figure 2A).
Figure 1.
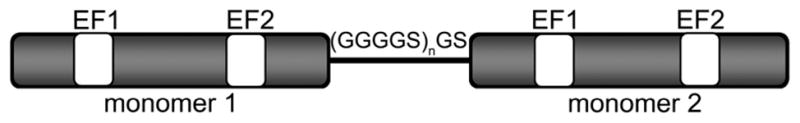
Cartoon of sc-S100A4 proteins. The C-terminus of monomer 1 was fused to the N-terminus of monomer 2 using the linker sequence (GGGGS)n, where n = 1, 2, or 3.
Figure 2.
Analytical gel filtration of sc-S100A4 proteins. (A) SDS–PAGE of purified wt-S100A4, sc-S100A4–5, and sc-S100A4–10 on a 12% Tris-Tricine gel. Molecular mass markers are indicated on the left. (B) Elution profile of wt-S100A4 (—) and protein standards (---) on a Superdex 75HR column in the presence of EGTA. The inset is a plot of protein standard molecular masses vs elution volume. (C and D) Elution profiles of sc-S100A4–5 and sc-S100A4–10 in the presence of EGTA: wt-S100A4 (—) and sc-S100A4–5 and sc-S100A4–10 (---).
Size-Exclusion Chromatography of the sc-S100A4 Proteins
To confirm that the proteins were properly folded in solution, they were examined by analytical size-exclusion chromatography. In the presence of EGTA or Ca2+, the elution profile of wt-S100A4 showed one major peak, which was coincident with the 27 kDa marker (Figure 2B). The apparent molecular mass of the native S100A4 dimer agreed well with the calculated mass (Table 1). Notably, at loading concentrations of 0.7 μM, we observed a single peak for wt-S100A4, demonstrating that the native S100A4 forms a dimer at submicromolar concentrations. Both sc-S100A4–5 and sc-S100A4–10 had elution profiles comparable to that of wt-S100A4, indicating that in solution both sc-S100A4 constructs adopted an overall organization similar to that of the noncovalent dimer of the wild-type protein (Figure 2C,D). In the presence of calcium, all three proteins eluted as more compact proteins (Table 1), consistent with other Ca2+-bound S100 proteins.47
Table 1.
Apparent Molecular Masses and Stokes Radii of S100A4 Proteins
| protein | condition | elution volume (mL)a | Stokes radius (Å)a | apparent molecular mass (×103 Da)a |
|---|---|---|---|---|
| wt-S100A4 | EGTA | 11.4 ± 0.1 | 22.4 ± 0.4 | 24.2 ± 1.2 |
| wt-S100A4 | Ca2+ | 12.0 ± 0.2 | 19.2 ± 1.1 | 19.4 ± 1.9 |
| sc-S100A4-5 | EGTA | 11.3 ± 0.0 | 22.4 ± 0.0 | 24.0 ± 0.0 |
| sc-S100A4-5 | Ca2+ | 12.0 ± 0.2 | 19.2 ± 1.1 | 19.4 ± 1.9 |
| sc-S100A4-10 | EGTA | 11.2 ± 0.2 | 23.1 ± 1.0 | 25.7 ± 2.5 |
| sc-S100A4-10 | Ca2+ | 12.0 ± 0.2 | 19.2 ± 1.1 | 19.4 ± 1.9 |
Mean ± standard deviation of two or three gel filtration runs in the presence of 2 mM EGTA or 2 mM CaCl2. Molecular masses and Stokes radii were calculated on the basis of the molecular masses and Stokes radii of the protein standards, respectively.
Chemical Denaturation of S100A4 Proteins
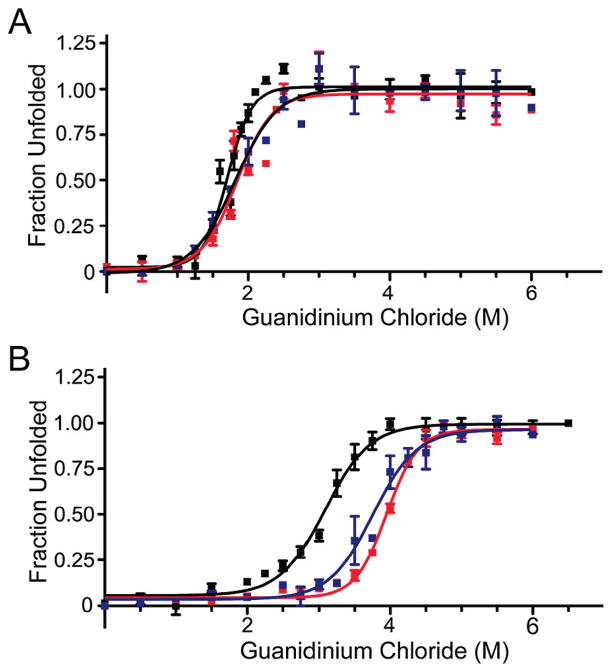
Previous studies demonstrated that some S100 family members are more stable to denaturation in the presence of calcium.48,49 To determine if S100A4 shares this characteristic and to establish if the flexible linker affected the stability of the single-chain proteins, we monitored unfolding in the presence of guanidine hydrochloride. In the absence of calcium, wt-S100A4, sc-S100A4–5, and sc-S100A4–10 had similar midpoints of denaturation, 1.71 ± 0.03, 1.84 ± 0.05, and 1.81 ± 0.06 M guanidine hydrochloride, respectively (Figure 3A). Consistent with other S100 proteins, the wt-S100A4, sc-S100A4–5, and sc-S100A4–10 proteins were more resistant to denaturant in the presence of calcium.48,50 Interestingly, the sc-S100A4 proteins were more stable than the wt-S100A4 protein with midpoints of denaturation of 3.95 ± 0.02 and 3.76 ± 0.06 M for sc-S100A4–5 and sc-S100A4–10, respectively, compared to 3.10 ± 0.04 M for wt-S100A4 (Figure 3B). These observations suggest that the addition of the flexible linker between the two S100A4 monomers has a stabilizing effect on the protein.
Figure 3.
Guanidine hydrochloride denaturation of sc-S100A4 proteins. (A) Unfolding of 5 μM wild-type S100A4 dimer or sc-S100A4 proteins in the presence of 2 mM EGTA. wt-S100A4, sc-S100A4–5, and sc-S100A4–10 exhibited similar midpoints of denaturation. (B) Unfolding in the presence of 2 mM CaCl2. The sc-S100A4 proteins were more resistant to denaturant-induced unfolding than wt-S100A4. wt-S100A4 (black), sc-S100A4–5 (red), and sc-S100A4–10 (blue). Values represent the mean ± standard error of the mean from three or four independent experiments.
Secondary Structural Analysis of S100A4 Proteins
Structural studies of wt-S100A4 showed that it is primarily composed of α-helices.14,16,51,52 Circular dichroism spectroscopy of sc-S100A4–5 and sc-S100A4–10 demonstrated that these proteins are mostly α-helical (Figure 4B,C). As for wt-S100A4, we observed a small increase in the helical content of sc-S100A4–5 and sc-S100A4–10 in the presence of calcium (Figure 4A). These data confirm that the addition of the linker has not grossly perturbed the secondary structure of S100A4 in either the apo or Ca2+-bound state.
Figure 4.
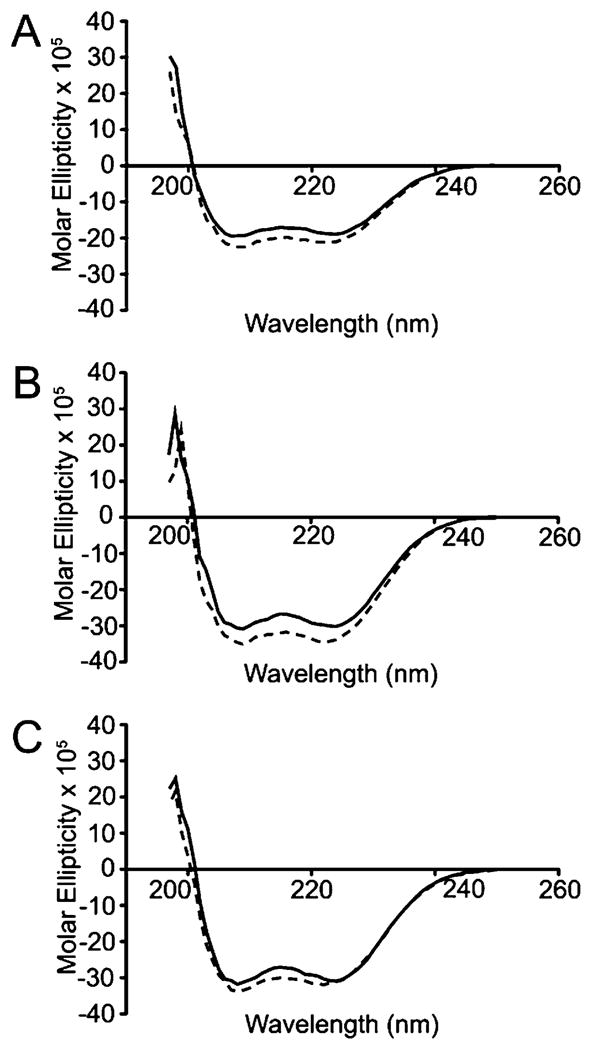
Circular dichroism spectroscopy of S100A4 proteins. Far-UV spectra of 6.8 μM S100A4 protein dimers of (A) wt-S100A4, (B) sc-S100A4–5, and (C) sc-S100A4–10. Solid and dotted lines correspond to spectra in the presence of 2 mM EGTA and 0.3 mM CaCl2, respectively. The α-helical structure of S100A4 is not altered by the addition of the flexible linker.
Biochemical Evaluation of sc-S100A4 Proteins
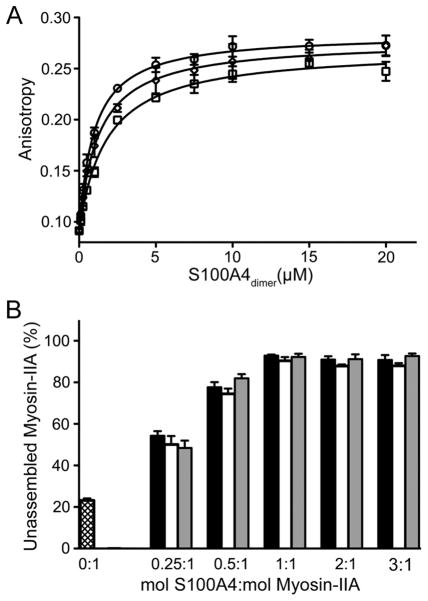
The ability of the sc-S100A4 proteins to bind myosin-IIA was assessed in an anisotropy assay using FITC-labeled MIIA1908–1923, which comprises the minimal binding site for S100A4.16,17 The measured dissociation constant for wt-S100A4 (1.8 ± 0.2 μM) was comparable to previously reported values.16,17 Both sc-S100A4-5 (1.0 ± 0.1 μM) and sc-S100A4–10 (1.3 ± 0.2 μM) demonstrated binding affinities for FITC-MIIA1908–1923 comparable to that of wt-S100A4 (Figure 5A). Moreover, binding was not observed in the presence of EGTA, demonstrating that the sc-S100A4 proteins exhibit Ca2+-dependent binding to myosin-IIA.
Figure 5.
Single-chain S100A4 proteins bind and regulate myosin-IIA assembly. (A) Affinities of S100A4 proteins for myosin-IIA were measured using a fluorescence anisotropy assay. wt-S100A4 (□), sc-S100A4–5 (○), and sc-S100A4–10 (⋄) exhibit similar affinities for myosin-IIA. (B) Promotion of myosin-IIA disassembly. wt-S100A4 (black bars), sc-S100A4–5 (white bars), and sc-S100A4–10 (gray bars) all promote maximal filament disassembly at a 1:1 molar ratio of S100A4 dimer to myosin-IIA dimer. Myosin-IIA disassembly in the absence of S100A4 (hatched bar). Values represent the mean ± standard error of the mean from three independent experiments.
Biochemical studies have shown that S100A4 regulates the monomer–polymer equilibrium of myosin-IIA by promoting the monomeric state.26,27 Using an assembly competent myosin-IIA construct, MIIA1338–1960, we monitored the ability of the sc-S100A4 proteins to promote the disassembly of preexisting myosin-IIA filaments. Similar to that of wt-S100A4, maximal promotion of myosin-IIA disassembly occurred at a ratio of 1 mol of sc-S100A4 protein dimer to 1 mol of MIIA1338–1960 dimer with disassembly of approximately 90% of the myosin-IIA filaments (Figure 5B).
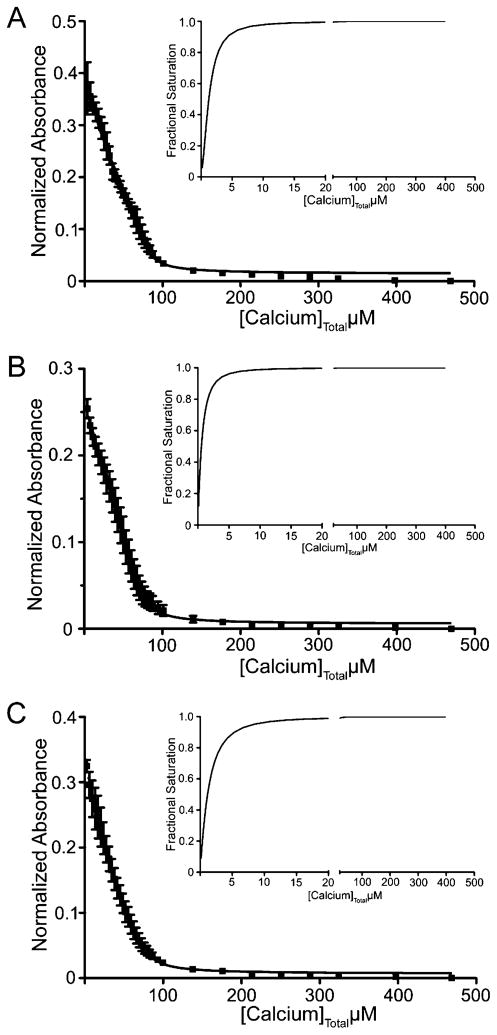
Dissociation constants for Ca2+ binding were determined using a competition assay with the chromophoric calcium chelator 5,5′Br2-BAPTA.17,39 In agreement with previous reports,16,17 wt-S100A4 exhibited relatively strong affinities for calcium in the presence of a 4-fold molar excess of myosin-IIA (EF1KD = 4.26 ± 0.04 μM; EF2KD = 0.44 ± 0.04 μM) (Table 2 and Figure 6A). The Ca2+ binding affinities for sc-S100A4–5 (EF1KD = 1.65 ± 0.05 μM; EF2KD = 0.74 ± 0.11 μM) and sc-S100A4–10 (EF1KD = 3.20 ± 0.30 μM; EF2KD = 1.10 ± 0.18 μM) in the presence of myosin-IIA were comparable to that of wt-S100A4 (Table 2 and Figure 6B,C).
Table 2.
Dissociation Constantsa for Binding of Ca2+ to S100A4
| protein | MIIA1851–1960 | EF1 (μM) | EF2 (μM) |
|---|---|---|---|
| wt-S100A4b | + | 4.26 ± 0.04 | 0.44 ± 0.04 |
| wt-S100A4c | − | >500 | 7.6 ± 0.4 |
| sc-S100A4–5b | + | 1.65 ± 0.05 | 0.74 ± 0.11 |
| sc-S100A4–10b | + | 3.20 ± 0.30 | 1.10 ± 0.18 |
| sc-S100A4–5M1b | + | 169 ± 23 | 0.51 ± 0.05 |
| sc-S100A4–5M1c | − | 366 ± 93 | 1.83 ± 0.34 |
| sc-S100A4–5M2b | + | 205 ± 41 | 0.24 ± 0.03 |
| sc-S100A4–5M2c | − | >500 | 0.75 ± 0.21 |
| sc-S100A4–10M1b | + | 244 ± 46 | 0.43 ± 0.06 |
| sc-S100A4–10M1c | − | 223 ± 26 | 1.46 ± 0.31 |
| sc-S100A4–10M2b | + | >500 | 0.92 ± 0.30 |
| sc-S100A4–10M2c | − | >500 | 1.26 ± 0.12 |
Values represent the average from two or three independent experiments.
Macroscopic Ca2+ binding constants determined for 12.5 μM S100A4 dimer in the presence of 100 μM MIIA1851–1960 monomer.
Macroscopic Ca2+ binding constants determined for 12.5 μM S100A4 dimer in the absence of target.
Figure 6.
Single-chain proteins and wt-S100A4 exhibit similar Ca2þ binding affinities in the presence of myosin-IIA. Ca2+ binding affinities were measured using a chromophoric calcium chelator 5,5′Br2-BAPTA competition assay. The absorbance at 263 nm was monitored as a function of the Ca2+ concentration: (A) wt-S100A4, (B) sc-S100A4–5, and (C) sc-S100A4–10. The data represent the average from three independent experiments. The insets show the saturation curve representation for the best fit in Caligator.
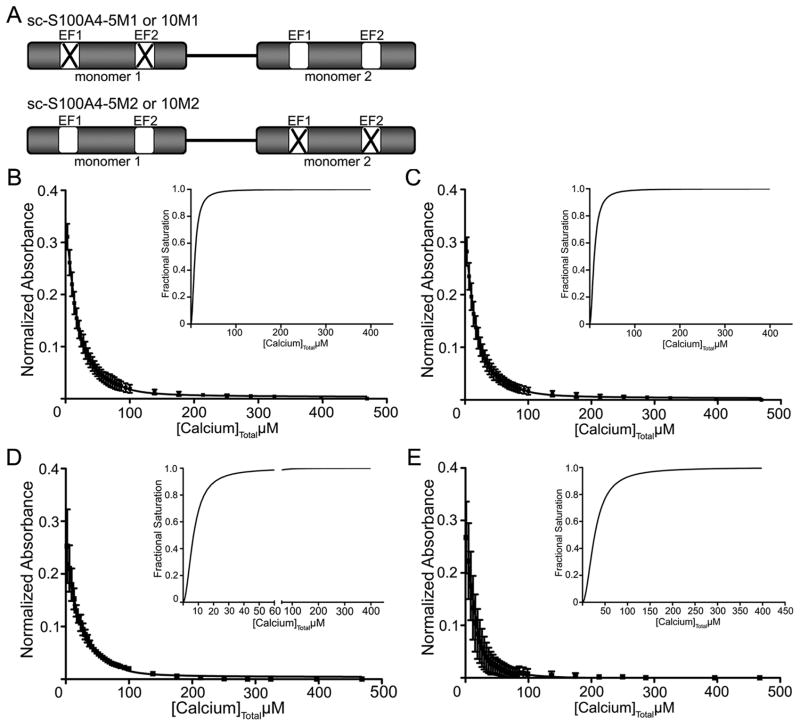
Development and Biochemical Evaluation of sc-S100A4 EF-Hand Mutant Proteins
To test whether both monomers in the S100A4 homodimer are required for function, we incorporated single-amino acid substitutions in both the pseudo (EF1) and canonical (EF2) EF-hands in either monomer 1 (M1) or monomer 2 (M2) of the sc-S100A4 constructs (Figure 7A). Previous studies with S100B demonstrated that substitution of E31 and E72 with alanine was sufficient to disrupt binding of Ca2+ to EF1 and EF2; therefore, we made the analogous substitutions in sc-S100A4 (E33A/E74A). Circular dichroism spectroscopy of the M1 and M2 proteins showed that the alanine substitutions did not disrupt the overall secondary structure of the single-chain proteins in the apo and Ca2+-bound states (Figure 1 of the Supporting Information).
Figure 7.
S100A4 single-chain EF-hand mutant proteins exhibit altered Ca2+ affinities. (A) Cartoon of sc-S100A4 EF-hand mutant constructs. Both E33 (EF1) and E74 (EF2) residues in either monomer 1 or monomer 2 were substituted with alanine. Calcium titrations of (B) sc-S100A4–5M1, (C) sc-S100A4–10M1, (D) sc-S100A4–5M2, and (E) sc-S100A4–10M2 in the presence of myosin-IIA and 5,5′Br2-BAPTA. The data represent the average from two or three independent experiments. The insets show the saturation curve representation for the best fit in Caligator.
Ca2+ binding affinities were measured for “wild-type” monomer 2 in sc-S100A4–5M1 and sc-S100A4–10M1 proteins and the wild-type monomer 1 in sc-S100A4–5M2 and sc-S100A4–10M2 proteins. In the absence of target, we obtained the following values: EF1KD = 366 ± 93 μM and EF2KD = 1.83 ± 0.34 μM for sc-S100A4–5M1, EF1KD = 223 ± 26 μM and EF2KD = 1.46 ± 0.31 μM for sc-S100A4–10M1, EF1KD > 500 μM and EF2KD = 0.75 ± 0.21 μM for sc-S100A4–5M2, and EF1KD > 500 μM and EF2KD = 1.26 ± 0.12 μM for sc-S100A4–10M2 (Table 2). Interestingly, in the presence of a 4-fold molar excess of myosin-IIA, the Ca2+ binding affinities for sc-S100A4–5M1 (EF1KD = 169 ± 23 μM; EF2KD = 0.51 ± 0.05 μM), sc-S100A4–10M1(EF1KD = 244 ± 46 μM; EF2KD = 0.43 ± 0.06 μM), sc-S100A4–5M2 (EF1KD = 205 ± 41 μM; EF2KD = 0.24 ± 0.03 μM), and sc-S100A4–10M2 (EF1KD > 500 μM; EF2KD = 0.92 ± 0.30 μM) were comparable to the KD values measured in the absence of target (Table 2 and Figure 7B-E). These observations indicate that mutation of either monomer 1 or monomer 2 in both sc-S100A4–5 and sc-S100A4–10 proteins affects binding of Ca2+ to the wild-type monomer for both proteins. Binding of Ca2+ to EF1 was weak both in the absence and in the presence of myosin-IIA, whereas EF2 exhibited a high affinity for Ca2+ irrespective of the presence of target.
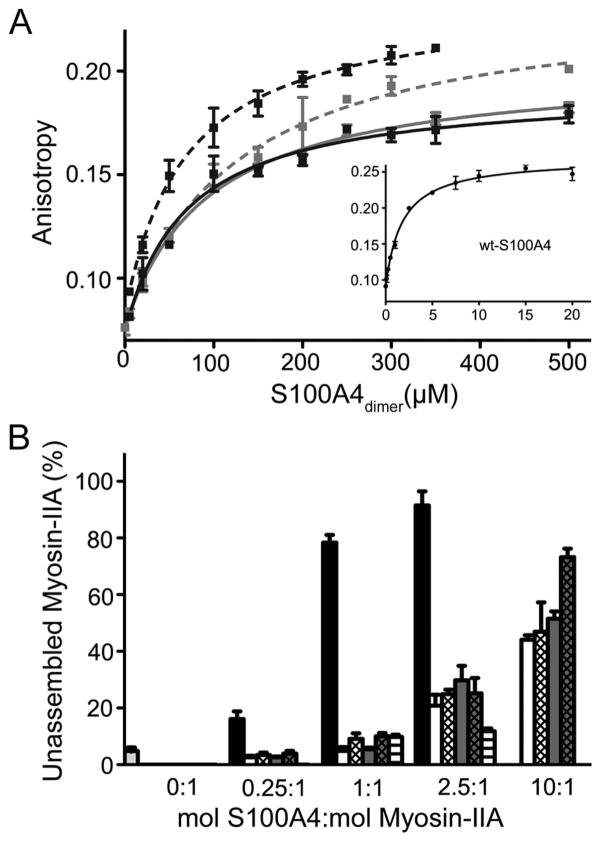
An examination of binding of sc-S100A4–5M1 and -M2 and sc-S100A4–10M1 and -M2 to FITC-MIIA1908–1923 in the anisotropy assay demonstrated that these proteins had 30–60-fold reduced affinities for myosin-IIA with dissociation constants of 93.4 ± 9.4, 125.1 ± 17.0, 69.3 ± 17.4, and 68.9 ± 9.7 μM, respectively (Figure 8A). These data demonstrate that neither the sc-S100A4-M1 nor -M2 proteins retained wild-type binding affinities for myosin-IIA.
Figure 8.
S100A4 single-chain EF-hand mutant proteins only weakly promote myosin-IIA disassembly. (A) Fluorescence anisotropy measurements of sc-S100A4–5M1 (solid gray line), sc-S100A4–10M1 (solid black line), sc-S100A4–5M2 (dashed gray line), and sc-S100A4–10M2 (dashed black line) demonstrated that sc-S100A4-M1 and -M2 EF-hand mutants have a 30–60-fold reduced affinity for FITC-MIIA1908–1923. Values represent the mean ± standard deviation from two or three independent experiments. (B) Myosin-IIA disassembly assays. Myosin-IIA disassembly in the absence of any added S100A4 (light gray bar). Ca2+-dependent myosin-IIA disassembly in the presence of wt-S100A4 (black bars) and EF-hand mutant proteins sc-S100A4–5M1 (white bars), sc-S100A4–5M2 (white-hatched bars), sc-S100A4–10M1 (dark gray bars), sc-S100A4–10M2 (dark gray-hatched bars), and wt-S100A4-E33A/E74A (white-lined bars). Values represent the mean ± standard error of the mean for three independent experiments.
To confirm that the combined E33A and E74A substitutions in EF1 and EF2 disrupt the Ca2+-dependent interactions of S100A4 with myosin-IIA, we examined the ability of a noncovalent S100A4 dimer that contained alanine substitution in all four EF hands to promote myosin-IIA disassembly. As expected, even at a molar ratio of 2.5:1 (S100A4 dimer:MIIA1338–1960 dimer), myosin-IIA disassembly was not observed (Figure 8B). Although the wild-type sc-S100A4 proteins promoted maximal disassembly of myosin-IIA at a 1:1 sc-S100A4 dimer:MIIA1338–1960 dimer molar ratio (Figure 6B), the sc-S100A4-M1 and -M2 proteins poorly regulated myosin-IIA disassembly (Figure 8B). We observed modest myosin-IIA disassembly at molar ratios of 2.5:1 (sc-S100A4-M1 or -M2 dimer to MIIA1338–1960 dimer). At ratios of 10:1, approximately 50% of the myosin-IIA had shifted to the supernatant fraction. These findings are consistent with the reduced affinities that the sc-S100A4-M1 and M2 proteins exhibit for myosin-IIA and suggest that two functional monomers are needed for binding and regulation of myosin-IIA.
Biological Characterization of sc-S100A4 Proteins
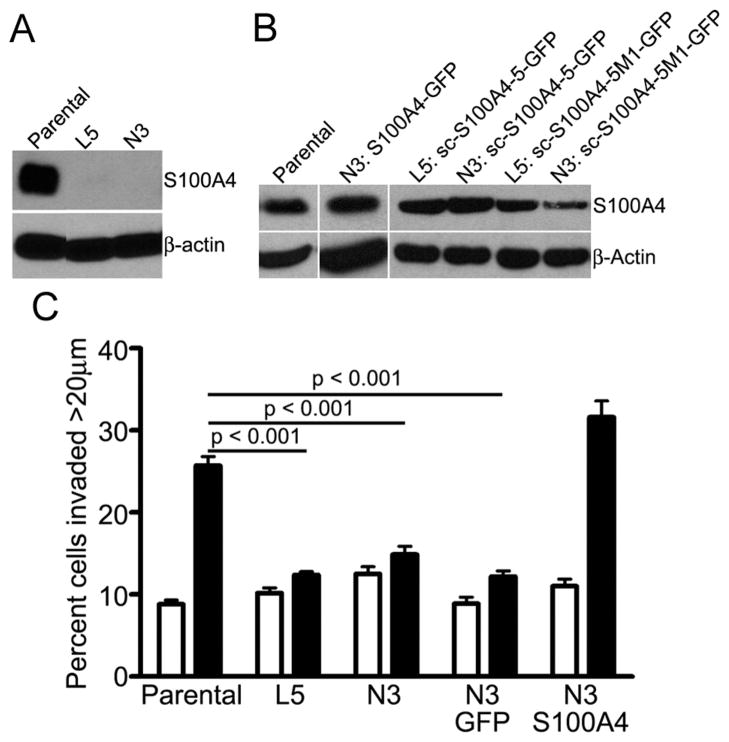
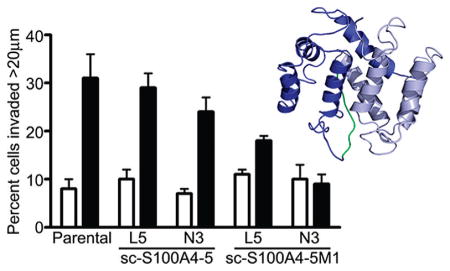
S100A4 overexpression promotes tumor cell migration and invasion.18,53 To evaluate the requirement for two functional S100A4 monomers to promote invasion, we used an in vitro assay that reconstitutes macrophage-dependent invasion of tumor cells into a 3D collagen gel43 using colorectal HCT116 carcinoma cells that are homozygous null for S100A4 (Figure 9A). When cultured with macrophages, 26% of the parental S100A4-expressing HCT116 cells invaded at least 20 μm into the collagen gel, whereas only 9% of parental HCT116 cells invaded the matrix when cultured alone (Figure 9C). The S100A4−/− HCT116 cell lines, L5 and N3, did not exhibit macrophage-dependent invasion (Figure 9C).
Figure 9.
HCT116 cells require S100A4 for macrophage-dependent invasion. (A) Immunoblot of S100A4 expression in parental and L5 and N3 S100A4−/− HCT116 cells. β-Actin was used as a loading control. (B) Immunoblot of S100A4 protein expression in L5 and N3 S100A4−/− HCT116 cells transfected with wild-type S100A4-GFP, sc-S100A4–5-GFP, or sc-S100A4–5M1-GFP proteins. β-Actin was used as a loading control. (C) Invasion of parental HCT116 cells, L5 and N3 S100A4−/− cells, and N3 S100A4−/− cells transfected with GFP or S100A4-GFP fusion protein in the absence (white bars) or presence (black bars) of macrophages. Values represent the mean ± standard error of the mean for three independent experiments.
To establish that HCT116 invasion required S100A4, we monitored the invasive capabilities of N3 S100A4−/− cells transfected with GFP alone or with wild-type S100A4-GFP fusion protein. To demonstrate that attachment of GFP to the native S100A4 did not disrupt protein function, we evaluated the ability of bacterially expressed and purified S100A4–GFP protein to promote myosin-IIA filament disassembly. Notably, identical purification protocols were used for S100A4–GFP protein and for the untagged wild-type S100A4, indicating that the addition of the GFP tag does not affect the biochemical properties of S100A4. At a 1:1 molar ratio of S1004–GFP dimer to MIIA1338–1960 dimer in the presence of calcium, we observed filament disassembly comparable to that elicited by wild-type S100A4 (Figure 2A of the Supporting Information). These observations demonstrate that S100A4–GFP protein is functionally equivalent to untagged wild-type S100A4. To compare the localization of the endogenous S100A4 and S100A4–GFP proteins, parental HCT116 cells were trans-fected with the S100A4–GFP construct. A comparison of the localization of S100A4–GFP protein with total S100A4 showed that the endogenous and the majority of the exogenously expressed S100A4 colocalize (Figure 2B of the Supporting Information). Moreover, the overall distribution of the S100A4–GFP protein is similar to the distribution of endogenous S100A4.
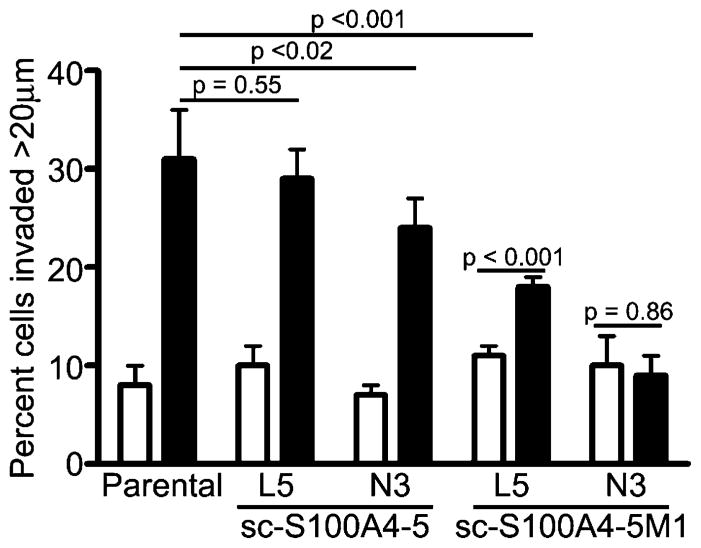
Transfection of N3 S100A4−/− cells with GFP alone did not rescue macrophage-dependent invasion; however, transfection with S100A4–GFP protein rescued the invasion of HCT116 cells with 32% of tumor cells invading more than 20 μm into the collagen gel (Figure 9C). The enhanced invasion observed for S100A4–GFP protein-expressing N3 cells likely results from an increased level of S100A4 expression (Figure 9B). Immunoblot analysis demonstrated that total S100A4 levels in the transfected N3 cells are comparable to levels of expression in parental HCT116 cells; however, the transfection efficiency was approximately 40–50% for N3 cells, and thus, the invading cells express higher S100A4 levels. The expression of sc-S100A4–5–GFP protein in L5 and N3 S100A4−/− cells, which had a transfection efficiency of 40–50%, also rescued the macrophage-dependent invasion of these null lines to varying extents (Figure 10). N3 cells expressing sc-S100A4–5M1–GFP protein did not exhibit macrophage-dependent invasion, whereas L5 cells exhibited a slight increase in the level of invasion in the presence of macrophages that was significantly reduced compared to that of parental HCT116 cells. The transfection efficiency for sc-S100A4–5M1–GFP protein was consistently 10–20% lower than that observed for sc-S100A4–5–GFP protein (Figure 9B). Taking into account the differences in transfection efficiency, we found the transfected cells express comparable levels of sc-S100A4–5–GFP and sc-S100A4–5M1–GFP proteins. Thus, it is unlikely that the inability of sc-S100A4–5M1–GFP protein to rescue HCT116 invasion results from a decreased level of expression of this construct.
Figure 10.
Single-chain S100A4–5 rescues HCT116 cell invasion. Invasion of parental HCT116 cells and L5 and N3 S100A4−/− HCT116 cells transfected with sc-S100A4–5-GFP or sc-S100A4–5M1-GFP fusion proteins in the absence (white bars) or presence (black bars) of macrophages. Values represent the mean ± standard deviation from two independent experiments.
Chemical Cross-Linking of S100A4-Myosin-IIA Complexes
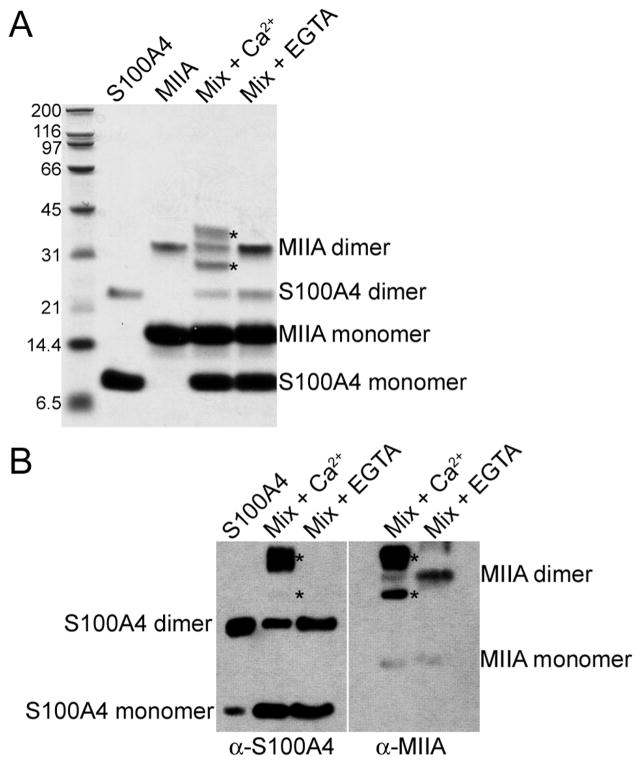
To experimentally evaluate the composition and stoichiometry of the S100A4–myosin-IIA complex, the native S100A4 dimer and MIIA1851–1960 were cross-linked in the presence of EGTA or calcium using disuccinimidyl tartrate (DST). To maximize binding, these studies were performed at S100A4 and myosin-IIA concentrations approximately 50-fold above the equilibrium dissociation constant for the S100A4–MIIA1851–1960 interaction.16 For both S100A4 and myosin-IIA alone, we observed both monomeric and dimeric species in the cross-linked sample (Figure 11A). Cross-linked S100A4 or MIIA1851–1960 dimers were not detected by mass spectrometry, which is likely due to the low abundance of these protein species in the sample. However, multiple monomeric species comprised of unmodified S100A4 or MIIA1851–1960 monomers (S100A4, 11599.4 Da; MIIA, 12399.5 Da) and monomers containing multiple intramolecular and hanging cross-links (S100A4, 11844.2 and 11977.2 Da; MIIA, 12530.4 and 12662.3 Da) were identified. Moreover, the S100A4 monomer was more extensively modified in the absence of MIIA1851–1960 than in the presence of MIIA1851–1960. The weak reactivity of the S100A4 and MIIA1851–1960 monomers on immunoblots is likely a consequence of modification by the DST cross-linker (Figure 11B).
Figure 11.
Composition of S100A4–myosin-IIA complexes. (A) Coomassie-stained SDS–PAGE of 12.5 μM wt-S100A4 dimer and 25μM MIIA monomer in the presence of DST, a 6.4 Å cross-linker. Asterisks denote complexes observed in the presence of calcium. (B) Western blots of cross-linked samples. Duplicate protein samples were separated by SDS–PAGE and transferred to a PVDF membrane, which was cut in half and probed with antibodies to either S100A4 or myosin-IIA.
In samples containing equimolar S100A4 and myosin-IIA, we observed two Ca2+-dependent species (Figure 11A, asterisks). Immunoblot analysis indicated that the upper band was comprised of both S100A4 and myosin-IIA, whereas the lower band contained myosin-IIA and exhibited very weak cross-reactivity with the S100A4 antibody (Figure 11B). Mass spectrometry showed that the upper band was comprised of two protein species with molecular masses of 36264 and 36130 Da; however, we did not identify any species associated with the lower Ca2+-dependent band. In the absence of any DST cross-links, the calculated mass of a S100A4 dimer bound to a single myosin-IIA polypeptide is 35592 Da, whereas the calculated mass of a MIIA1851–1960 dimer complexed to a S100A4 monomer is 36392 Da. The observed 36264 and 36130 Da species are thus consistent with a complex comprised of an S100A4 dimer and one MIIA1851-1960 polypeptide with several cross-links. Although we were not able to identify the lower Ca2+-dependent band by mass spectrometry, its migration via SDS-PAGE is consistent with a complex of a S100A4 monomer bound to a single myosin-IIA polypeptide (calculated mass of 23995 Da) with several cross-links.
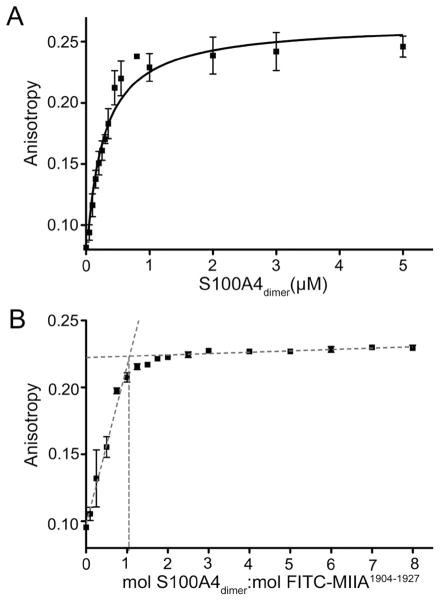
Because the cross-linking data suggested that one S100A4 dimer binds a single myosin-IIA polypeptide chain, we performed anisotropy titration measurements to directly measure the stoichiometry of binding. The measured dissociation constant for the binding of FITC-MIIA1904–1927 to wild-type S100A4 was 0.26 ± 0.03 μM, which is approximately 7-fold tighter than the affinity observed for FITC-MIIA1908–1923 (1.8 ± 0.2 μM) (Figure 12A). Consistent with the cross-linking data, titration of 1 μM FITC-MIIA1904–1927 with wild-type S100A4 revealed a stoichiometry of one S100A4 dimer per myosin-IIA peptide (Figure 12B).
Figure 12.
Stoichiometry of binding of FITC-MIIA1904–1927 to wt-S100A4. (A) Fluorescence anisotropy measurements of wt-S100A4 binding to MIIA1904–1927 demonstrated that wt-S100A4 has an approximately 7-fold increased affinity for FITC-MIIA1904–1927 as compared to FITC-MIIA1909–1923. Values represent the mean ± standard deviation from two or three independent titrations. (B) Stoichiometry of binding of wt-S100A4 to FITC-MIIA1904–1927. wt-S100A4 binds to FITC-MIIA1904–1927 at a 1:1 molar ratio (wt-S100A4 dimer:peptide). Gray dotted lines represent the linear regression of the rise and plateau of the titration curve. Values represent the mean ± standard deviation from two or three independent titrations.
DISCUSSION
Here we show that S100A4, a known metastasis factor, requires both monomers to effectively interact with nonmuscle myosin-IIA. Our single-chain S100A4 proteins, sc-S100A4, allowed us to disrupt Ca2+ binding in one monomer while leaving the other intact. Similar approaches involving the linking of subunits to evaluate or enhance protein function have been used previously to increase the blood circulation time of CuZn superoxide dismutase,54 to create bispecific antibodies,55 and to assess the DNA binding affinity for lambda Cro repressor.56
The biophysical and biochemical characterization of the wild-type sc-S100A4 proteins revealed that the sc-S100A4–5 and sc-S100A4–10 proteins are true mimics of the wt-S100A4 noncovalent dimer. One exception is that the Ca2+-bound sc-S100A4 proteins were more resistant to chemical denaturants than the wild-type noncovalent S100A4 dimer, suggesting that the flexible linker stabilizes the S100A4 dimer against chemical denaturation. The precise mechanism responsible for the enhanced stability of the sc-S100A4 proteins is likely to be complex, though it is likely to benefit at least in part from entropic contributions. For example, the condensation of two distinct chains into a dimeric species results in the loss of six degrees of freedom (three rotational and three translational), while in the case of the sc-S100A4 proteins, there is no analogous entropic penalty. The absence of this entropic penalty in the folding of the sc-S100A4 proteins may contribute to the observed enhancement in stability relative to that of the wild-type noncovalent dimer.
Increased resistance to denaturation has been documented for other single-chain proteins, including thermal denaturation of the single-chain variant of monellin and urea denaturation of the Arc-L1-Arc single-chain protein.57–59 This stabilizing effect is not due to gross structural changes as circular dichroism spectroscopy of the sc-S100A4 proteins showed that these proteins remain mostly α-helical with spectra similar to those of wt-S100A4. Similar to other S100 proteins, this single transition suggests that the native S100A4 dimer unfolds via a two-state mechanism, where unfolding of the dimer proceeds to the denatured state without an intermediate.48,49 Although folding intermediates have been detected for S100B, S100A11, and S100A12, our data do not support the existence of a folding intermediate.48,60,61 While it remains possible that weakly populated intermediates, which are beyond the detection limits of this study, contribute to the folding of S100A4, additional studies will be required to evaluate the mechanism of S100A4 unfolding.
The selection of the EF1 E33 and EF2 E74 residues for mutation was based on analogous Ca2+-disrupting substitutions examined in S100B.62 Other studies assessing binding of Ca2+ to S100A4 used the EF-hand substitutions EF1 E33Q and EF2 D63N, which also disrupt Ca2+ binding.63,64 An examination of the canonical EF-hand sequence for all S100 proteins, as well as other EF-hand-containing proteins, shows that both D63 and E74 are invariant, indicating that these positions are necessary for Ca2+ ligation. This is also supported by the X-ray structure of the Ca2+-bound S100A4, which shows that D63 and E74 are critical for Ca2+ ion coordination in EF2.16,51,52 For the native S100A4 dimer, Ca2+ binding affinities increase in the presence of myosin-IIA 10- and 100-fold for EF2 and EF1, respectively.16 For the sc-S100A4-M1 and -M2 proteins, inactivation of monomer 1 or monomer 2 yielded S100A4 proteins whose Ca2+ affinities were the same in the absence and presence myosin-IIA. It is not surprising that EF1 did not exhibit an increased affinity for Ca2+ in the presence of myosin-IIA as the sc-S100A4-M1 and -M2 proteins have 30-60-fold reduced affinities for myosin-IIA and the calcium measurements were taken at concentrations below the KD for myosin-IIA. However, EF2 in sc-S100A4-M1 and M2 proteins exhibited affinities for Ca2+ in the absence of target that are typically only observed for the wild-type protein in the presence of myosin-IIA. These data suggest that there is communication between the two S100A4 monomers in which the inactive conformation of one monomer effects binding of Ca2+ to the second wild-type monomer.
Mutagenesis studies demonstrated that binding of Ca2+ to EF2 alone is sufficient for binding of S100A4 to myosin-IIA.63 On the basis of this finding, the wild-type monomer in the sc-S100A4-M1 and -M2 mutant proteins should retain myosin-IIA binding activity; however, these proteins exhibited significantly decreased myosin-IIA binding activity and only weakly promoted myosin-IIA filament assembly. The inability of the sc-S100A4 mutant proteins to bind myosin-IIA is reminiscent of studies with the single-chain tetracycline repressor (sc-TetR).65 TetR binds with high affinity to the tet operator as a dimer. Tetracycline binding induces a conformational rearrangement in the dimerization domain that is transmitted to the DNA binding domain and reduces the affinity of TetR for DNA.65 Covalent tethering of the two TetR monomers revealed that a functional tetracycline binding site in each TetR monomer was necessary to promote the conformational rearrangements required for DNA release.65 S100A4 may function in a similar manner, where the open, active conformation of both monomers is required for interactions with myosin-IIA.
This idea is supported by our cross-linking data that suggest that the S100A4 dimer binds a single MIIA1851–1960 polypeptide. Moreover, these data are consistent with our observation that S100A4 binds a 24mer myosin-IIA peptide with a stoichiometry of one peptide per S100A4 dimer and a recent report that a 32mer myosin-IIA peptide binds to S100A4 with a stoichiometry of one peptide per S100A4 dimer.64 Given the length of the MIIA1851–1960 construct, it may be surprising that only one polypeptide chain is cross-linked to the S100A4 dimer; however, recent NMR studies of residues 1850-1960 of myosin-IIA demonstrated that the 17 N-terminal amino acids and the 35 C-terminal residues are unstructured.64 We proposed previously that S100A4 binding promotes myosin-IIA filament disassembly by inducing local unwinding of the myosin-IIA coiled coil.27 Because a significant portion of the MIIA1851–1960 construct is unstructured, S100A4-mediated conformational rearrangements of the coiled coil could result in dissociation of the two myosin-IIA polypeptides, resulting in the cross-linking of a single MIIA1851–1960 polypeptide chain. Structural studies indicate that most S100 protein dimers bind the target peptide in the hydrophobic cleft between helices 3 and 4, which is exposed upon Ca2+ binding, thus allowing two target peptides to bind per S100 dimer.66–69 However, the recent structure of S100A6 bound to the C-terminal domain of the Siah-1 interacting protein showed that although the S100A6 dimer binds two target peptides, each peptide binds across the S100A6 dimer interface and forms interactions with the canonical target binding cleft.70 On the basis of these observations, one may envision that a target peptide, which binds across the dimer interface and is of sufficient length, could make contacts with both canonical target binding clefts. Such a configuration would result in a complex of one target peptide bound per S100 dimer.
On the basis of our biochemical analysis of the sc-S100A4 proteins with myosin-IIA, our cross-linking data, and the determination of S100A4:myosin-IIA stoichiometries, we propose a model in which the myosin-IIA heavy chain wraps around the S100A4 dimer to form interactions in the vicinity of the canonical target binding cleft of each monomer in such a manner that both target binding sites are required for the efficient interaction with myosin-IIA.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health Grants CA129598 (A.R.B.) and GM58888 (D.J.W.).
We thank Dr. Michael Brenowitz and Dr. Steven Almo for helpful discussions and Ms. Janine Stanford and Mr. Christopher Pudney for technical assistance.
ABBREVIATIONS
- MMTV
mouse mammary tumor virus
- PyMT
polyoma middle T
- sc-S100A4
single-chain S100A4
- wt-S100A4
native S100A4 dimer
- GdnHCl
guanidine hydrochloride
- BAPTA
1,2-bis(o-aminophenoxy)ethane-N,N,N’,N’-tetraacetic acid
- MIIA
non-muscle myosin-IIA
- FITC
fluorescein isothiocyanate
- M1
monomer 1
- M2
monomer 2
- 3D
three-dimensional
- DST
disuccinimidyl tartrate
- MS
mass spectrometry
Footnotes
Circular dichroism spectra of sc-S100A4-5M1 and -M2 EF-hand mutant proteins, Ca2+-dependent myosin-IIA promotion of disassembly by S100A4-GFP protein, and immunofluor-escent localization of S100A4 in untransfected parental HCT116 cells and parental HCT116 cells transfected with S100A4-GFP protein. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 2.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 3.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: The HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persechini A, Moncrief ND, Kretsinger RH. The EF-hand family of calcium-modulated proteins. Trends Neurosci. 1989;12:462–467. doi: 10.1016/0166-2236(89)90097-0. [DOI] [PubMed] [Google Scholar]
- 5.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins. 1999;37:499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Ravasi T, Hsu K, Goyette J, Schroder K, Yang Z, Rahimi F, Miranda LP, Alewood PF, Hume DA, Geczy C. Probing the S100 protein family through genomic and functional analysis. Genomics. 2004;84:10–22. doi: 10.1016/j.ygeno.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schafer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 9.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: History, function, and expression. Brain Res Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 10.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 11.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B(β β) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 12.Koch M, Diez J, Fritz G. Crystal structure of Ca2+-free S100A2 at 1.6-Å resolution. J Mol Biol. 2008;378:933–942. doi: 10.1016/j.jmb.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Otterbein LR, Kordowska J, Witte-Hoffmann C, Wang CL, Dominguez R. Crystal structures of S100A6 in the Ca2+-free and Ca2+-bound states: The calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure. 2002;10:557–567. doi: 10.1016/s0969-2126(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 14.Vallely KM, Rustandi RR, Ellis KC, Varlamova O, Bresnick AR, Weber DJ. Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41:12670–12680. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]
- 15.Wright NT, Cannon BR, Zimmer DB, Weber DJ. S100A1: Structure, Function, and Therapeutic Potential. Curr Chem Biol. 2009;3:138–145. doi: 10.2174/187231309788166460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, Ramagopal UA, Almo SC, Weber DJ, Bresnick AR. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrett S, Hodgson L, Rybin A, Toutchkine A, Hahn KM, Lawrence DS, Bresnick AR. A Biosensor of S100A4 Metastasis Factor Activation: Inhibitor Screening and Cellular Activation Dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies BR, Davies MP, Gibbs FE, Barraclough R, Rudland PS. Induction of the metastatic phenotype by transfection of a benign rat mammary epithelial cell line with the gene for p9Ka, a rat calcium-binding protein, but not with the oncogene EJ-ras-1. Oncogene. 1993;8:999–1008. [PubMed] [Google Scholar]
- 19.Davies MP, Rudland PS, Robertson L, Parry EW, Jolicoeur P, Barraclough R. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–1637. [PubMed] [Google Scholar]
- 20.Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- 21.Rudland PS, Platt-Higgins A, Renshaw C, West CR, Winstanley JH, Robertson L, Barraclough R. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595–1603. [PubMed] [Google Scholar]
- 22.van Dieck J, Brandt T, Teufel DP, Veprintsev DB, Joerger AC, Fersht AR. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29:2024–2035. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 23.Takenaga K, Nakamura Y, Sakiyama S, Hasegawa Y, Sato K, Endo H. Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J Cell Biol. 1994;124:757–768. doi: 10.1083/jcb.124.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriajevska M, Fischer-Larsen M, Moertz E, Vorm O, Tulchinsky E, Grigorian M, Ambartsumian N, Lukanidin E. Liprin β1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1) J Biol Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- 25.Kriajevska MV, Cardenas MN, Grigorian MS, Ambartsumian NS, Georgiev GP, Lukanidin EM. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J Biol Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- 26.Murakami N, Kotula L, Hwang YW. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: Phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry. 2000;39:11441–11451. doi: 10.1021/bi000347e. [DOI] [PubMed] [Google Scholar]
- 27.Li ZH, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 28.Mitsuhashi M, Sakata H, Kinjo M, Yazawa M, Takahashi M. Dynamic assembly properties of nonmuscle myosin II isoforms revealed by combination of fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy. J Biochem. 2011;149:253–263. doi: 10.1093/jb/mvq134. [DOI] [PubMed] [Google Scholar]
- 29.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 30.Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 31.Tarabykina S, Scott DJ, Herzyk P, Hill TJ, Tame JR, Kriajevska M, Lafitte D, Derrick PJ, Dodson GG, Maitland NJ, Lukanidin EM, Bronstein IB. The dimerization interface of the metastasis-associated protein S100A4 (Mts1): In vivo and in vitro studies. J Biol Chem. 2001;276:24212–24222. doi: 10.1074/jbc.M009477200. [DOI] [PubMed] [Google Scholar]
- 32.Helfman DM, Kim EJ, Lukanidin E, Grigorian M. The metastasis associated protein S100A4: Role in tumour progression and metastasis. Br J Cancer. 2005;92:1955–1958. doi: 10.1038/sj.bjc.6602613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ismail TM, Zhang S, Fernig DG, Gross S, Martin-Fernandez ML, See V, Tozawa K, Tynan CJ, Wang G, Wilkinson MC, Rudland PS, Barraclough R. Self-association of calcium-binding protein S100A4 and metastasis. J Biol Chem. 2010;285:914–922. doi: 10.1074/jbc.M109.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashtchian A. Novel methods for cloning and engineering genes using the polymerase chain reaction. Curr Opin Biotechnol. 1995;6:30–36. doi: 10.1016/0958-1669(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 35.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Pace CN, Shirley BA, Thomson JA. Measuring the conformational stability of a protein. In: Creighton T, editor. Protein Structure: A Practical Approach. IRL Press. Oxford University Press; New York: 1997. pp. 299–321. [Google Scholar]
- 38.Schmid F. Optical spectroscopy to characterize protein conformation and conformational change. In: Creighton T, editor. Protein Structure: A Practical Approach. IRC Press at Oxford University Press; New York: 1997. pp. 289–297. [Google Scholar]
- 39.Linse S. Calcium binding to proteins studied via competition with chromophoric chelators. In: Vogel HJ, editor. Calcium Binding Protein Protocols. Humana Press; Totowa, NJ: 2002. pp. 15–24. [DOI] [PubMed] [Google Scholar]
- 40.Andre I, Linse S. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Anal Biochem. 2002;305:195–205. doi: 10.1006/abio.2002.5661. [DOI] [PubMed] [Google Scholar]
- 41.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 43.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 44.Akke M, Drakenberg T, Chazin WJ. Three-dimensional solution structure of Ca2+-loaded porcine calbindin D9k determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1992;31:1011–1020. doi: 10.1021/bi00119a009. [DOI] [PubMed] [Google Scholar]
- 45.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 46.Hagemeyer CE, von Zur Muhlen C, von Elverfeldt D, Peter K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb Haemostasis. 2009;101:1012–1019. [PubMed] [Google Scholar]
- 47.Drohat AC, Nenortas E, Beckett D, Weber DJ. Oligomerization state of S100B at nanomolar concentration determined by large-zone analytical gel filtration chromatography. Protein Sci. 1997;6:1577–1582. doi: 10.1002/pro.5560060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia AF, Garcia W, Nonato MC, Araujo AP. Structural stability and reversible unfolding of recombinant porcine S100A12. Biophys Chem. 2008;134:246–253. doi: 10.1016/j.bpc.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Botelho HM, Koch M, Fritz G, Gomes CM. Metal ions modulate the folding and stability of the tumor suppressor protein S100A2. FEBS J. 2009;276:1776–1786. doi: 10.1111/j.1742-4658.2009.06912.x. [DOI] [PubMed] [Google Scholar]
- 50.Ferguson PL, Shaw GS. Role of the N-terminal helix I for dimerization and stability of the calcium-binding protein S100B. Biochemistry. 2002;41:3637–3646. doi: 10.1021/bi0118052. [DOI] [PubMed] [Google Scholar]
- 51.Pathuri P, Vogeley L, Luecke H. Crystal structure of metastasis-associated protein S100A4 in the active calcium-bound form. J Mol Biol. 2008;383:62–77. doi: 10.1016/j.jmb.2008.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gingras AR, Basran J, Prescott A, Kriajevska M, Bagshaw CR, Barsukov IL. Crystal structure of the Ca2+-form and Ca2+-binding kinetics of metastasis-associated protein, S100A4. FEBS Lett. 2008;582:1651–1656. doi: 10.1016/j.febslet.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 53.Bjornland K, Winberg JO, Odegaard OT, Hovig E, Loennechen T, Aasen AO, Fodstad O, Maelandsmo GM. S100A4 involvement in metastasis: Deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res. 1999;59:4702–4708. [PubMed] [Google Scholar]
- 54.Hallewell RA, Laria I, Tabrizi A, Carlin G, Getzoff ED, Tainer JA, Cousens LS, Mullenbach GT. Genetically engineered polymers of human CuZn superoxide dismutase. Biochemistry and serum half-lives. J Biol Chem. 1989;264:5260–5268. [PubMed] [Google Scholar]
- 55.Mallender WD, Voss EW., Jr Construction, expression, and activity of a bivalent bispecific single-chain antibody. J Biol Chem. 1994;269:199–206. [PubMed] [Google Scholar]
- 56.Jana R, Hazbun TR, Fields JD, Mossing MC. Single-chain lambda Cro repressors confirm high intrinsic dimer-DNA affinity. Biochemistry. 1998;37:6446–6455. doi: 10.1021/bi980152v. [DOI] [PubMed] [Google Scholar]
- 57.Kim SH, Kang CH, Kim R, Cho JM, Lee YB, Lee TK. Redesigning a sweet protein: Increased stability and renaturability. Protein Eng. 1989;2:571–575. doi: 10.1093/protein/2.8.571. [DOI] [PubMed] [Google Scholar]
- 58.Robinson CR, Sauer RT. Equilibrium stability and sub-millisecond refolding of a designed single-chain Arc repressor. Biochemistry. 1996;35:13878–13884. doi: 10.1021/bi961375t. [DOI] [PubMed] [Google Scholar]
- 59.Aghera N, Earanna N, Udgaonkar JB. Equilibrium Unfolding Studies of Monellin: The Double-Chain Variant Appears To Be More Stable Than the Single-Chain Variant. Biochemistry. 2011;50:2434–2444. doi: 10.1021/bi101955f. [DOI] [PubMed] [Google Scholar]
- 60.Shaw GS, Marlatt NM, Ferguson PL, Barber KR, Bottomley SP. Identification of a dimeric intermediate in the unfolding pathway for the calcium-binding protein S100B. J Mol Biol. 2008;382:1075–1088. doi: 10.1016/j.jmb.2008.07.083. [DOI] [PubMed] [Google Scholar]
- 61.Pan J, Rintala-Dempsey AC, Li Y, Shaw GS, Konermann L. Folding kinetics of the S100A11 protein dimer studied by time-resolved electrospray mass spectrometry and pulsed hydrogen-deuterium exchange. Biochemistry. 2006;45:3005–3013. doi: 10.1021/bi052349a. [DOI] [PubMed] [Google Scholar]
- 62.Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, Horrocks WD, Weber DJ. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 63.Kim EJ, Helfman DM. Characterization of the metastasis-associated protein, S100A4. Roles of calcium binding and dimerization in cellular localization and interaction with myosin. J Biol Chem. 2003;278:30063–30073. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 64.Badyal SK, Basran J, Bhanji N, Kim JH, Chavda AP, Jung HS, Craig R, Elliott PR, Irvine AF, Barsukov IL, Kriajevska M, Bagshaw CR. Mechanism of the Ca2+-dependent interaction between S100A4 and tail fragments of nonmuscle myosin heavy chain IIA. J Mol Biol. 2011;405:1004–1026. doi: 10.1016/j.jmb.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamionka A, Majewski M, Roth K, Bertram R, Kraft C, Hillen W. Induction of single chain tetracycline repressor requires the binding of two inducers. Nucleic Acids Res. 2006;34:3834–3841. doi: 10.1093/nar/gkl316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(β β) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 67.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: Insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 68.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J Biol Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) J Mol Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee YT, Dimitrova YN, Schneider G, Ridenour WB, Bhattacharya S, Soss SE, Caprioli RM, Filipek A, Chazin WJ. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: A novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–10932. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.