Abstract
The V122I mutation that alters the stability of transthyretin (TTR) affects 3–4% of African Americans and leads to amyloidogenesis and development of cardiomyopathy. In addition, 10–15% of individuals over the age of 65 develop senile systemic amyloidosis (SSA) and cardiac TTR deposits due to wild-type TTR amyloidogenesis. As no approved therapies for TTR amyloid cardiomyopathy are available, the development of drugs that prevent amyloid-mediated cardiotoxicity is desired. To this aim, we developed a fluorescence polarization-based HTS screen, which identified several new chemical scaffolds targeting TTR. These novel compounds were potent kinetic stabilizers of TTR and prevented tetramer dissociation, unfolding and aggregation of both wild type and the most common cardiomyopathy-associated TTR mutant, V122I-TTR. High-resolution co-crystal structures and characterization of the binding energetics revealed how these diverse structures bound to tetrameric TTR. Our study also showed that these compounds effectively inhibited the proteotoxicity of V122I-TTR towards human cardiomyocytes. Several of these ligands stabilized TTR in human serum more effectively than diflunisal, which is one of the best known inhibitors of TTR aggregation, and may be promising leads for the treatment and/or prevention of TTR-mediated cardiomyopathy.
INTRODUCTION
The misassembly of soluble proteins into toxic amyloid aggregates underlies a large number of human degenerative diseases (1–3). TTR is one of more than 30 human amyloidogenic proteins whose misassembly can cause a variety of degenerative gain-of-toxic-function diseases. TTR is a tetrameric protein (54 kDa), secreted from the liver into the blood where, using orthogonal sites, it transports thyroxine (T4) and holo-retinol binding protein (4). However, 99% of the TTR T4 binding sites remain unoccupied in humans owing to the presence of two other T4 transport proteins in blood (3). Familial TTR amyloid diseases, which are associated with one of more than 80 mutations in the TTR gene, include the systemic neuropathies (familial amyloid polyneuropathy [FAP]), cardiomyopathies (familial amyloid cardiomyopathy [FAC]), and central nervous system amyloidoses (CNSA) (5–8).
Cardiac amyloidosis is most commonly caused by deposition of immunoglobulin light chains or TTR in the cardiac interstitium and conducting system. It is a chronic and progressive condition, which can lead to arrhythmias, biventricular heart failure, and death (8–10). Two types of TTR-associated amyloid cardiomyopathies are clinically important. Wild-type (WT) TTR aggregation underlies the development of senile systemic amyloidosis (SSA). Cardiac TTR deposits can be found in 10 to 15% of the population over the age of 65 at autopsy (10,11). Many of these patients are asymptomatic, but there is little doubt that SSA is an underdiagnosed disease. In addition, a number of TTR mutations, including V122I, lead to amyloidogenesis and familial amyloid cardiomyopathy (FAC) (12–15). Population studies show that the V122I mutation is found in 3–4% of African Americans (~1.3 million people) and contributes to the increased prevalence of heart failure among this population segment (14,15). The mutant TTR allele behaves as an autosomal dominant allele with age-dependent penetrance and the frequency of cardiac amyloidosis from TTR in African-American individuals above age 60 is four times that seen in Caucasian-Americans of comparable age.
All of the TTR mutations associated with familial amyloidosis decrease tetramer stability, and some decrease the kinetic barrier for tetramer dissociation (3, 16). The latter is important because tetramer dissociation is the rate-limiting step in the TTR amyloidogenesis cascade (3). Kinetic stabilization of the native, tetrameric structure of TTR by interallelic trans suppression (incorporation of mutant subunits that raise the dissociative transition state energy) prevents post-secretory dissociation and aggregation, as well as the related disease familial amyloid polyneuropathy (FAP), by slowing TTR tetramer dissociation (17). Occupancy of the TTR T4 binding sites with rationally designed small molecules is known to stabilize the native tetrameric state of TTR over the dissociative transition state, raising the kinetic barrier, imposing kinetic stabilization on the tetramer and preventing amyloidogenesis (3, 16, 18).
Previous studies have focused on rational ligand design and as a result most of the TTR stabilizers reported to date are halogenated biaryl analogues of T4, many resembling non-steroidal anti-inflammatory drugs (NSAIDs). Some of these compounds, such as the NSAID diflunisal, which is currently tested in clinical trials in FAP patients for its efficacy to ameliorate peripheral neuropathy resulting from TTR deposition, (19) have anti-inflammatory activity (20, 21). The pharmacological effects of NSAIDs are due to inhibition of cyclo-oxygenase (COX) enzymes (22). Inhibition of COX-1 can produce side effects such as gastrointestinal irritation, leading to ulcers and bleeding (23). Inhibition of COX-2 has been associated with an increased risk of severe cardiovascular events, including heart failure, particularly in patients with preexisting cardiorenal dysfunction (20, 21, 24, 25). Therefore, heart and kidney impairment are exclusion criteria for participation of patients in the diflunisal clinical trials to treat TTR-mediated FAP (19). Genomic variations can increase the sensitivity of individuals to adverse side effects of NSAIDs. Serum concentrations of NSAIDs depend on CYP2C9 and/or CYP2C8 activity. CYP2C9 polymorphism might play a significant role in the profile of adverse side effects of NSAID and alleles that affect the activity of CYP2C9 are found at different frequency in subjects of Caucasian, African or Asian descent (26, 27). Hence, the long-term therapy with drugs that have inhibitory effect on COX activity to prevent TTR aggregation is especially problematic in patients who suffer from TTR-mediated cardiomyopathy. The design and development of drugs to treat/prevent FAC or SSA thus presents the challenge not only to find compounds with a greater variety of chemical scaffolds that accomplish stabilization, but do so without the adverse side effects due to inhibition of COX activity.
For these reasons, the development of a rapid and robust screen for compounds that bind to and stabilize TTR could be useful. To date, no high-throughput screening (HTS) methodology is available for the discovery of TTR ligands (28,29). Therefore, we developed a versatile fluorescence polarization (FP) based HTS assay that can detect binding of small molecules to the T4 binding pocket of TTR under physiological conditions.
RESULTS
Design and synthesis of the TTR FP probe
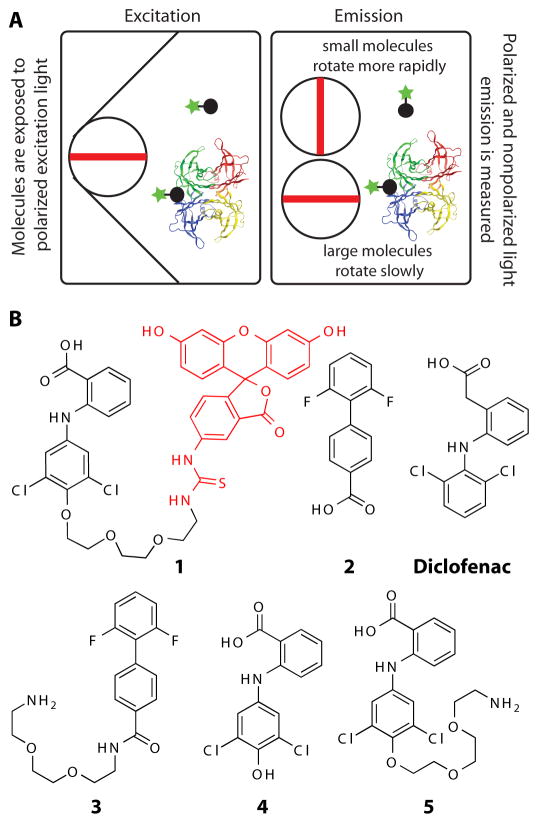
FP is used to study molecular interactions by monitoring changes in the apparent size of a fluorescently labeled molecule. Binding is measured by an increase in the FP signal, which is proportional to the decrease in the rate of tumbling of a fluorescent ligand upon association with macromolecules such as proteins (Fig. 1A). To synthesize a fluorescent TTR ligand 1, we initially started with the NSAID diflunisal analogue 2 (Fig. 1B) (30). The product of attaching a linker to 2, compound 3, had very low binding affinity to TTR (Kd1 > 3290 nM, fig. S1A and fig. S1B). The crystal structure of the diclofenac analog 4 showed that the phenolic hydroxyl flanked by the two chlorine atoms is oriented out of the binding pocket into the solvent (31). We reasoned that attaching a PEG amine linker to the phenol group of 4 would generate compound 5 which would bind to TTR (Fig. 1B and fig. S1C). 5 was coupled to fluorescein isothiocyanate (FITC) to produce the FITC-coupled TTR FP probe (1, Fig. 1B). The binding characteristics of the probe (Kd1 = 13 nM and Kd2 = 100 nM) were assessed with ITC (Fig. 2A).
Fig. 1.
Schematic of the FP assay and structures of TTR ligands and FP probe 1. (A) The rate of tumbling of fluorescent probe 1 decreases upon binding to TTR, which results in increasing its fluorescence polarization signal. (B) Chemical structures of the TTR ligands 2–5 used to design probe 1.
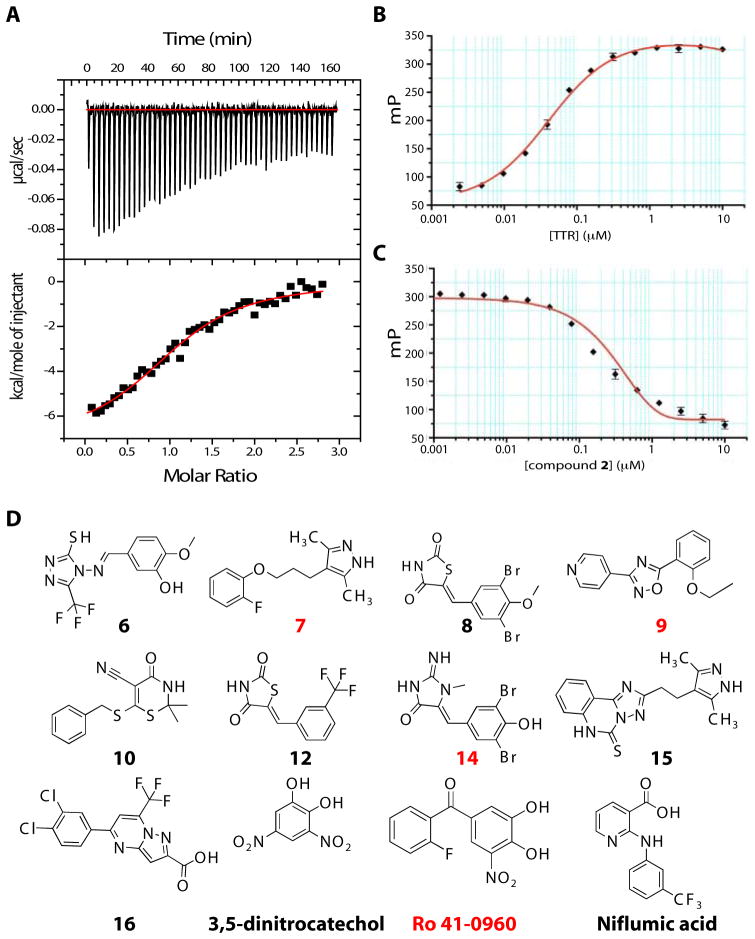
Fig. 2.
Assessment of FP probe 1 binding to TTR and structures of TTR ligands. (A) Assessment of the binding affinity of probe 1 to TTR by ITC. Calorimetric titration of probe 1 against TTR (Kd,1 = 13 nM and Kd,2 = 100 nM). Raw data (top) and integrated heats (bottom) from the titration of TTR (2 μM) with probe (25 μM). The solid red line represents the best fit binding isotherm to a two-site binding model. (B) FP saturation binding between probe 1 (0.1 μM) and increasing concentration of TTR (0.005 to 10 μM) (C) Competition of probe 1 from TTR by increasing concentrations (0.01 μM – 50 μM) of ligand 2 (Kapp = 0.231 μM, R2 = 0.997). FP Assays were performed in triplicate. Error bars: SD. (D) Structures of the newly identified TTR ligands. Compounds with co-crystal structures in Fig. 5 are labeled in red in all figures.
Evaluation of the FP assay
The binding of 1 to TTR was evaluated to test its suitability for the FP assay with a standard saturation binding experiment. A fixed concentration of probe 1 (0.1 μM) was incubated with increasing concentrations of TTR (0.005 μM to 10 μM) and the formation of 1•TTR complex was quantified by the increase in FP signal (excitation λ 485 nm, emission λ 525 nm) relative to the concentration of TTR (Fig. 2B). The fluorescence polarization increased with the concentration of TTR until saturation was reached. A large dynamic range (70 – 330 mP) was measured for the assay. To validate the FP assay, we tested known TTR binders in a displacement assay (for detailed information see Supplemental Material). Compound 2 (Kapp = 231 nM, R2 = 0.997), Thyroxine (T4) (Kapp = 186 nM, R2 = 0.998) and diclofenac (Kapp = 4660 nM, R2 = 0.999) decreased the FP signal in a dose-dependent manner (Fig. 2C, fig. S2B and S2C). The FP assay is a competitive displacement assay and therefore it provides apparent binding constants (Kapp). However, these apparent binding constants correlate well with the data obtained by ITC which measures direct interactions in solution and gives an actual (Kd) value (table S1).
Adaptation of the FP assay for HTS
Next, we optimized the FP assay for HTS and screened a ~130,000 small molecule library for compounds that displaced probe 1 from the T4 binding sites of TTR. The FP assay was performed in 384-well plates with low concentrations of probe 1 (1.5 nM) and TTR (50 nM) in a 10 μL assay volume. Detergent (0.01% Triton X-100) was added to the assay buffer to avoid false positive hits from aggregation of the small molecules. The assay demonstrated robust performance, with a, large dynamic range (~70–230 mP) and a Z′ factor (32, 33) in the range of 0.57–0.78 (fig. S3A and S3B).
Hits were defined as compounds, which resulted in at least 50% decrease in FP and demonstrated relative fluorescence between 70 and 130%. Many fluorescence quenchers and enhancers, which have less than 70% and greater than 130% total fluorescence relative to a control (compound without TTR), were excluded from the hit list. The excluded compounds have native fluorescence that is similar to fluorescein, which would interfere with the FP measurements and result in false positive hits. Two hundred compounds were designated as positive hits (0.167% hit rate). The top 33 compounds (compounds with lowest FP IC50) were assayed in a 10-point duplicate dose-response FP assay and displayed an IC50 (concentration that resulted in 50% decrease in the FP signal) between 0.277 and 10.957 μM (table S2).
Validation of the HTS hits
The top 33 compounds were retested with the FP assay (table S2) and with surface plasmon resonance (SPR) as another independent biophysical method. Solutions of the 33 hits were passed over immobilized, biotinylated TTR on a streptavidin coated chip. The binding of a small molecule to TTR on the sensor chip produces a SPR response signal (RU). The RU signal after addition of the top 33 compounds was measured and compared to a negative, solvent only, control. All compounds identified by the screen as hits were confirmed as TTR binders using SPR (fig. S4). We also found known TTR binders, such as NSAIDs (diclofenac, meclofenamic acid, and niflumic acid) and isoflavones (apigenin) in our screen (3, 34) (table S2). Among the best ligands (Fig. 2D) were the NSAID, niflumic acid, two catechol-O-methyl-tranferase (COMT) inhibitors, 3,5-dintrocatechol and Ro 41-0960 (35) and a number of compounds not previously known to bind to TTR. The chemical structures of these ligands were confirmed by 1H NMR and high-resolution mass spectrometry (HRMS) and the chemical purity was determined to be >95% (fig. S5).
Inhibition of TTR amyloidogenesis by the HTS hits
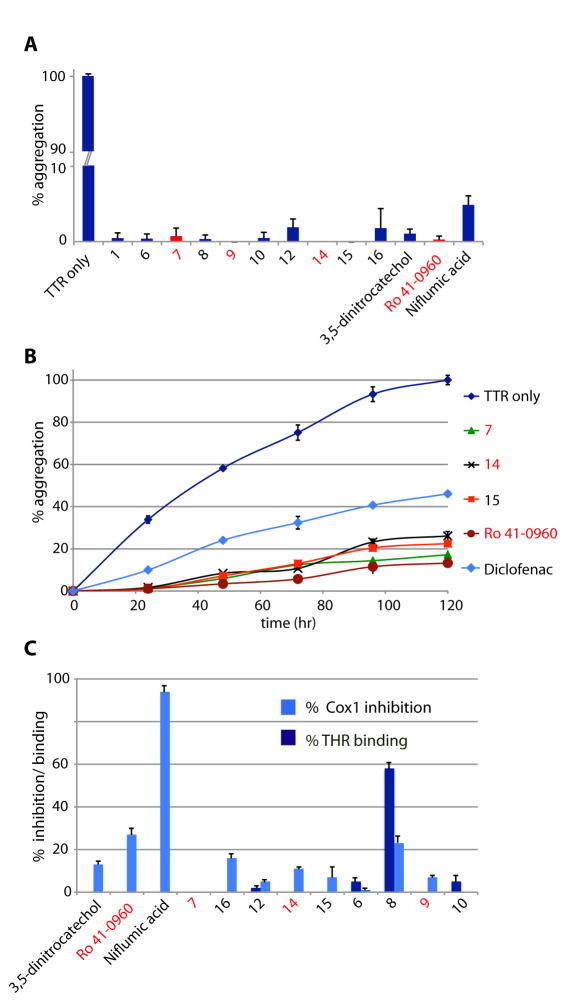
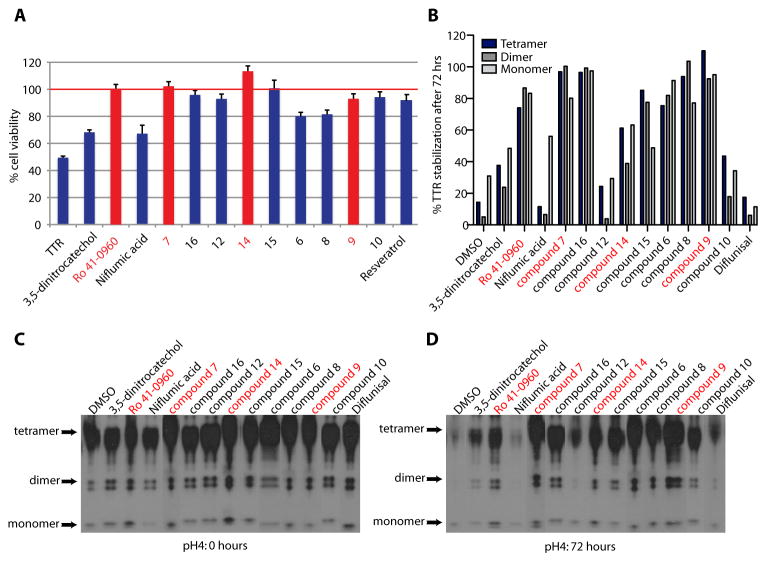
To test whether the new TTR ligands (7.2 μM) could function as kinetic stabilizers, we measured their ability to inhibit TTR (3.6 μM) amyloidogenesis at 72 hrs at pH 4.4 (fig. S6) (29). All 33 compounds inhibited TTR aggregation (<50% fibril formation, table S2). Of these, 23 were very good (<20% fibril formation) and 11 were excellent (<2% fibril formation) TTR kinetic stabilizers (Fig. 3A). All of the potent TTR stabilizers, except niflumic acid, and the two COMT inhibitors 3,5-dintrocatechol and Ro 41-0960, were chemical entities with no previously reported biological activity. Since occupancy of only one T4 binding site within TTR is sufficient for kinetic stabilization of the tetramer (3), we tested the most potent ligands at substoichiometric concentrations (2.4 fold molar excess of TTR relative to ligand) in a kinetic aggregation assay monitored over 5 days (Fig. 3B). Under these conditions ligands 7, 14, 15 and Ro 41-0960 dramatically slowed fibril formation and outperformed the known TTR stabilizer, diclofenac, which blocked only ~55% of TTR aggregation.
Fig. 3.
Evaluation of inhibition of TTR aggregation and binding to THR and COX-1 (A) Percentage of TTR (3.6 μM) fibril formation in the presence of ligands (7.2 μM) relative to aggregation in the absence of ligands (denoted 100%) after 72 hours. (B) Comparison of TTR (3.6 μM) aggregation inhibition in the presence of substoichiometric amounts of ligands (3.0 μM) compared to diclofenac. (C) THR binding and COX-1 inhibition data for the most potent TTR ligands. THR binding is expressed as % displacement of 125I-labeled triiodothyronine (T3) by test compounds (10 μM). COX-1 inhibition is shown as % inhibition of COX-1 mediated conversion by test compounds (10 μM). Error bars: SEM
Evaluating the TTR ligands for COX-1 enzymatic inhibition and binding to thyroid hormone receptor
A successful clinical candidate against TTR amyloid cardiomyopathy should have minimal off-target toxicity due to the potential need for life-long use of these drugs. Specifically, the TTR ligands should exhibit minimal binding to COX and the nuclear thyroid hormone receptor (THR). Inhibition of COX is contraindicated for treating FAC patients, since COX inhibition can not only lead to renal dysfunction and blood pressure elevation, but may precipitate heart failure in vulnerable individuals (20, 21, 24, 25). Therefore, the most potent TTR ligands were evaluated for their ability to inhibit COX-1 activity, as well as, for binding to THR, in comparison with the NSAID niflumic acid. Although niflumic acid exhibited substantial (94%) COX-1 inhibition, three of the 12 new compounds evaluated (7, 6 and 10) displayed less than 1% inhibition of COX-1. Only one ligand (compound 8) showed significant (58%) and two compounds (6 and 10) minor (5%) binding to THR (Fig. 3C).
Characterization of the binding energetics to TTR
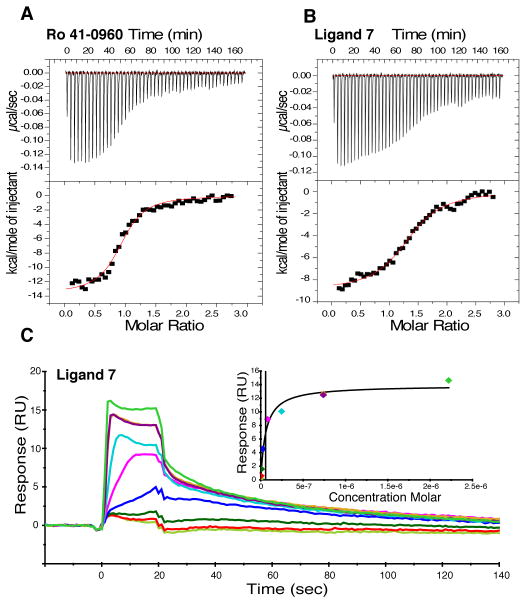
Many reported TTR ligands, including T4, bind TTR with negative cooperativity, which appears to arise from subtle conformational changes in TTR upon ligand binding to the first T4 site (3, 16, 36). We used ITC to determine the binding constants and to evaluate cooperativity between the two TTR T4 sites (Fig. 2A, Fig. 4A, Fig. 4B and fig. S1 and fig. S7). The ITC data for compounds 1, 7, 14, and Ro 41-0906 binding to TTR were fit to a two-site binding model and show that these potent ligands bind TTR with low nanomolar affinity. The dissociation constants for these ligands indicated that they bound TTR with negative cooperativity (table S3). Analysis of the free energies associated with ligand binding to TTR indicates that binding was driven both by burial of the hydrophobic ligand in the TTR binding site (which leads to the favorable binding entropies) and specific ligand-TTR interactions (which leads to the favorable binding enthalpies) (Fig. 2A, Fig. 4A, Fig. 4B, and fig. S7B) (37). The binding of compounds 7 (Kd1 = 58 nM and Kd2 = 500 nM) and 14 (Kd1 = 26 nM and Kd2 = 1800 nM) to TTR did not cause major conformational changes to the TTR tetramer structure (Fig. 5).
Fig. 4.
Assessment of the binding of ligands Ro 41-0960 and 7 to TTR using ITC and SPR. Calorimetric titration of (A) Ro 41-0960 (Kd,1 = 15 nM and Kd,2 = 2000 nM) and (B) 7 (Kd,1 = 58 nM and Kd,2 = 500 nM) against TTR. Raw data (top) and integrated heats (bottom) from the titration of TTR (2 μM) with ligand (25 μM). The solid red line through the integrated heats represents the best fit binding isotherm to a two-site binding model. (C) SPR sensogram showing concentration-dependent binding of ligand 7 to TTR (0.001 μM to 2.2 μM). Normalized RUs are plotted over a time course. Equilibrium binding analysis (inset) indicates an apparent Kd of 57.91 ± 13.2 nM (SD).
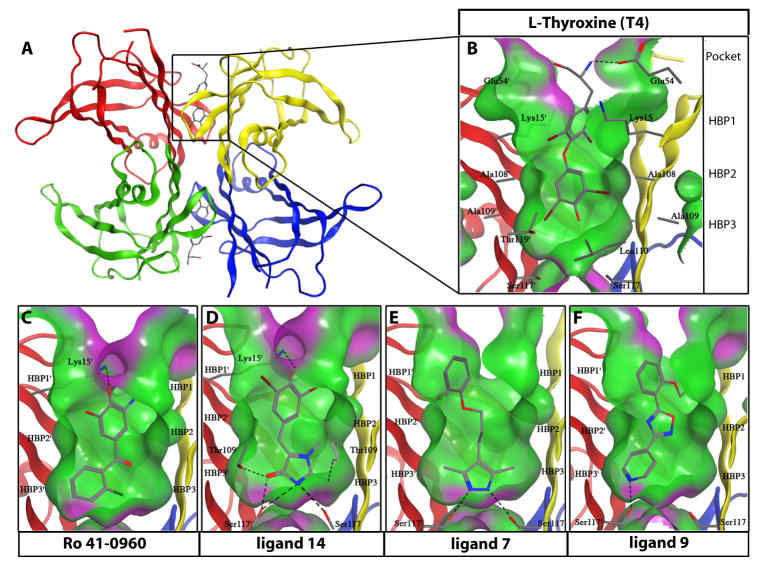
Fig. 5.
Crystal structures of WT-TTR bound to T4, Ro 41-0960, 14, 7, and 9 (A) Ribbon diagram of WT-TTR with T4 bound to each of the two T4 binding pockets within the tetramer (monomer subunits individually colored). (B) Expanded view of T4 within the binding site with a “Connelly” analytical molecular surface applied to residues within 8 Å of ligand (green = hydrophobic, purple = polar). (C to F) Expanded views of Ro 41-0960, 14, 7, and 9 bound to TTR. Hydrogen bonds are represented as dashed lines between functional groups. The innermost halogen binding pockets (HBPs) 3 and 3′ are composed of the methyl and methylene groups of Ser117/117′, Thr119/119′, and Leu110/110′. HBPs 2 and 2′ are assembled from the side chains of Leu110/110′, Ala109/109′, Lys15/15′, and Leu17/17′. The outermost HBPs 1 and 1′ (labeled in a) are lined by the methyl and methylene groups of Lys15/15′, Ala108/108′, and Thr106/106 ′ (Thr 106 not shown).
We explored the binding kinetics of the potent TTR ligands 7 and Ro 41-0960 by SPR (Fig. 4C and fig. S4B). Ligand 7 exhibited concentration-dependent binding to TTR with a Kd of 57.91± 13.2 nM, determined by fitting steady state data (Fig. 4C) (38). This affinity was similar to the one obtained by ITC (Kd,1 = 58 nM). Measurement of the binding kinetics of ligand 7 to TTR gave a similar binding constant (Kd = 20.22 ± 2.04 nM) to the one determined from the steady state data. Target residence time (τ), the reciprocal of koff, differentiates between transient and long-lived ligand-protein complexes. Compounds that dissociate from their target slowly have a longer τ, allowing less frequent administration. Ligand 7 had favorable binding kinetics to TTR with rapid on-rate and a slow off-rate (longer τ) (kon = 1.29 × 106 ± 1.3 × 105 M−1s−1 and koff = 0.026 ± 0.001 s−1). Ro 41-0960 also showed similar binding kinetics as ligand 7 (Kd = 56.05 ± 4.14 nM, kon = 2.35 × 106 ± 1.5 × 105 M−1s−1 and koff = 0.132 ± 0.009 s−1), although the koff was ~5x faster (fig. S4B). In contrast, NSAIDs such as niflumic acid and diclofenac displayed a much faster dissociation from TTR (koff = 0.523 s−1 and 1.01 s−1, respectively) compared to ligand 7 or Ro 41-0960 (~ 20x or 40x lower target residence time than ligand 7) (fig. S4C and fig. S4D).
Crystal structures of WT • Ligand complexes
The crystal structures of WT-TTR in complex with Ro 41-0960, 7, 9 and 14 were determined to 1.25, 1.45, 1.50 and 1.60-Å resolutions, respectively (Fig. 5). Known TTR kinetic stabilizers are composed of two substituted aryl rings connected by a linker (39), but the FP-based HTS screen identified molecules with distinct ring systems (Fig. 2D). The 2-imino-1-methylimidazolidin-4-one ring of 14, the 3,5-dimethyl-1H-pyrazole ring of 7, and the pyridine ring of 9, all sat within the inner cavity of the T4 binding site, and were able to hydrogen bond with the Ser117 and Thr109 that comprise the innermost boundary of the T4 binding site (Fig. 5C–F). The 3,5-methyl groups of 7 extended into the two symmetrical HBPs, 3 and 3′. In doing so, the two amino nitrogens of the pyrazole ring formed hydrogen bonds with Ser117 and 117′ similar to the single H-bond made by the pyridine ring of 9, while the 2-imino-1-methylimidazolidin-4-one ring of 14 formed the most extensive, hydrogen-bond network with Thr109 and 109′ and Ser117 and 117. ′ The aforementioned hydrogen-bonds tethered these ring systems in position (Fig. 5C–F). Although it has been reported that hydrophobic linkers are preferred for ligand binding within HBP2 and 2′, the compounds identified here had diverse linkers that enable the adjoined rings to have sufficient flexibility to make optimal interactions with HBPs 1,1′ and 3,3′ (39). The 2-fluoro substituted phenyl ring of 7 and the ethoxy benzyl ring of 9 occupied the outer binding cavity, placing a portion of the aryl ring into HBP1 or 1′. The 3,4-dihydroxy-5-nitrophenyl and 3,5-dibromo-phenyl rings of Ro 41-0960 and 14, respectively, displayed an acidic phenol, which was able to make electrostatic interactions with Lys15 and 15′ through a putative phenolate -LysNH3+ bridge (30). This interaction closed the T4 pocket around the inhibitor (Fig. 5C and 5D).
This combination of the electrostatic and hydrophobic interactions bridged and stabilized the two adjacent TTR subunits across the weaker TTR dimer–dimer interface, resulting in native state stabilization (3), which subsequently increased the activation energy for tetramer dissociation, the rate-limiting step in TTR amyloidogenesis cascade (3, 17, 18). The structural analysis of WT-TTR in complex with 14 (Fig. 5D) suggested why this compound was a very strong TTR kinetic stabilizer, as it made multiple hydrogen bonds with TTR within the T4 binding site, while engaging in electrostatic interactions with Lys15 and Lys15′ at the periphery of the pocket.
Kinetic stabilizers prevent TTR-induced cytotoxicity against human cardiac and neuronal cell lines
Because, there are no mouse models of V122I-TTR FAC, we turned to a recently developed in vitro cell-based assay, which employs the human cardiac cell line AC16 (40). The AC16 cell line was generated from adult ventricular cardiomyocytes (41), which is one of the sites of TTR amyloid deposition. This cellular model is currently the patho-physiologically most relevant tissue culture model for SSA and FAC. Treatment of AC16 cells with the destabilized V122I TTR variant decreased cell viability to 49±1% relative to vehicle treated cells (100% viability). Most of the ligands identified by our HTS effectively prevented the proteotoxicity of V122I TTR towards AC16 cells (Fig. 6A). We also assessed the toxicity of WT-TTR towards human neuroblastoma cell line IMR-32, as an in vitro a model for FAP (42), which provided similar results (fig. S8B). Ro 41-0960 and ligands 7, 14 and 15 completely rescued AC16 cell viability (Fig. 6A). None of the compounds identified in the HTS were cytotoxic to AC16 cells or to IMR-32 cells at the concentration tested (fig. S8A and fig. S8C). Unexpectedly, despite their potency as TTR stabilizers and lack of cellular toxicity (Fig. 3A and fig. S8A and fig. S8C), the COMT inhibitor, 3,5-dinitrocatechol, and the NSAID, niflumic acid, performed poorly in the cell culture system (Fig. 6A and fig. S8B).
Fig. 6.
Inhibition of V1221I TTR cytotoxicity and stabilization of human serum TTR. (A) Inhibition of V1221I TTR cytotoxicity towards AC16 cells. TTR and ligands concentration: 8 μM. Resveratrol is a positive control. Cell viability is shown relative to cells treated with vehicle only. Error bars: SEM (B) Quantitative analysis of TTR serum stability after 72 hours. (C&D) Stabilization of human serum TTR. Serum TTR in the presence and absence 50 μM compounds was subjected to acid denaturation. TTR immunoblot at 0 (C) and 72 (D) hours after acidification of serum
Stabilization of serum TTR against acid-mediated dissociation and amyloidogenesis
To stabilize the TTR tetramer and thus prevent amyloid fibril formation in FAC or SSA, small molecules must be able to selectively bind to TTR in human serum over other serum proteins. Compounds that bind to TTR selectively in serum are the best candidates for further evaluation. The 12 most potent ligands, identified in this HTS, were tested for their ability to stabilize TTR in human serum (Fig. 6B–D). Since TTR must first dissociate into monomers in order to denature and aggregate, we tested how much intact TTR tetramer remained after 72 hours of acid treatment in the presence and absence of 50 μM TTR ligands (TTR concentration in human serum is ~ 3–5 μM). We compared our TTR ligands to diflunisal, which had previously been shown to inhibit acid-mediated tetramer dissociation and aggregation of serum wild type and FAP-mutant TTR (43). Seven of the twelve newly identified TTR ligands almost fully stabilized serum TTR (% tetramer at 72 hours: 75.5 - 100% of tetramer present at 0 hours) at 50 μM (Fig. 6B–D). At 50 μM, diflunisal (%tetramer at 72hrs: 17.5%) and niflumic acid (%tetramer at 72hrs: 11.5%), as well as 3,5-dinitrocatechol (%tetramer at 72hrs: 37.6%), compound 10 (%tetramer at 72hrs: 43.6%) and compound 12 (%tetramer at 72hrs: 24.3%), were unable to completely stabilize TTR. At 200 μM, which is within the therapeutic concentration range of diflunisal (100–200 μM), all tested compounds stabilized TTR (fig. S9).
DISCUSSION
Amyloidosis is a systemic disease caused by structural alteration of proteins that tend to precipitate in the extracellular space as insoluble fibrils. FAC is a hereditary TTR-related systemic amyloidosis with predominant cardiac involvement resulting from myocardial infiltration of amyloid protein. There are currently no FDA-approved drugs for prevention or treatment of TTR-induced amyloid cardiomyopathy and the therapy for most patients is confined to symptomatic relief. Liver transplantation, which removes the source of the pathologic protein, has been the treatment of choice for hereditary TTR amyloidoses. Heart transplantation is performed as a palliative measure for a subset of FAC patients who display predominately cardiac pathology and only mild systemic involvement (44,45).
Small molecules targeting the formation, clearance, or assembly of toxic aggregates provide a promising strategy to treat amyloidoses (3, 16, 46). A number of disease causing mutations (e.g., L55P, V30M, and V122I) influence the thermodynamic stability and the kinetics of dissociation of the TTR tetramer in vitro, and these properties of the variant tetramers appear to be correlated to the severity of the resulting disease (47,48). Moreover, suppressor mutations in TTR (e.g., T119M) that kinetically stabilize the native tetramer and protect against the development of pathology in compound heterozygotes carrying a disease-associated mutation, have been identified (49).
Binding of small molecule ligands to the unoccupied thyroxine (T4) binding sites of TTR stabilizes the TTR tetramer and slows tetramer dissociation and amyloidogenesis in vitro. These kinetic stabilizers could potentially be used as small molecule therapeutics for the treatment of TTR amyloidoses (for review see: (16, 46,50).
Recently clinical trials of TTR kinetic stabilizers have been initiated in FAP patients. The two compounds, which are being tested are the NSAID, diflunisal (19), and the benzoxazole, tafamidis (51). Due to the lack of animal models that faithfully reproduce the pathology of human TTR-mediated familial amyloidoses, the efficacy of TTR kinetic stabilizers was assessed in human FAP patients. Several transgenic mouse models that overexpress human mutant TTR (V30M) do not reproduce the tissue distribution of TTR deposits observed in peripheral nerves of human FAP patients carrying the same mutation (52,53). No mouse models that express transgenic human V122I TTR have been generated. In addition, the level of TTR overexpression required to observe TTR amyloid deposits in mice is very high. Because of the stoichiometric requirements of small molecule to TTR tetramer the concentrations of drug required to stabilize transgenic human TTR are difficult to achieve in these mouse models (2,3). Almost all existing TTR kinetic stabilizers were identified by structure-based drug designs and time consuming assays, performed under non-physiological binding conditions. One such assay uses the displacement of radioactively-labeled T4 from TTR (28). However, the use of radioactivity makes it unsuitable for HTS. Another assay is based on the ability of TTR binders to stabilize TTR during acid-mediated aggregation (29). This is a simple but cumbersome assay (72 hours), which requires long incubation times, large amounts of protein and little kinetic information about ligand binding. In contrast, the FP-based assay we developed provided a simple, sensitive, and robust method that detected the binding of small molecules to the T4 binding pocket of TTR under physiological conditions.
Preliminary results indicate that probe 1 bound to TTR in human serum (fig. S10). The measurement of TTR concentrations in blood samples is clinically used as a sensitive indicator of the nutritional status of patients. The half-life of TTR in humans in vivo is about 2–4 days, which is much shorter than that of other nutritional markers such as albumin (approximately 20 days). Therefore, TTR concentration is more sensitive to changes in protein-energy status than albumin as it reflects recent dietary intake rather than overall nutritional status. In addition to being an excellent small molecule screening tool, this new FP assay has the potential to be developed as an alternative to the more error-prone immunoprecipitation-turbidity assay that is used by many clinical laboratories to determine plasma TTR concentrations (54).
The FP assay was adapted for HTS and enabled us to identify a variety of highly potent and structurally diverse TTR kinetic stabilizers. Most TTR ligands up to date were identified by rational design. Consequently their chemical diversity is limited and many have COX-inhibitory activity. Genomic variations in the human population are known to influence the response to pharmacotherapy. Hence, in the current era of pharmacogenomics it seems desirable not to limit disease therapy to a “one drug fits all” system. Unlike many previously reported halogenated TTR ligands, most of the new chemical TTR binding scaffolds we discovered using this HTS, had minimal COX-1 or THR activity. Many of the known kinetic stabilizers of TTR, including diflunisal, which has been in clinical trials for FAP since 2006 (19), show inhibitory activity towards COX. While there is never a simple way to predict whether a compound will be effective in all patients or whether individuals will suffer from adverse side effects, it is well known that COX inhibitors are associated with a number of adverse side effects, especially in patients with already compromised cardiac function, which makes their use in patients suffering from cardiomyopathy problematic. These adverse reactions include renal dysfunction and elevated blood pressure and they may precipitate heart failure in vulnerable individuals (20, 21, 24, 25, 55).
Our newly identified TTR kinetic stabilizers were able to rescue cardiomyocytes from the proteotoxicity of TTR aggregates thought to cause FAC and SSA in patients. Thus these compounds offer additional classes of highly effective TTR tetramer stabilizers. Several of the new TTR ligands we identified appeared to be very selective for TTR in human serum, where they stabilized human TTR even in the presence of all other serum proteins such as albumin. Eight of the twelve ligands we tested were more potent TTR stabilizers (at 50 μM) than diflunisal, which is currently in clinical trials for the treatment of FAP. Binding to serum proteins is an important factor in determining the overall distribution, metabolism, activity, and toxicity of a drug. When ligands exhibit excellent TTR amyloid fibril inhibition data in vitro, yet display poor serum selectivity, it is assumed that they prefer to bind to the drug-binding sites in albumin and/or similar sites in other proteins found in plasma. It is unlikely that such promiscuous inhibitors will prevent TTR misfolding and amyloidosis in a complex environment like human blood or CSF.
The ligands we identified should be effective against SSA and FAC, because they bound both WT and V122I mutant TTR to kinetically stabilize tetramers. In conclusion, we discovered a number of new chemical scaffolds that were able to stabilize wt and mutant TTR and our data provide the incentive to further evaluate the efficacy of the most selective and potent ligands against FAC and SSA. No FDA-approved drugs or accepted therapeutic strategies are at this time available for treating SSA and FAC. Hence, we envision that several of the new TTR kinetic stabilizers described in this manuscript might be promising leads to be developed as therapeutics for FAC and SSA.
Supplementary Material
Fig. S1. Calorimetric titration of ligands against TTR.
Fig. S2. Displacement FP assay of TTR ligands.
Fig. S3. Dynamic range and Z′ factor for the FP assay.
Fig. S4. Confirmation and assessment of ligands binding to TTR using SPR.
Fig. S5. 1H NMR (chemical shifts and spectra), HRMS, and HPLC traces for HTS hits.
Fig. S6. Cartoon of TTR fibril formation assay.
Fig. S7. Calorimetric titration of ligands against TTR.
Fig. S8. Cytotoxicity of TTR ligands (8 μM) towards AC16 cardiac cells and inhibition of WT-TTR cytotoxicity towards IMR-32 neuroblastoma cells by the TTR ligands.
Fig. S9. Stabilization of serum TTR against acid-mediated denaturation.
Fig. S10. Fluorescence polarization of probe binding to human serum TTR.
Fig. S11. Structure and crystal packing of TTR in complex with 14.
Table S1. Affinity of ligands as determined by ITC and FP.
Table S2. Chemical structures of TTR ligands. aTTR amyloidogenesis inhibition activity (%fibril formation). bIC50 (compound concentration that resulted in 50% decrease in the FP signal).
Table S3. Isothermal Titration Calorimetry (ITC) analysis of ligands binding to human wild type TTR.
Table S4. X-ray data collection and refinement statistics.
Table S5. Summary of small molecule screening data.
Tables S6. Information summary about TTR FP Probe 1.
Acknowledgments
We thank; D. Solow-Cordero for help with the HTS, G. Crabtree for critical reading of the manuscript, C. Garcia for use of the Biacore T100, Alexandra Esteras-Chopo for advice and M. Davidson for providing the AC16 cell line.
Funding: This work was supported by the Hillblom Foundation (IAG), NIH Grants 5PN2EY016525 (IAG), DK 46335 (JWK), AI 42266 (IAW), by the Skaggs Institute of Chemical Biology (JWK, IAW).
Footnotes
Author Contributions: M.M.A. designed and performed most experiments, S.C. performed crystallographic structure determination, A.C peformed the serum TTR stabilization. N.R. performed the cell-based assays. E.T.P. analyzed the ITC data. D.W.P. helped with probe synthesis. I.A.W. supervised the crystallographic work. J.W.K. supervised the work, S.C., N.R., I.A.W. and J.W.K. edited the paper. I.A.G supervised the work, M.M.A. and I.A.G prepared the manuscript.
Competing interests: The authors declare that they have no competing interests.
Accession codes: Atomic coordinates have been deposited in the RCSB Protein Data Bank and are available under accession codes 3P3R (Ro-41-0960), 3P3S (14), 3P3T (7) and 3P3U (9).
REFERENCES AND NOTES
- 1.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 2.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 3.Johnson SM, et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 4.Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 5.Falk RH, Comenzo RL, Skinner M. The systemic amyloidoses. N Engl J Med. 1997;337:898–909. doi: 10.1056/NEJM199709253371306. [DOI] [PubMed] [Google Scholar]
- 6.Connors LH, Lim A, Prokaeva T, Roskens VA, Costello CE. Tabulation of human transthyretin (TTR) variants. Amyloid. 2003;10:160–184. doi: 10.3109/13506120308998998. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson DR, Gorevic PD, Buxbaum JN. A homozygous transthyretin variant associated with senile systemic amyloidosis: evidence for a late-onset disease of genetic etiology. Am J Hum Genet. 1990;47:127–136. [PMC free article] [PubMed] [Google Scholar]
- 8.Shah KB, Inoue Y, Mehra MR. Amyloidosis and the heart: a comprehensive review. Arch Intern Med. 2006;166:1805–1813. doi: 10.1001/archinte.166.17.1805. [DOI] [PubMed] [Google Scholar]
- 9.Desai HV, Aronow WS, Peterson SJ, Frishman WH. Cardiac amyloidosis: approaches to diagnosis and management. Cardiol Rev. 2010;18:1–11. doi: 10.1097/CRD.0b013e3181bdba8f. [DOI] [PubMed] [Google Scholar]
- 10.Westermark P, Sletten K, Johansson B, Cornwell GG., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci USA. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapezzi C, et al. Transthyretin-related amyloidoses and the heart: a clinical overview. Nat Rev Cardiol. 2010;7:398–408. doi: 10.1038/nrcardio.2010.67. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson DR, et al. Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black Americans. N Engl J Med. 1997;336:466–473. doi: 10.1056/NEJM199702133360703. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X, Buxbaum JN, Kelly JW. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc Natl Acad Sci USA. 2001;98:14943–14948. doi: 10.1073/pnas.261419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connors LH, et al. Cardiac amyloidosis in African Americans: comparison of clinical and laboratory features of transthyretin V122I amyloidosis and immunoglobulin light chain amyloidosis. Am Heart J. 2009;158:607–614. doi: 10.1016/j.ahj.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Buxbaum J, et al. Significance of the amyloidogenic transthyretin Val 122 Ile allele in African Americans in the Arteriosclerosis Risk in Communities (ARIC) and Cardiovascular Health (CHS) Studies. Am Heart J. 2010;159:864–870. doi: 10.1016/j.ahj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr Opin Struct Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammarstrom P, Schneider F, Kelly JW. Trans-suppression of misfolding in an amyloid disease. Science. 2001;293:2459–2462. doi: 10.1126/science.1062245. [DOI] [PubMed] [Google Scholar]
- 18.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 19.http://clinicaltrials.gov/ct2/show/NCT00294671
- 20.Kearney PM, Baigent C, Godwin J, Halls H, Emberson JR, Patrono C. Do selective cyclo-oxygenase-2 inhibitors and traditional non-steroidal anti-inflammatory drugs increase the risk of atherothrombosis? Meta-analysis of randomised trials. BMJ. 2006;332:1302–1308. doi: 10.1136/bmj.332.7553.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein M. J. Non-steroidal anti-inflammatory drugs and the continuum of renal dysfunction. Hypertens. 2002;(Suppl 20):S17–23. [PubMed] [Google Scholar]
- 22.Marnett LJ, Kalgutka AS. Cyclooxygenase 2 inhibitors: discovery, selectivity and the future. Trends Pharmacol Sci. 1999;20:465–469. doi: 10.1016/s0165-6147(99)01385-1. [DOI] [PubMed] [Google Scholar]
- 23.Wallace JL. Pathogenesis of NSAID-induced gastroduodenal mucosal injury. Best Pract Res Clin Gastroenterol. 2001;15:691–703. doi: 10.1053/bega.2001.0229. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. JAMA. 2001;286:954–959. doi: 10.1001/jama.286.8.954. [DOI] [PubMed] [Google Scholar]
- 25.Page J, Henry D. Consumption of NSAIDs and the development of congestive heart failure in elderly patients: an underrecognized public health problem. Arch Intern Med. 2000;160:777–784. doi: 10.1001/archinte.160.6.777. [DOI] [PubMed] [Google Scholar]
- 26.Kirchheiner J, Brockmöller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues AD. Impact of CYP2C9 genotype on pharmacokinetics: are all cyclooxygenase inhibitors the same? Drug Metab Dispos. 2005;33:1567–1575. doi: 10.1124/dmd.105.006452. [DOI] [PubMed] [Google Scholar]
- 28.Prapunpoj P, Leelawatwatana L, Schreiber G, Richardson SJ. Change in structure of the N-terminal region of transthyretin produces change in affinity of transthyretin to T4 and T3. FEBS J. 2006;273:4013–4023. doi: 10.1111/j.1742-4658.2006.05404.x. [DOI] [PubMed] [Google Scholar]
- 29.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 30.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. Diflunisal analogues stabilize the native state of transthyretin. Potent inhibition of amyloidogenesis. J Med Chem. 2004;47:355–374. doi: 10.1021/jm030347n. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman RL, et al. Kinetic stabilization of an oligomeric protein by a single ligand binding event. J Am Chem Soc. 2005;127:5540–5551. doi: 10.1021/ja042929f. [DOI] [PubMed] [Google Scholar]
- 32.Threshold value: mean signal of negative/positive control plus/minus three times their standard deviation; Separation band (S): difference between the positive and negative thresholds values; Dynamic range (R): difference between the positive and negative means. Z-factor (= S/R): a value between 0.5 and 1 indicates an excellent screening assay.
- 33.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 34.Green NS, Foss TR, Kelly JW. Genistein, a natural product from soy, is a potent inhibitor of transthyretin amyloidosis. Proc Natl Acad Sci USA. 2005;102:14545–14550. doi: 10.1073/pnas.0501609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borgulya J, Bruderer H, Bernauer K, Zürcher G, Da Prada M. Catechol-O-methyltransferase-Inhibiting Pyrocatechol Derivatives: Synthesis and Structure-Activity Studies. Helvetica Chimica Acta. 1989;72:952–968. [Google Scholar]
- 36.Trivella DB, et al. Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: Implications to tetramer stability and ligand-binding. J Struct Biol. 2010;170:522–531. doi: 10.1016/j.jsb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 38.We note here that the ability of TTR to bind to two ligands complicates the analysis of our SPR data: in principle, there should be two on-rates and two off-rates for ligand binding, requiring a double exponential model to fit the data. Since we used monoexponentials, our reported rate constants should be considered estimates for the slower of the two binding and dissociation events.
- 39.Johnson SM, Connelly S, Wilson IA, Kelly JW. Toward optimization of the linker substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies. J Med Chem. 2008;51:6348–6358. doi: 10.1021/jm800435s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourgault S, Choi S, Buxbaum JN, Kelly JW, Price JL, Reixach N. Mechanisms of transthyretin cardiomyocyte toxicity inhibition by resveratrol analogs. Biochem Biophys Res Commun. 2011 doi: 10.1016/j.bbrc.2011.04.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson MM, et al. Novel cell lines derived from adult human ventricular cardiomyocytes. J Mol Cell Cardiol. 2005;39:133–147. doi: 10.1016/j.yjmcc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Reixach N, Deechongkit S, Jiang X, Kelly JW, Buxbaum JN. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc Natl Acad Sci USA. 2004;101:2817–2822. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller SR, Sekijima Y, Kelly JW. Native state stabilization by NSAIDs inhibits transthyretin amyloidogenesis from the most common familial disease variants. Lab Invest. 2004;84:545–552. doi: 10.1038/labinvest.3700059. [DOI] [PubMed] [Google Scholar]
- 44.Jonsen E, Suhr OB, Tashima K, Athlin E. Early liver transplantation is essential for familial amyloidotic polyneuropathy patients’ quality of life. Amyloid. 2001;8:52–57. doi: 10.3109/13506120108993814. [DOI] [PubMed] [Google Scholar]
- 45.Hamour IM, et al. Heart transplantation for homozygous familial transthyretin (TTR) V122I cardiac amyloidosis. Am J Transplant. 2008;8:1056–1059. doi: 10.1111/j.1600-6143.2008.02162.x. [DOI] [PubMed] [Google Scholar]
- 46.Almeida MR, Gales L, Damas AM, Cardoso I, Saraiva MJ. Small transthyretin TTR) ligands as possible therapeutic agents in TTR amyloidoses. Curr Drug Targets CNS Neurol Disord. 2005;4:587–596. doi: 10.2174/156800705774322076. [DOI] [PubMed] [Google Scholar]
- 47.McCutchen SL, Colon W, Kelly JW. Transthyretin mutation Leu-55-Pro significantly alters tetramer stability and increases amyloidogenicity. Biochemistry. 1993;32:12119–12127. doi: 10.1021/bi00096a024. [DOI] [PubMed] [Google Scholar]
- 48.Quintas A, Saraiva MJ, Brito RM. The amyloidogenic potential of transthyretin variants correlates with their tendency to aggregate in solution. FEBS Lett. 1997;418:297– 300. doi: 10.1016/s0014-5793(97)01398-7. [DOI] [PubMed] [Google Scholar]
- 49.Coelho T, et al. A strikingly benign evolution of FAP in an individual found to be a compound heterozygote for two TTR mutations: TTR Met 30 and TTR Met 119. J Rheumatol. 1993;120:179. [Google Scholar]
- 50.Kolstoe SE, Wood SP. Drug targets for amyloidosis. Biochem Soc Trans. 2010;38:466–470. doi: 10.1042/BST0380466. [DOI] [PubMed] [Google Scholar]
- 51.http://clinicaltrials.gov/ct2/show/ NCT00409175, NCT00791492
- 52.Yi S, Takahashi K, Naito M, Tashiro F, Wakasugi S, Maeda S, Shimada K, Yamamura K, Araki S. Systemic amyloidosis in transgenic mice carrying the human mutant transthyretin (Met30) gene. Pathologic similarity to human familial amyloidotic polyneuropathy, type I. Am J Pathol. 1991;138:403–12. [PMC free article] [PubMed] [Google Scholar]
- 53.Kohno K, Palha JA, Miyakawa K, Saraiva MJ, Ito S, Mabuchi T, Blaner WS, Iijima H, Tsukahara S, Episkopou V, Gottesman ME, Shimada K, Takahashi K, Yamamura K, Maeda S. Analysis of amyloid deposition in a transgenic mouse model of homozygous familial amyloidotic polyneuropathy. Am J Pathol. 1997;150:1497–1508. [PMC free article] [PubMed] [Google Scholar]
- 54.Ingenbleek Y, Young V. Transthyretin (prealbumin) in health and disease: nutritional implications. Annu Rev Nutr. 1994;14:495–533. doi: 10.1146/annurev.nu.14.070194.002431. [DOI] [PubMed] [Google Scholar]
- 55.Fauci AS, Braunwald E, Kasper DL, Hauser SL, Longo DL, Jameson JL, Loscalzo J. Harrison’s Principles of Internal Medicine. 17. Vol. 1448. 2008. p. 1763.p. 1770. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Calorimetric titration of ligands against TTR.
Fig. S2. Displacement FP assay of TTR ligands.
Fig. S3. Dynamic range and Z′ factor for the FP assay.
Fig. S4. Confirmation and assessment of ligands binding to TTR using SPR.
Fig. S5. 1H NMR (chemical shifts and spectra), HRMS, and HPLC traces for HTS hits.
Fig. S6. Cartoon of TTR fibril formation assay.
Fig. S7. Calorimetric titration of ligands against TTR.
Fig. S8. Cytotoxicity of TTR ligands (8 μM) towards AC16 cardiac cells and inhibition of WT-TTR cytotoxicity towards IMR-32 neuroblastoma cells by the TTR ligands.
Fig. S9. Stabilization of serum TTR against acid-mediated denaturation.
Fig. S10. Fluorescence polarization of probe binding to human serum TTR.
Fig. S11. Structure and crystal packing of TTR in complex with 14.
Table S1. Affinity of ligands as determined by ITC and FP.
Table S2. Chemical structures of TTR ligands. aTTR amyloidogenesis inhibition activity (%fibril formation). bIC50 (compound concentration that resulted in 50% decrease in the FP signal).
Table S3. Isothermal Titration Calorimetry (ITC) analysis of ligands binding to human wild type TTR.
Table S4. X-ray data collection and refinement statistics.
Table S5. Summary of small molecule screening data.
Tables S6. Information summary about TTR FP Probe 1.