Abstract
A Papua New Guinea collection of the marine cyanobacterium cf. Lyngbya sordida yielded three known compounds as well as a new PKS-NRPS-derived malyngamide with anti-inflammatory and cytotoxic activity. Malyngamide 2 features an extensively oxidized cyclohexanone ring. Resolution of the ring core as a 6,8,9-triol rather then a 7,8,9-triol and relative configuration was based on chemical shift and bond geometry modeling in conjunction with homonuclear and heteronuclear coupling constants, NOE and ROE correlations, and other structural information. Malyngamide 2 exhibited anti-inflammatory activity in LPS-induced RAW macrophage cells (IC50 = 8.0 μM) with only modest cytotoxicity to the mammalian cell line.
Various species of the mostly marine cyanobacterial genus Lyngbya are renowned for their capacity to produce secondary metabolites of many different structural classes.1 One such natural product class with many members is the malyngamides, a group easily recognized by their combination of an unsaturated and methoxy-bearing fatty acid with a highly crafted and functionalized amine portion. The majority of the malyngamides isolated to date have been obtained from collections of Lyngbya species, although a few were isolated from the sea hares Bursatella leachii (malyngamides X, S) and Stylocheilus longicauda (malyngamides O, P).2,3 In addition, malyngamides M and N were isolated from the red alga Gracilaria coronopifolia.4 However, it is likely that the original natural product sources in these latter cases were also Lyngbya species as these cyanobacteria are reported to be eaten by Bursatella and Stylocheilus,2,3 and form epiphytic associations with Gracilaria.4
Lyngbic acid, the methoxy-containing unsaturated fatty acid portion of most malyngamides, likely derives from a PKS pathway with only a few variations having been observed which involve its chain length (12-20 carbons).5 Thus, the principal variable domain of the malyngamides is the polar head group, usually a multiply functionalized cyclohexyl ring, which is attached to the lyngbic acid portion through an amide or N-methyl amide linkage. Additionally, a branching vinyl group with chlorine as a substituent, as found in the jamaicamides and recently shown to derive from a novel β-branch forming reaction,6 is often present in the head group. The malyngamides possess a broad spectrum of biological activities, including antimicrobial, anti-inflammatory, cytotoxicity, and anti-mycobacterial.5 Here, we report our discovery of a new member of the malyngamide family, malyngamide 27 (1), from a Papua New Guinea collection of cf. L. sordida (synonym: L. polychroa), a relatively unstudied species of Lyngbya.8 Malyngamide 2 exhibits promising anti-inflammatory activity in the nitric oxide production assay (IC50 = 8.0 μM) with only modest cytotoxic properties (IC50 = 27.3 μM).
Pale red tufts of the benthic cyanobacterium cf. Lyngbya sordida were collected from a depth of 3 to 5 m by SCUBA near Dutchess Island, Papua New Guinea, in 2002. The lipophilic extract was separated by normal phase silica vacuum liquid chromatography to generate nine subfractions. The eighth fraction harbored compounds exhibiting signficanct activity in a cytotoxicity assay and was subsequently subjected to RP C18 SPE cartridge purification and reversed-phase HPLC. A mixture of majusculamides A and B, wewakazole and malyngamide 2 (1) were eluted as partially purified compounds; the two latter compounds were further purified using analytical HPLC. The known compounds were identified by comparison of their respective analytical data sets in comparison with literature values,9,10 whereas malyngamide 2 (1) was determined to be a new compound through dereplication using the MarinLit database program.
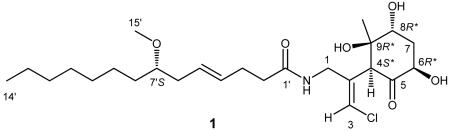
Malyngamide 2 (1) gave an [M+H]+ m/z 488.2757 which established the molecular formula as C25H42 ClNO6 with five degrees of unsaturation. The 1H NMR spectrum was well dispersed and contained several proton resonances that are signatures for the malyngamide structure class. Specifically, a singlet proton resonance at δH 6.31 (δC 121.1) was diagnostic of a vinyl chloride, a two proton multiplet at δH 5.49 (δC = 128.3, 130.6) indicated a disubstituted olefin, a three proton singlet at δH 3.32 (δC 56.4) suggested a methoxy group, and resonances at δH 6.54 and δC 173.5 indicated a secondary amide, and were all in accord with published spectroscopic data for the malyngamide series. Ensuing 1D and 2D NMR experiments confirmed the presence of lyngbic acid, 7S-methoxytetradec-4(E)-enoic acid, as a substructure of compound 1.2-4,11
Having established the lyngbic acid portion of compound 1, the polar head group was defined as a fragment of C10H15ClNO4 composition, and thus was more highly oxidized than any previously described malyngamide. The vinyl chloride appendage was connected in proximity to the amide functionality, consistent with other malyngamides, via a COSY correlation between the amide proton and the methylene protons at C-1 in combination with reciprocal H-1/C-3 and H-3/C-1 HMBC correlations. Thus, with the linear portion of the molecule accounting for three degrees of unsaturation, the remaining two degrees were assigned to a carbonyl C-5 (δC =209.6) and a ring structure, again consistent with the malyngamide class. Moreover, this atom accounting required the final three oxygen atoms to be present as hydroxy groups, and defined the ring as a cyclohexanone. From consideration of the unassigned remaining carbon shifts, the substituents on the ring were thus one methine, one methylene, two oxygenated methines, a quaternary oxygenated carbon, and a ketone. A final substituent that needed accommodation on this ring structure was a singlet methyl group at a chemical shift (δH 1.39) consistent with its attachment to an oxygenated carbon. Location of these groups about this cyclohexanone ring, especially the oxygenation pattern, was a more difficult task than initially envisioned because of ambiguity over the number of bonds for which HMBC and COSY correlations were observed.
A unique resonance associated with this cyclohexanone ring was a fine doublet proton (J = 1.0 Hz) at δ 4.37, allylically coupled with the H-3 vinyl proton, and associated by HSQC spectroscopy with a carbon at 54.3 ppm. This proton was highly coupled by HMBC, showing six prominent correlations, and defined this methine as the attachment point between the cyclohexanone and linear portions of malyngamide 2. Notably H-4 was HMBC correlated to C-1, C-2 and C-3 in the linear portion of 1, and to the carbonyl, quaternary oxygenated carbon, and singlet methyl group associated with the cyclohexanone portion, thus locating the carbonyl to one side of the attachment point and a quaternary carbon with hydroxy and methyl substituents to the other side. HMBC correlations from the quaternary methyl group (H3-10) confirmed these assignments (Table 1) and also positioned the methyl group on a carbon adjacent to oxygenated methine (C-8). The proton attached to C-8 showed a 3.2 Hz coupling to one of the methylene protons, thus locating the CH2 group at C-7. H-8 was weakly coupled to the other proton at C-7, suggesting that H-8 was an equatorial proton with a nearly 90° dihedral angle to H-7ax. Consistent with this latter assignment, a 12 Hz coupling was observed between this H-7ax proton and the last oxymethine proton at C-6, a coupling value only consistent with an axial-axial orientation. This was confirmed by observation of a 7.0 Hz coupling between H-7eq and H-6ax. Thus, a cyclohexanone ring was defined with a ketone at C-5, hydroxy groups at C-6, C-8 and C-9, a methyl group at C-9, and a juncture to the remainder of the molecule at C-4.
Table 1.
1H and 13C NMR assignments for malyngamide 2 (1) in CDCl3.
| position | δCb | δH (J in Hz)a | HMBCa | ROEa |
|---|---|---|---|---|
| 1a | 41.7 | 4.02, ddd (1.9, 6.2, 16.4) | 2, 3, 1′ | 3, 10, 2′, NH |
| 1b | 4.28, dd (6.2, 16.4) | 2, 3, 1′ | 3, 10, 2′, 9-OH, NH | |
| 2 | 136.4 | |||
| 3 | 121.1 | 6.31, d (1.9) | 1, 2, 4 | 1, 2′ |
| 4 | 54.3 | 4.37, d (1.0) | 1, 2, 3, 5, 9, 10 | 7b, 10, 8-OH |
| 5 | 209.6 | |||
| 6 | 70.7 | 4.55, dd (7.0, 12.0) | 7 | 7b, 10, 2′, 6-OH, 8-OH |
| 7a | 37.3 | 2.30, m | 5, 6, 8 | 1, 8, 9-OH |
| 7b | 2.47, ddd (3.2, 7.0, 13.2) | 5, 6, 8, 9 | 1, 4, 6, 7a, 8 | |
| 8 | 74.2 | 3.89, br s | 6 | 7a, 7b, 10, 8-OH, 9-OH |
| 9 | 80.8 | |||
| 10 | 23.9 | 1.39, s | 4, 5, 8, 9 | 1, 3, 4, 6, 8, 9-OH |
| NH | 6.54, t (6.2) | 1′ | 1, 2′ | |
| 1′ | 173.5 | |||
| 2′ | 36.2 | 2.30, m | 1′, 3′, 4′ | 1, 3, 6, 4′, 5′, NH |
| 3′ | 28.6 | 2.34, m | 2′, 4′, 5′ | 4′, 5′, 7′ |
| 4′ | 130.6 | 5.49, m | 2′, 3′, 5′, 6′ | 2′, 3′, 6′, 7′, 8′, 15′ |
| 5′ | 128.3 | 5.49, m | 2′, 3′, 4′, 6′ | 2′, 3′, 6′, 7′, 8′, 15′ |
| 6′ | 36.1 | 2.20, m | 4′, 5′, 7′, 8′ | 4′, 5′, 7′, 8′, 15′ |
| 7′ | 80.6 | 3.18 | 5′, 8′, 9′ | 3′, 4′, 5′, 6′, 8′, 14′ |
| 8′ | 33.2 | 1.42, m | 9′ | 4′, 5′, 6′, 7′, 14′ |
| 9′ | 25.4 | 1.27, br s | ||
| 10′ | 29.7 | 1.27, br s | ||
| 11′ | 29.3 | 1.27, br s | ||
| 12′ | 31.8 | 1.27, br s | ||
| 13′ | 22.7 | 1.27, br s | ||
| 14′ | 14.1 | 0.88, t (6.8) | 12′, 13′ | 2′, 4′, 5′, 7′, 8′ |
| 15′ | 56.4 | 3.32, s | 7′ | 4′, 5′, 6′ |
| 6-OH | 3.38, s | 5 | 6 | |
| 8-OH | 2.11, br s | 8 | 4, 8 | |
| 9-OH | 5.27, s | 4, 8, 9, 10 | 1, 7a, 8, 10 |
500 MHz for 1H NMR, HMBC, and ROE
75 MHz for 13C NMR
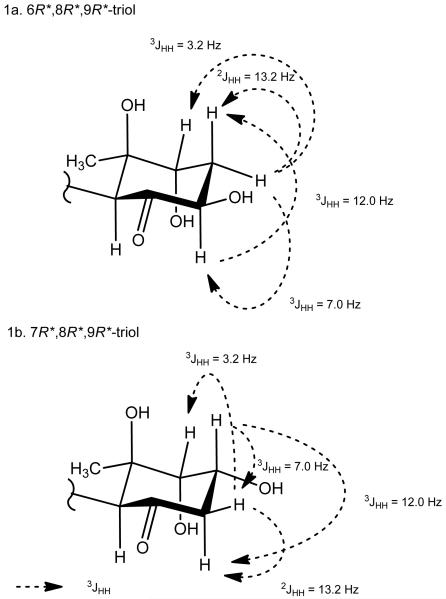
However, upon completion of these assignments, a second possibility was considered which placed hydroxy groups at C-7, C-8 and C-9. Alternative assignments were reasoned as follows. The proposed methylene group at C-6 showed large (12.0 Hz) and medium (7.0 Hz) sized couplings to H-7, and thus H-7 must be axial. That H-7 and H-8 do not show any coupling could be explained by a 0 Hz axial-equatorial arrangement. The 3.2 Hz coupling between H-6eq and H-8eq could arise through W-type coupling about the cyclohexanone ring. Thus, this alternate arrangement was consistent, in principle, with the observed homonuclear couplings (Figure 1).
Figure 1.
Candidate structures for the cyclohexanone ring core with key homonuclear coupling constants. 1a. 6R*,8R*,9R*-triol; 1b. 7R*,8R*,9R*-triol.
To distinguish between these two possible arrangements, data from heteronuclear couplings, ROE measurements, and conformational and chemical shift analysis were considered, and led to a firm conclusion that the original formulation was correct, namely, a C-6 hydroxy rather than a C-7 hydroxy substituent was present in malyngamide 2 (1). Structure possibility 1b was inconsistent with HMBC results which measured correlations between the methylene protons and C-9 as well as from the proposed C-6 hydroxy proton to C-5, both of which would require 4-bond HMBC correlations in 1b. Observation of ROE from the C-6 methylene protons to H-8 was also inconsistent with structure possibility 1b, as these protons would be predicted to be separated by greater than 4 Å. In contrast, structure possibility 1a (figure 1) with a C-6 hydroxy group is fully consistent with the HMBC results with all observed correlations being either 2- or 3-bonds, and ROE results consistent with the vicinal nature of H2-7 and H-8 (Figure 1). Finally, computer modeling was used to calculate the best four conformations of the 6,8,9-triol (1a) and the 7,8,9-triol (1b). Semi-empirical calculation of the four best conformers for 1a and 1b, ab initio geometry optimization, and single point energy calculation resulted in a single best conformer at C-6/C-7 for each; the predicted coupling constants for each were compared with the observed couplings, and again favored structure 1a (see Supporting Information). Finally, 13C NMR chemical shift modeling of the two structural hypotheses were compared with the observed shifts, and interestingly, each possessed shifts at variance with the calculation; however, the 7,8,9-triol was less favored having a greater average Δδ for the carbon atoms of the cyclohexanone ring as well as the single largest carbon atom shift disparity (a calculated shift for C-8 of δ 88.8; Δδ = 14.6 from observed).
With the planar structure complete, the relative configuration of the cyclohexanone ring was assigned using 1D NOE and 2D ROESY correlations as well as homonuclear coupling constants. 1D NOEs between the H-4ax and H-6ax methines as well as a strong 2D ROESY correlation between H-4ax and H-10 indicated that these substituents were on the same face of the cyclohexyl ring (Figure 1). Thus, in combination with the relative relationships of H-6 and H-8 as described above, these data led to a relative configuration of 4S*, 6R*, 8R*, 9R*. Analysis of the absolute configuration of these stereocenters was precluded due to a lack of sufficient amount of compound for NMR or other methods of stereochemical analysis. Characterization of the olefin geometries and the lyngbic acid configuration were next examined. The E geometry of the vinyl chloride was established from 1D NOE correlations between H-3 and H2-1. The geometry of the C-4′ olefin was assigned as E based upon the close comparison of 13C NMR shifts of C-3′ to C-6′ with those of other malygnamides with a C-4′/C-5′ trans geometry. The configuration at C-7′ is predicted to be 7′S because the lyngbic acid isolated from the same extract possessed a negative optical rotation and was consistent with the specific rotation of synthetic 7(S)-methoxytetradec-4(E)-enoic acid (lyngbic acid).12
The biogenesis of malyngamide 2 (1) has several noteworthy features. Formation of the vinyl chloride is predicted to proceed in a similar fashion to that of the jamaicamides.6,13 Specifically, an HMGCoA synthase is predicted to add acetate to the intermediate carbonyl formed by ketide extension of a glycyl unit attached to lyngbic acid, thereby forming a β-branch. Next, the C-2 position of the β-branch acetate unit is predicted to be halogenated via a non-heme (Fe II), α-ketoglutarate dependent halogenase. Subsequent dehydration and decarboxylation in a regiospecific manner would then yield the vinyl chloride functionality.6 Intriguing biosynthetic features of malyngamide 2′s PKS-derived cyclohexanone core are methylation (C-10) at a C-1-derived position (C-9) and hydroxylation at a C-2-derived position of the putative acetate subunits. Methylation at the C-1 position of acetate implies incorporation of a second HMG-CoA synthase cassette within the biosynthetic gene cluster whereas hydroxylation at the C-2 position suggests the possible utilization of a hydroxymalonyl extender unit, as featured in the biosynthetic cluster of zwittermicin A.14 While malyngamide 2 shares biosynthetic features with several other malyngamides reported to date, particularly with hydroxylation at the C-6 position, only malygnamide G shares methylation at a proposed C-1 position in the cyclohexyl ring, and thus the possible utilization of a second HMG-CoA synthase cassette in the biosynthetic cluster of each.15
Malyngamide 2 was tested in a panel of assays to explore its biological properties. In the murine RAW264.7 macrophage cell line treated with lipopolysaccharide (LPS), malyngamide 2 (1) displayed anti-inflammatory properties by inhibiting induced nitric oxide production with an IC50= 8.0 μM (95% confidence interval: 4.6-13.9 μM). Recently, several other malyngamides have been shown to possess anti-inflammatory activity in this assay, and initial SAR features are emerging, such as the requirement for a C-6 hydroxy or acetoxy group.16 Only modest cytotoxicity was observed for compound 1 in both the RAW macrophage cell line (94% cell survival at 21 μM) and H-460 human lung carcinoma cells (IC50= 27.3 μM; 95% confidence interval: 22.4-32.9 μM), indicating a reasonable therapeutic window between inhibition of nitric oxide and cytotoxicity. However, the known compounds re-isolated in this study, wewakazole and a mixture of majusculamides A and B, were also tested in the H-460 cytotoxicity assay; the mixture of majusculamides A and B exhibited the strongest activity and most likely accounts for the cytotoxic activity of the parent extract.9 Wewakazole, originally reported without biological activity,10 was active in the H-460 cytotoxicity assay (IC50= 10.1 μM; 95% confidence interval: 8.4-12.1 μM), but was inactive in a Neuro-2A-based Voltage Gated Sodium Channel (VGSC) assay at 17.5 μM.17
Experimental Section
General Experimental Procedures
Optical rotation measurements were recorded using a Jasco P1010 polarimeter, UV measured on a Beckman-Coulter DU-800 spectrophotometer and IR spectra were obtained using a Nicolet IR-100 FT-IR spectrophotometer. NMR spectra were obtained using Varian Unity 300 and 500 MHz spectrometers. CDCl3 (δH = 7.26; δC = 77.0) was used as an internal reference. High resolution mass spectra were obtained using an Agilent ESI-TOF mass spectrometer. Extracts were processed using a HPLC Waters 515 pump, Waters 996 photodiode array detector, and Millenium software for acquisition and analysis of data. All solvents were either distilled or of HPLC quality.
Collection
The cf. Lyngbya sordida sample (PNG-06/02/02-3) was collected in June 2002 at a depth of 3-5 m by SCUBA near Dutchess Island in Papua New Guinea with GPS coordinates of 9° 57.228′ S and 150° 51.054′ E. The specimens, measuring 1.5 L in total biomass, were stored in 70% EtOH at −20°C until extraction.
Morphological Identification
The PNG-06/02/02-3 cyanobacterial collection was morphologically identified as Lyngbya sordida Gomont ex Gomont (order: Oscillatoriales). The cf. L. sordida collection was composed of brownish, long (>1 cm) and straight or slightly waved filaments measuring 40.5 ± 5.4 μm (n = 3) wide with distinct visible, thick, and colorless sheaths. The cells were disk-shaped, 33.0 ± 4.3 μm wide and 4.5 ± 0.4 μm long (n = 30) with distinct (7.5% ± 0.7) constrictions at the cross-walls. The terminal cells were rounded, non-capitated, and lacked calyptra (i.e. thicker outer cell walls). Taxonomic identification was performed in accordance with bacteriological systems,18 traditional as well as current phycological systems,19 and relevant taxonomic literature. Morphological characterizations were performed using an Olympus IX51 epifluorescent microscope (100x objective) equipped with an Olympus U-CMAD3 camera. Measurements were an average of 10 neighboring cells from three different filaments and calculated with standard deviations. It should be noted that the morphology of Lyngbya resembles that of marine Oscillatoria species and that these two genera cannot be distinguished without phylogenetic analysis. Taxonomic identification of these marine cyanobacteria on the basis of morphology alone has resulted in extensive misclassification, hence the usage of the ‘cf’ designation.20 Furthermore, Lyngbya is a polyphyletic group and tropical marine specimens are unrelated to the genus type-strain PCC 7419T.20 Thus, the taxonomy of tropical marine Lyngbya need to be revised and separated from the genus Lyngbya.
Extraction and Isolation
The collection was extracted six times with heat using CH2Cl2:MeOH (2:1) and yielded an organic extract of mass 1.28 g. This extract was further processed on silica gel using vacuum liquid chromatography with a 100% hexanes-EtOAC-MeOH gradient to generate nine subfractions A-I. Fraction H eluted with 75/25 EtOAc:MeOH and harbored the most potent cytotoxic activity. Fraction H (259 mg) was subsequently filtered using a Waters RP C18 SPE cartridge with 100% MeOH and further processed using reversed-phase HPLC. Malyngamide 2, wewakazole, and majusculamides A/B (5.0 mg) eluted with 60:40 CH3CN/H2O (Phenomenex Synergi Fusion 10 μm, 250 × 10 mm). Malyngamide 2 (C18 Kromasil, 5 μm, 250 × 4.6 mm; 55:45 CH3CN/H2O) and wewakazole (Phenomenex Synergi Fusion, 4 μm, 250 × 4.6 mm; 50:65 CH3CN/H2O) were further purified to yield 1.0 mg and 0.7 mg, respectively. Lyngbic acid was obtained by HPLC purification of fraction F of the same extract using 70:30 CH3CN/H20 containing 0.01% trifluoroacetic acid (Phenomenex Jupiter 10 μm, 250 × 10 mm) yielding 28.5 mg and showing [α]D −22.9 (c 2.0, CHCl3); Lit. [α]D −13.3 (c 2.5, CHCl3).12
Malyngamide 2 (1)
pale yellow oil; [α]D 1.1 (c 2.5, CHCl3); UV (MeOH) λmax (log ε) 202 (4.34); IR (neat) vmax 3300, 2927, 2855, 1720, 1641, 1546, 1458, 1372, 1251, 1128, 1063 cm−1; 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 75 MHz) data, see Table 1; (+)-HRESIMS [M+H]+ m/z 488.2757 (calcd for C25H43 35ClNO6, 488.2779).
Conformer Distributions, Ab Initio Geometry Optimization, and Single Point Energy Calculations
Spartan 2004 software (Wavefunction, Inc) was used to perform all molecular modeling. The best four conformers were obtained for each candidate structure using a semi-empirical method (PM3), and Hartree-Fock 3-21G* ab initio calculations were used to obtain optimized geometries and single point energies for each conformer. Dihedral angles of the optimized structures were measured and coupling constants subsequently calculated using the Altona21 and Karplus22 equations with the aid of Sweet J 2.1 software (Nucleomatica).
Biological Activity
Malyngamide 2 (1), majusculamides A/B, and wewakazole were all tested in the H-460 cytotoxicity assay and the Neuro-2A sodium channel activation and blocking assays using methods previously described.17,23 Malyngamide 2 and wewakazole were submitted for further testing in an anti-inflammatory assay as described below.
Anti-inflammatory activity was evaluated using the mouse macrophage cell line RAW 264.7 (ATCC) cultured in DMEM with 4 mM L-glutamine and 4.5 g/L glucose.16 Media was further supplemented with 10% FBS, penicillin, and streptomycin. RAW 264.7 cells were seeded in 96-well plates (5 × 104 cells/well) and after one day were stimulated in triplicate with 3 μg/mL LPS in the absence or presence of various pure compounds (1 to 30 μg/mL) for 24 h at 37 °C with 5% CO2. The generation of NO was assessed in the supernatant of cell cultures by quantification of nitrite using the Griess reaction. In brief, 50 μL of each supernatant was added to 96-well plates together with 50 μL 1% sulfanilamide in 5% phosphoric acid and 50 μL 0.1% N-(1-naphthyl)-ethylendiamine (NED) in H2O. All assays were run in triplicate with lipopolysaccharide (LPS) as the positive control (assigned as 100%) and cells alone as the negative control (average = 2.5 ± 0.3%). Optical density was measured at 570 nm. IC50 values, the sample concentrations which resulted in 50% inhibition of NO production, were determined using non-linear regression analysis (percent nitrite versus concentration).
Supplementary Material
Acknowledgment
We thank the government of Papua New Guinea for permission to make these cyanobacterial collections, T. Suyama and T.L. Simmons for helpful advice, J. Wingerd for the H-460 cytotoxicity screening, and the TSRI and UCSD mass spectrometry facilities for their analytical services. This work was supported by NIH grants CA 52955 and NS 053398.
Footnotes
Supporting Information Available: 1H NMR, 13C NMR, COSY, TOCSY, HSQC and HMBC spectra in CDCl3 for malyngamide 2 (1). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Tidgewell K, Clark BT, Gerwick WH. In: Comprehensive Natural Products Chemistry. 2nd Ed. Moore B, Crews P, editors. Elsevier Limited; Oxford: 2010. in press. [Google Scholar]
- (2).Suntornchashwej S, Khanit S, Kazushi K, Minoru I. Chem. Asian J. 2007;2:114–122. doi: 10.1002/asia.200600219. [DOI] [PubMed] [Google Scholar]; Appleton DR, Sewell MA, Berridge MV, Copp BR. J. Nat. Prod. 2002;65:630–631. doi: 10.1021/np010511e. [DOI] [PubMed] [Google Scholar]
- (3).Gallimore WA, Scheuer PJ. J. Nat. Prod. 2000;63:1422–1424. doi: 10.1021/np0000365. [DOI] [PubMed] [Google Scholar]
- (4).Kan Y, Fujita T, Nagai H, Sakamoto B, Hokama Y. J. Nat. Prod. 1998;61:152–155. doi: 10.1021/np970423n. [DOI] [PubMed] [Google Scholar]
- (5).Gerwick WH, Tan LT, Sitachitta N. Alkaloids Chem. Biol. Vol. 57. 2001. pp. 75–184. [DOI] [PubMed] [Google Scholar]
- (6).Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Junyang H, editor. Ph.D. Dissertation. National Dong Hwa University; 2008. This newest representative of the malyngamide family is named ‘malyngamide 2’ to follow a recent report of ‘malyngamide Z’ and ‘malyngamide 1’. [Google Scholar]
- (8).Gutierrez M, Suyama TL;, Engene N, Wingerd JS;, Matainaho T, Gerwick WH. J. Nat. Prod. 2008;71:1099–1103. doi: 10.1021/np800121a. [DOI] [PubMed] [Google Scholar]
- (9).Marner F-J, Moore RE, Hirotsu K, Clardy J. J. Org. Chem. 1977;42:2815–2819. [Google Scholar]
- (10).Nogle LM, Marquez BL, Gerwick WH. Org. Lett. 2003;5:3–6. doi: 10.1021/ol026811k. [DOI] [PubMed] [Google Scholar]
- (11).Cardellina JH, Dalietos D, Marner FJ, Mynderse JS, Moore RE. Phytochemistry. 1978;17:2091–2095. [Google Scholar]
- (12).Suntornchashwej S, Suwanborirux K, Isobe M. Tetrahedron. 2007;63:3217–3226. [Google Scholar]
- (13).Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem. Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- (14).Chan YA, Boyne MT, II, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Praud A, Valls R, Piovetti L, Banaigs B. Tetrahedron Lett. 1993;34:5437–5440. [Google Scholar]
- (16).Villa FA, Lieske K, Gerwick L. Eur. J. Pharmacol. 2010;629:140–146. doi: 10.1016/j.ejphar.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Manger RL, Leja LS, Lee SY, Hungerford JM, Hokama Y, Dickey RW, Granade HR, Lewis R, Yasumoto T, Wekell MM. J. AOAC Int. 1995;78:521–527. [PubMed] [Google Scholar]
- (18).Castenholz RW, Rippka R, Herdman M. In: Bergey’s Manual of Systematic Bacteriology. Boone DR, Castenholz RW, editors. Vol. 1. Springer; New York: 2001. pp. 473–599. [Google Scholar]
- (19).Komárek J, Anagnostidis K. Süsswasserflora von Mitteleuropa. 19/2. Elsevier/Spektrum; Heidelberg, Germany: 2005. [Google Scholar]
- (20).Engene N, Coates RC;, Gerwick WH. J. Phycol. 2010;46:591–601. [Google Scholar]
- (21).Haasnoot CAG, de Leeuw FAAM, Altona C. Tetrahedron. 1980;36:2783–2792. [Google Scholar]
- (22).Karplus M. J. Am. Chem. Soc. 1963;85:2870–2871. [Google Scholar]
- (23).Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.