Summary
Newborns are colonized with an intestinal microbiota shortly after birth but the factors governing the retention and abundance of specific microbial lineages are unknown. Nursing infants consume human milk oligosaccharides (HMOs) that pass undigested to the distal gut where they may be digested by microbes. We determined that the prominent neonate gut residents, Bacteroides thetaiotaomicron and Bacteroides fragilis, induce the same genes during HMO consumption that are used to harvest host mucus glycans, which are structurally similar to HMOs. Lacto-N-neotetraose, a specific HMO component, selects for HMO-adapted species such as Bifidobacterium infantis, which cannot use mucus, and provides a selective advantage to B. infantis in vivo when bi-associated with B. thetaiotaomicron in the gnotobiotic mouse gut. This indicates that the complex oligosaccharide mixture within HMOs attracts both mutualistic mucus-adapted species and HMO-adapted bifidobacteria to the infant intestine that likely facilitate both milk and future solid food digestion.
Introduction
The infant gut undergoes a complex and unpredictable process of colonization during the first months of life characterized by extreme fluctuations in overall density and membership. (Koening et al., 2010; Palmer et al., 2007). Factors that drive microbiota assembly remain poorly understood. Several studies indicate that breast-milk versus formula feeding can play a large role in infant intestinal microbiota composition (Adlerberth and Wold, 2009). Human milk oligosaccharides (HMOs), a class of carbohydrates within human milk, may promote intestinal colonization of specific subsets of microbes (Engfer et al., 2000). HMOs are composed of more than 200 structurally distinct oligosaccharides that occur at high concentrations (~20 g/L in colostrums; ~5–12 g/L in mature milk). The wide diversity of structures includes linear or branched lactosamine chains (Galβ1-3/4,GlcNAcβ1-3/6) extended from a lactose (Galβ1-4Glc) core. Structural variability is due to the addition of the terminating sugars N-acetylneuraminic acid (Neu5Ac) in α2-3 or α2-6 linkages and/or fucose in α1-2, α1-3 or α1-4 linkages (reviewed in Chichlowski et al., 2011). The complete degradation of HMOs requires an extensive set of glycoside hydrolases and/or membrane transporters that humans lack in the small intestine. In the lower part of the intestine, HMOs theoretically can be used by members of the infant intestinal microbiota and thereby promote HMO-utilizing microbes. The ability of certain Bifidobacterium species to consume HMOs has been demonstrated and genome sequencing of HMO-utililizing Bifidobacterium isolates has revealed genes involved in HMO uptake, degradation and metabolism (Sela et al., 2008; LoCascio et al., 2007). Recently, it has been described that HMO utilization is not exclusive to certain Bifidobacterium, since members from the genus Bacteroides can use milk glycans as a sole carbon source (Marcobal et al. 2010).
Bacteroides species are variably abundant in the infant gastrointestinal microbiota, but by the first year of life they are consistently present in the gut (Palmer et al., 2007). These species are well adapted to use a multitude of both dietary polysaccharides and host-derived glycans (e.g., mucus) due to specialized machinery encoded by polysaccharide utilization loci (PULs) (Martens et al., 2008). PULs have been widely expanded within the genomes of Bacteroides and each appears to be specialized in the use of a particular class of carbohydrates (Martens et al., 2008; Sonnenburg et al., 2010). The ability of Bacteroides to utilize HMO suggests that specific PULs within these Bacteroides genomes are involved in HMO use.
Using Bacteroides thetaiotaomicron (Bt) and Bacteriodes fragilis (Bf), two of the predominant Bacteroides spp. found in the gut of neonates (Rotimi et al., 1981), we have demonstrated that each species induces a common set of genes when growing in host mucin glycans or HMOs. However, these genes are not conserved between Bt and Bf. These data suggests that HMO use is either a convergent functionality between members of the genus or relies upon pathways that have been differentially shaped by other selective forces, such as mucus utilization. The ability of the short HMO, lacto-N-neotetraose (LNnT), to selectively expand the HMO-using Bifidobacterium infantis (Bi) relative to Bt in gnotobiotic bi-associated mice suggests that milk oligosaccharides play a dual role in selecting for species that are HMO or mucus adapted.
RESULTS
Bt exhibits an expansive glycoside hydrolase response during consumption of HMOs in vitro
Bt possesses a repertoire of predicted glycoside hydrolases (GH) capable of accommodating the structural diversity found in milk oligosaccharides. Among Bt’s >260 glycoside hydrolases, 67 make up six GH families with predicted activities required to process linkages found in HMOs (Figure 1A). Bt grows efficiently when cultured in minimal medium containing HMO (1.5% w/v; MM-HMO) as the sole carbon source. Five additional sequenced Bacteroides species, Bf, B. caccae, B. vulgatus, B. ovatus, and B. stericoris, all grow in the presence of HMOs. Growth of Bf, B. vulgatus and B. caccae are comparable to that of Bt (doubling times of 2.9h, 3.3h, and 2.8h, respectively; saturating OD 600>0.9 for each strain). B. ovatus and B. stercoris do not exhibit exponential growth in MM-HMO, indicating that efficient use of milk oligosaccharides is not universal in the gut resident Bacteroides (Figure 1B).
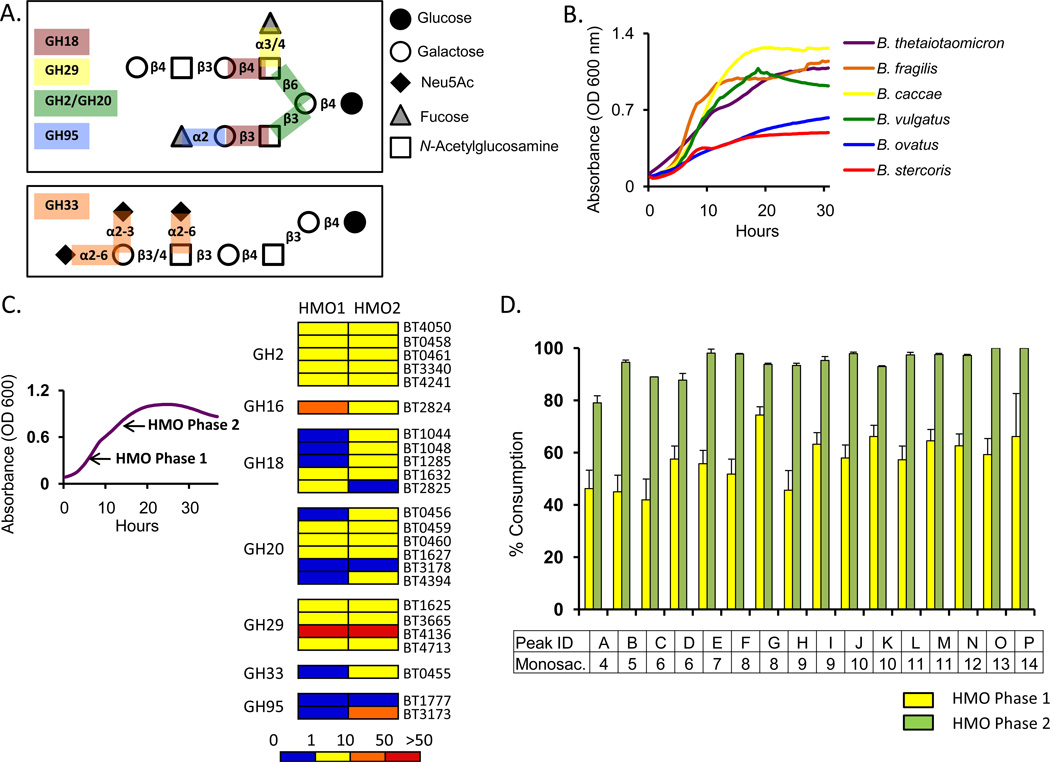
Figure 1. Bt up regulates numerous glycoside hydrolases during consumption of HMO.
A) Schematic of HMO linkages (branched,top box; linear,bottom box) and putative HMO active glycoside hydrolase families (GH) (Wu et al., 2010). Linkages are to the 1-carbon of the underlying sugar unless otherwise noted.
B) In vitro growth of Bacteroides species in MM-HMO.
C) Bt gene expression at two time points (HMO1, HMO2) in MM-HMO relative to MM-glucose for 24 putative HMO active GHs (predicted to hydrolyze linkages found in milk glycans in panel A). See Table S1 for full list of genes up-regulated in all tested carbon sources.
D) Bt HMO consumption at two time points (HMO1, HMO2), determined by MALDI-FTICR-MS. Peak IDs correspond to a characteristic oligosaccharide, with the following discrete mass to charge ratios (m/z): A, 732.25; B, 878.31; C, 1024.36; D, 1097.38; E, 1243.44; F, 1389.50; G, 1462.51; H, 1535.55; I, 1608.57; J, 1754.63; K, 1827.64; L, 1900.69; M, 1973.70; N, 2119.76; O, 2265.82; P, 2484.89. Number of monosaccharides for each mass is indicated. See Figure S1 for detailed structural information for each oligosaccharide. Error bars represent standard deviation for three biological replicates.
We identified the Bt genes induced by HMOs by transcriptional profiling Bt during growth in MM-HMO at the midpoints of the two logarithmic growth phases using a Bt GeneChip (n=2 biological replicates/growth phase, four datasets total) (Figure 1C). As a baseline for comparison, we used previously reported data of Bt grown in MM-glucose (Martens et al., 2008). A total of 156 genes are significantly up regulated during the first phase, and 230 genes are up regulated during the second phase, relative to MM-glucose (see Experimental Procedures for criteria used to define significance; see Table S1 for complete list). Forty-six genes of the 253 gene response to HMOs are predicted to encode glycoside hydrolases and over half of those (24 genes) belong to the seven GH families that target the most common linkages found within HMOs (Figure 1A and C).
The biphasic growth of Bt in MM-HMO suggests a sequential, ordered degradation of glycans. We used laser desorption/ionization coupled with mass spectrometry to characterize the consumption of 16 structurally defined neutral milk oligosaccharides, which represents >85% of the total HMO pool (LoCascio et al., 2007). After the completion of each exponential phase, HMOs were purified from culture supernatants, reduced and profiled (Marcobal et al., 2010). During the first growth phase, Bt consumes the full spectrum of HMOs; a slight preference for larger oligosaccharides is apparent (Figure 1D). After the second phase all the glycans are depleted >80%, with the exception of the smallest oligosaccharide mass (Figure 1D. Peak A), which corresponds to two isomers present at high concentration in the HMO pool: lacto-N-tetraose (LNT, Galβ1-4GlcNAcβ1-3Galβ1-4Glc) and LNnT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) (see Figure S1). Bt exhibits no preference for fucosylated or non- fucosylated glycans. These data demonstrate Bt’s capacity to utilize of a broad range of HMOs, adding to its previously described saccharolytic capacity for mucus glycans and plant polysaccharides (Sonnenburg et al., 2005).
Bt HMO use is coupled to up regulation of mucin glycan degradation pathways
To identify which portion of the Bt transcriptional response in MM-HMO was due to the complex oligosaccharides versus the simple core sugars, lactose and galactose, we performed transcriptional profiling of Bt grown in MM-galactose and MM-lactose (n=2 mid-log phase for each monophasic growth; Figure S2A). Comparison of these data with the baseline MM-glucose revealed 40 and 34 Bt genes are significantly up regulated (>5-fold) in MM-galactose and MM-lactose, respectively. Thirty-two of the 137 genes that were up regulated at least 5-fold in the MM-HMO response were also up regulated in the presence of galactose and/or lactose, consistent with the presence of these simple sugars in the core structures of all HMOs (Figure S2B). The 105 genes that were up regulated in either phase of HMO growth and not in MM-galactose or MM-lactose were highly enriched in the COG functional group “carbohydrate metabolism and transport” (32.1% compared with 11% representation across the genome) (Figure S2C). These data suggest that this 105-gene signature captures the Bt response to the structural complexity within HMOs.
Eighty of the 105 HMO-specific genes are found within 10 PULs, (loci involved in oligosaccharide acquisition and degradation). The larger 253-gene group that is up regulated in both phases of HMO growth included regions of three additional PULs, which resulted in 13 PULs or partial PULs up regulated in one or both phases of HMO growth (Figure 2A). Three of these PULs encode putative fucosidases, key enzymes that hydrolyze the terminal fucose residues from HMOs (BT1624–BT1632, BT3172–BT3173, BT4132–BT4136) and three endo-β-N-acetylglucosaminidases. Average fold changes for genes within each polysaccharide utilization locus (PUL) or partial PUL ranged from 8- to 173-fold (Table S2).
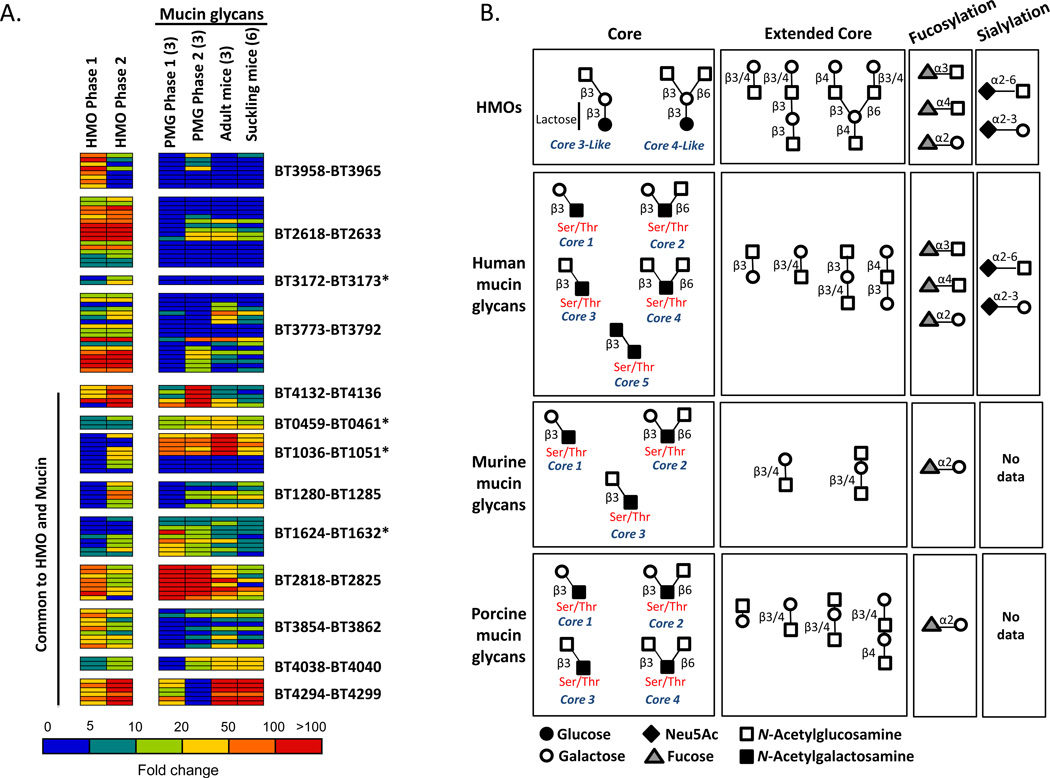
Figure 2. Bt up regulates mucus-utilization loci during HMO consumption.
A) Gene expression profile of Bt’s induced PULs or partial PULs (*) in MM-HMO, MM-porcine mucin glycans (PMG), or host intestinal glycans from adult or suckling mice relative to MM-glucose. Parentheses denote sample number per condition. See also Figure S2 and Table S2.
B) Schematic of mucin glycans based on previous reports (Hurd et al., 2005; Karlsson et al., 1997; Robbe et al., 2004; Robbe et al., 2003b; Thomsson et al., 2002). Structural information includes the core, extended core, and terminal (fucosylation, sialylation) motifs.
Bt profiles in previously defined conditions were examined to search for overlap with the HMO response. Substrate specificities of several Bt PULs have been inferred using growth conditions in which Bt is reliant upon host-derived gut mucus glycans. We compared our in vitro Bt HMO growth expression data with that from three different experimental paradigms in which Bt is reliant upon host mucus glycans: (i) in vitro growth in purified porcine mucus glycans (PMG) (n=3 replicates/growth phase, 2 time points, during exponential phases from biphasic growth) (Martens et al., 2008); (ii) in vivo Bt-colonized 17-day-old gnotobiotic suckling mice (n=6 samples) (Bjursell et al., 2006); and (iii) in vivo Bt-colonized adult gnotobiotic mice that were fed a diet lacking complex glycans (n=3 samples) (Sonnenburg et al., 2005). Transcriptional profiles were analyzed using Bt grown in MM-glucose in vitro as a baseline (Table S2 lists genes up regulated in at least one condition).
Nine of the 13 upregulated PULs or partial PULs in MM-HMO were also highly up regulated (≥ 10 fold-induction) in one or more of the conditions of Bt grown in mucin glycans (Figure 2A). This overlap between HMO- and mucin-induced genes presents the possibility that Bt responds to common structural motifs found in oligosaccharides from mother’s milk and intestinal mucin glycans. For instance, in all datasets we observed increased expression of the PUL BT2818–BT2826, which encodes glycoside hydrolases predicted to cleave linkages from Galβ-GlcNAcβ-Gal, a structure common to HMOs and mucins. Alternatively, four Bt PULs exhibited increased expression specific to HMOs: three complete PULs (BT2618–BT2633, BT3172–BT3173 and BT3958–BT3965) and one partial PUL (BT3172–BT3173) (Figure 2A). These data indicate that Bt responds to aspects of the milk-derived glycans that are not found appreciably in the mucin preparations.
Comparing the structures of HMOs and human intestinal mucin glycans, it is apparent that HMOs exhibit less structural complexity (Karlsson et al., 1997; Robbe et al., 2004; Robbe et al., 2003a; Robbe et al., 2003b; Thomsson et al., 2002) (Figure 2B). Mucin glycans are typically built upon an N-acetyl-galactosamine that is O-linked to serine and threonine residues of the mucin protein and the most abundant are based on five different core structures. Milk oligosaccharides are elaborated from a galactose of the “core” lactose disaccharide which is analogous to the reducing GalNAc of mucin O-linked glycans; structures similar to core 3 [GlcNAcβ1-3Gal] and core 4 [GlcNAcβ1-3(GlcNAcβ1-6)Gal] are present in HMOs. Structures very similar to human mucin glycans are found in the porcine and mouse mucin glycans, which have been used experimentally to define Bt’s mucus use capability (Martens et al., 2008) (Figure 2B). In both the intestinal mucins and in HMOs, repeated motifs containing galactose and N-acetylglucosamine are present and terminate with fucose and sialic acid residues. The extensive structural similarity between HMOs and mucins provides a parsimonious explanation for Bacteroides species up regulating the same PULs for the utilization of glycans from these two different sources.
We tested if the HMO-induced PULs are required for HMO consumption by creating Bt mutants in four of the HMO-specific loci. In-frame deletions for the respective susC-like genes, involved in carbohydrate binding, and the fucosidase BT4136 containing PUL (the most up-regulated HMO-responsive glycoside hydrolases in Bt) were constructed, but showed no defect in growth in HMOs in vitro (see Figure S2D). These data indicate that extensive degeneracy exists within Bt’s HMO response, which contrasts to the strict requirement of genes within the Bt’s fructan-utilization locus (Sonnenburg et al., 2010).
B. fragilis up regulates a distinct set of mucin-use genes when consuming HMOs
The HMO-induced Bt susC/susD homologs and genes located adjacent to the susC/susD genes were used as markers of HMO-utilization genes to identify the presence or absence of orthologs across five sequenced Bacteroides (Bf, B. caccae, B. ovatus, B. ovatus and B. stercoris). In cases where a species shares an orthologous susC/susD pair with Bt, adjacent genes within the PUL generally display a lack of conservation (Figure S3). Bf grows efficiently in MM-HMOs (Figure 1B), but does not show conservation of any of the HMO-utilization PULs identified in Bt. These results suggest that Bacteroides species have developed diverse strategies for using HMOs with varying levels of efficiency.
Whole genome transcriptional profiling of Bf at mid-log phase of its monophasic growth in either MM-HMO, MM-lactose, MM-galactose or MM-glucose was used to identify the genes up regulated by Bf during HMO consumption. MM-glucose served as a baseline to define the genes that were up regulated in HMO, but were not up regulated in MM-lactose or MM-galactose (n=2 per condition). These data revealed a Bf response to HMO composed of a much smaller set of genes compared to Bt. We identified 21 genes specifically up regulated (>5-fold) by Bf in HMO (See Table S3) compared to 105 genes by Bt. Just four Bf susC/susD-homolog-containing PULs were up regulated in HMO compared to 13 in Bt (Figure 3A).
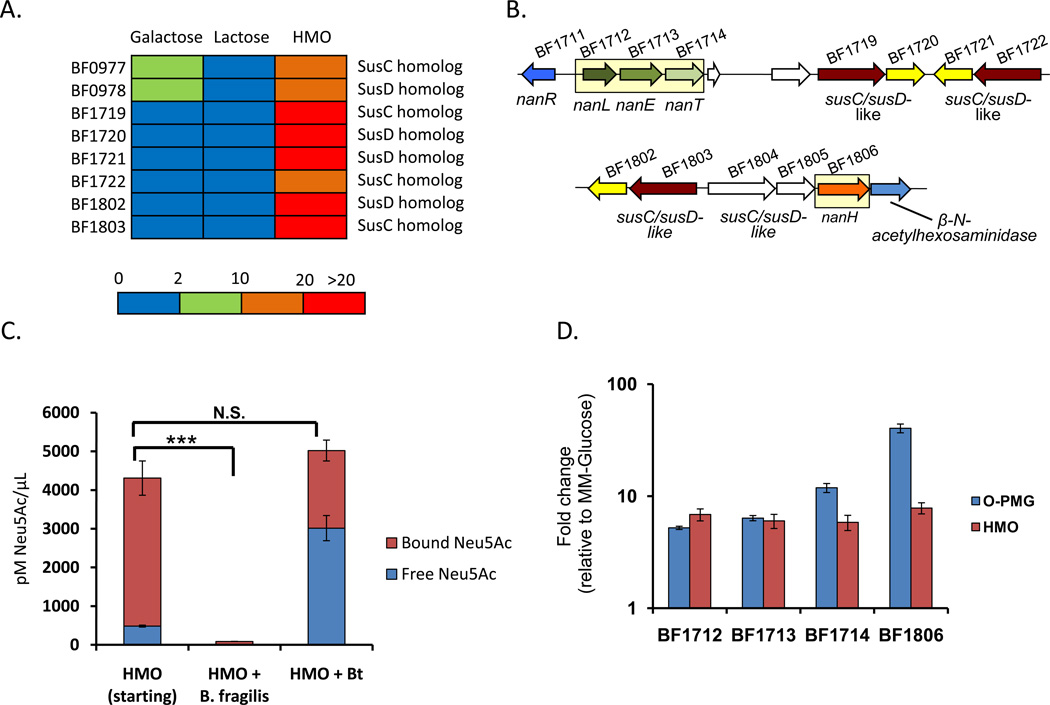
Figure 3. Bf response to HMOs includes sialic acid catabolism.
A) Bf susC/susD homologs up regulated (fold change) in vitro in MM-galactose, MM-lactose, and MM-HMO relative to MM-glucose. See also Figure S3.
B) Genomic organization of Bf genes with >5-fold induction in MM_HMO relative to MM-glucose. Yellow boxes frame genes related to sialic acid consumption. White genes are up regulated <5 fold. Table S3 lists all significant HMO up regulated genes.
C) HMO-bound versus liberated Neu5Ac content in MM-HMO and in MM-HMO after Bf and Bt growth. Error bars represent standard error for three biological replicates.
D) Fold-induction of sialic acid-related genes from Bf grown in MM-O-PMG and MM-HMO relative to growth in MM-glucose as measured by qRT-PCR. Error bars represent standard error for three biological replicates.
Twelve of the 21 HMO-specific Bf genes were distributed within two loci that are dedicated to sialic acid acquisition and catabolism (Brigham et al., 2009) (Figure 3B). These genes include a neuraminidase (nanH, BF1806), an N-acetyl neuraminate permease (nanT, BF1724), an N-acetylneuraminate lyase (nanL, BF1712), and an N-acetylmannosamine 2-epimerase (nanE, BF1713) (upregulated 7.8-fold, 5.8-fold, 6.9-fold, and 6.0-fold, respectively) and provide the machinery necessary to cleave and catabolize Neu5Ac from sialylated glycans. To confirm that Bf does in fact consume sialic acids from HMOs, sialic acid content in the MM-HMO after growth was measured before and after acid hydrolysis using derivatization with 1,2-diamino-4,5-methylenedioxybenzene followed by reverse phase HPLC. The sialic acid Neu5Ac was completely depleted by Bf after growth in HMO. In contrast, Bt can cleave Neu5Ac from sialylated HMOs, presumably to access underlying sugars, but is unable to catabolize it. (Figure 3C).
We wondered if Bf exhibited an overlap in the general strategies used for accessing HMOs and mucin glycans similar to that observed in Bt. The expression of genes that represent the Bf response to HMOs (BF1712, BF1713, BF1714 and BF1806) in MM supplemented with the O-glycan fraction from porcine mucin (MM-O-PMG) and MM-glucose was measured by qRT-PCR. We found that all four genes are up regulated in the presence of mucin, confirming that Bf up regulates sialic acid-use pathways in the consumption of both HMOs and intestinal mucin (Figure 3D). Therefore, while the specific PULs employed for HMO use by Bf and B. theta differ, the mobilization of mucin-use PULs for HMO consumption is conserved between these Bacteroides species.
Bt and Bifidobacterium infantis differentially consume the structurally similar mucin glycans and HMO
Previous work has demonstrated that Bifidobacterium species are well-adapted to HMO use (Sela and Mills, 2010). We grew an HMO-consuming Bifidobacterium strain that is abundant in the microbiota of breast-fed infants, B. infantis, in purified porcine mucin glycans (MM-O-PMG) to elucidate its competence in mucin consumption (Haarman and Knol, 2006). Despite its proficiency at HMO use, B. infantis fails to grow in the MM-O-PMG (Figure 4A). These data suggest that structures unique to HMOs (i.e., not found in mucin glycans) are responsible for supporting growth of the HMO-adapted Bifidobacterium strain. B. infantis has previously been shown to exhibit a preference for the smaller oligosaccharides found in HMOs, some of which are structurally distinct from mucin glycans (LoCascio et al., 2007). One such simple oligosaccharide is the four-sugar lacto-N-neotetraose (LNnT: Galβ1-3GlcNAcβ1-3Galβ1-4Glc). Therefore, we grew both Bt and B. infantis in a pure preparation of this single human milk oligosaccharide (MM-LNnT). B. infantis grew efficiently reaching a high OD, while Bt did not (Figure 4A).
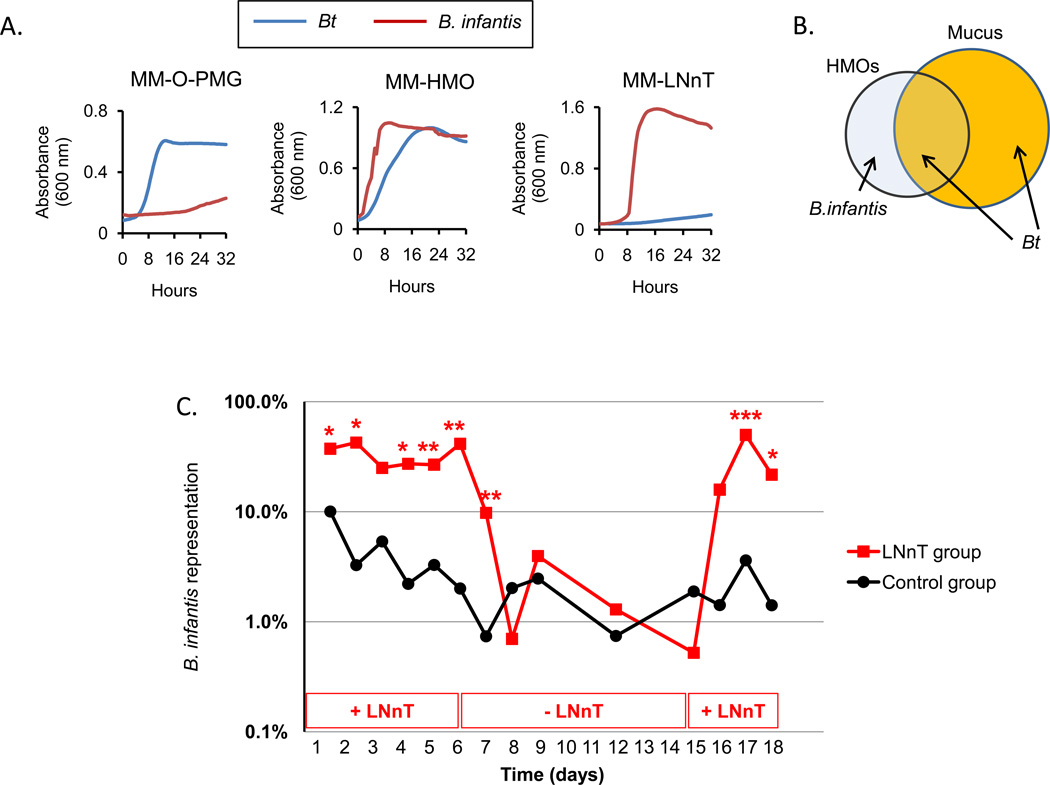
Figure 4. Selective use of the HMO lacto-N-neotetraose by B. infantis provides in vivo advantage.
A) In vitro growth of Bt and B. infantis in the presence of MM-O-PMG, MM-HMO or MMLNnT.
B) Venn diagram representing the structural relationship of mucin and milk glycans. HMOs include a subset of structures found in mucus that can be consumed by mucus-adapted mutualists (e.g., Bacteroides). B. infantis is adapted to use simple structures within HMOs (e.g., LNnT) and is unable to use the structures found in mucin glycans.
C) Bt and B. infantis bi-association of adult germ-free mice fed a polysaccharide-deficient diet without (black circles) or with LNnT (red squares). Values represent average of fecal communities within each group (n=4 mice/group).
These results suggest a model in which the structural complexity of HMOs includes structures that represent mucin glycans that may promote colonization of mucin-adapted symbionts, like Bacteroides, early in life. HMOs also contain structures distinct from mucins, such as LNnT, which may provide a niche for species that are important in microbiota assembly but that are not well adapted to mucus, like B. infantis, and would otherwise be out-competed by mucus-adapted species like Bacteroides. The incomplete overlap of these glycan structural features allows HMOs to attract both HMO-adapted Bifidobacterium species and mucus-adapted Bacteroides species simultaneously (Figure 4B).
We performed an in vivo experiment to test whether LNnT could exhibit selectivity in vivo, enabling the expansion of a Bifidobacterium species in the presence of a Bacteroides species. Two groups of 6-week-old germ free mice were fed a polysaccharide-deficient diet, forcing a reliance on host mucus glycans for carbon and energy. One group received LNnT supplementation in the water (1% w/v; average consumption of 75 mg of LNnT daily), the control group received plain water. After one day of LNnT feeding, both groups were colonized with equivalent quantities of Bt and B. infantis (108 CFU each). At day six post-inoculation, the group receiving LNnT was switched to normal water; at day 15 post-inoculation, LNnT was re-administered for three additional days. Total bacterial colonization density was determined by assessing the CFUs in feces over the course of the 18-day experiment.
The administration of LNnT resulted in an expanded population of B. infantis relative to Bt throughout the first period (day 1–6) compared to control (41.5 ± 6.4 % verses 2 ± 0.5 % on day 6; p=0.009, day 6, n=4 mice) (Figure 4C). LNnT supplementation was withdrawn on day six and colonization density was determined on days 7,8, 9, 12 and 15 post colonization (1, 2, 3, 6, and 9 days after removing LNnT from the water). Removal of LNnT resulted in a drop in B. infantis to levels similar to controls within two days. At day three post withdrawal of LNnT, B. infantis representation was 3.9 ± 0.2 % compared to 41.5 ± 6.4 % just prior to withdrawal (p=0.006, day 6 versus day 9, n=4 mice). LNnT was re-introduced into the water on day 15 and bacterial colonization was monitored at days 16, 17 and 18 post-colonization. Two days after LNnT re-introduction, B. infantis expanded from 0.5 ± 0.5 % at day 15 to 49.9 ± 2 % at day 17 (p=0.012, day 15 versus day 17, n=4 mice). By day 18, B. infantis represented 25.9 + 9.6 % of the community in LNnT-fed mice, compared to 1.9 ± 1.5 % in the control group (p=0.014, n=4 mice) (Figure 4C). These results confirm that this short milk oligosaccharide provides a selective advantage to a Bifidobacterium species over a Bacteroides species in vivo.
Discussion
HMO use by Bt and Bf results in the induction of PULs involved in mucin consumption. One important question is whether HMOs or mucin was the primary selective force shaping locus functionality. Strains that are strictly adapted to one of the glycan sources, the selective pressure can be more confidently assigned, such as B. infantis’ adaptation to HMOs. However for Bt, it is less clear whether (i) adaptation to mucin glycans provides extant pathways that may be mobilized for HMO use, or (ii) if HMOs play a significant role in Bt propagation and provide a selective force, in addition to mucin, that shapes multifunctional loci adapted to accommodate the unique structural aspects of HMOs and mucin glycans. The inability of Bt to readily consume LNnT argues against the latter. If Bt’s entire HMO response is a result of HMO glycans co-opting extant pathways, it is unclear why a subset of Bt’s transcriptional response to HMOs includes loci that are not induced by mucin use. Considering the more diverse carbohydrate structures that human mucus contains, compared to the pig or mouse mucin glycans, it is possible that the set of Bt genes identified as mucin-glycan responsive is not exhaustive. Specifically, the complete set of Bt genes responsive to human mucus, once defined, may include the loci that now appear to be ‘HMO-specific’.
The lack of requirement of the HMO-specific loci, for efficient growth in HMOs by Bt, as demonstrated by our gene deletion studies, may appear to support that these loci are not HMO-adapted, but rather are ‘co-opted’ by the structurally similar HMOs. However, mutation of five mucin-glycan-use PULs in a quintuple KO strain of Bt did not reveal a profound growth phenotype during use of mucin glycans in vitro (Martens et al., 2008). Rather, the requirement of these PULs became apparent upon competition with the wt parent strain in vivo. Therefore, our results highlight the caveats associated with assessing the importance of a gene or locus of a gut resident in a non-competitive environment. Due to the paucity of pure and complex HMOs, in vivo competition experiments are not currently possible.
One hypothesis that arises from this work is that HMO structures mimic mucus glycans to attract mucin-adapted resident mutualists to an infant microbiota. Attraction of Bacteroides may provide the additional benefit of seeding the community with species that are also well-adapted to dietary glycan use, thus preparing the infant microbiota for a smooth transition upon the introduction of solid food. This possibility is supported by recent metagenomic studies that have revealed an abundance of plant polysaccharide-degrading glycoside hydrolases within the gut microbiomes of breast-fed infants (Koening et al., 2010; Vaishampayan et al., 2010). At the same time, unique structural features of HMOs, such as LNnT, are potentially important in shaping the infant microbiota in ways that are independent of mucus use.
Experimental Procedures
Bacterial Strains and Culture Conditions
Bacterial strains used are listed in Supplementary Experimental Procedures. Type strains were used unless otherwise indicated. Bacteroides species were grown in tryptone-yeast extract-glucose (TYG) medium and minimal medium (MM) (Martens et al., 2008). B. infantis was grown in Reinforced Clostridial Medium (RCM, Becton Dickinson and Company, MD, United States) and minimal medium consisting of modified Man-Rogosa-Sharpe medium (MRS, Basingstoke, Hampshire, England), which lacks glucose. Carbon sources were added at 0.5% (w/v) to the respective MM for Bacteroides or Bifidobacterium with the exception of HMO, LNnT and PMG, which were added at 1.5% (w/v). OD600 was monitored using a BioTek PowerWave 340 plate reader (BioTek, Winoosky, VT, United States) every 30 min, at 37° C anaerobically (6% H2, 20% CO2, 74% N2).
Whole Genome Transcriptional Profiling
Transcriptional profiling of Bt and Bf was performed in MM supplemented with milk oligosaccharides, glucose, galactose and lactose (Sigma-Aldrich). Bt samples profiled in HMO were collected at OD600=0.5 and OD600=0.8. Cultures grown on galactose, lactose and glucose were collected at OD600=0.5. RNA targets were prepared and hybridized to custom Bt Affymetrix GeneChips or Bf GeneChips (Sonnenburg et al., 2005). (See Supplementary Experimental Methods for B.fragilis GeneChip generation and validation and GeneChip data analysis).
Genetic Manipulation of Bt
Genetic manipulation of Bt was performed using counter-selectable allele exchange resulting in “in-frame” gene deletion (Martens et al., 2008).
Glycoprofile analysis of HMO consumption
Bacteria culture supernatants in MM-HMO were collected by centrifugation. Remaining oligosaccharides were recovered and profiled by HiRes matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry (MALDI-FTICR-MS) (Marcobal et al., 2010).
Sialic acid content determination
Culture supernatant was isolated after growth in MM-HMO and clarified by centrifugation and filtration. Sialic acid concentrations were determined by the Glycotechnology Core Facility UCSD (University of California, San Diego, California, United States), using the Sigma DMB labeling kit protocol (Prozyme, San Leandro, CA, United States).
Competitive colonization of gnotobiotic mouse
Germ-free Swiss-Webster mice were reared in gnotobiotic isolators and fed an autoclaved polysaccharide-deficient diet (BioServ, http://bio-serv.com) in accordance with A-PLAC, the Stanford IACUC. Mice were bi-associated using oral gavage (108 CFU of each species). Subsequent community enumerations from mice were determined from freshly collected feces, by selective plating of serial dilutions in RCM agar and BHI-blood agar supplemented with gentamicin (200 mg/ml). Significant differences between sample groups were determined using Student’s t-test. Synthetic LNnT (Glycom A/S, Denmark) was purified by crystallization to a final purity of >99%. Characterization was performed using multiple methods including NMR (1D and 2D) mass-spectrometry, and HPLC.
Highlights.
Bacteroides are adept at consuming human milk oligosaccharides
Bacteroides induce mucus-utilization genes to consume human milk oligosaccharides
Lacto-N-neotetraose selects for Bifidobacteria relative to Bacteroides in vivo
Supplementary Material
Acknowledgements
We thank members of the Sonnenburg Lab for helpful discussions and Sara Fisher for editing the manuscript. GeneChip data files have been submitted to the Gene Expression Omnibus under accession numbers GSE26772 and GSE32207. We thank the UCSD Glycotechnology Core Facility for technical assistance and Glycom for their generous contribution of LNnT. Some Bacteroides genomic data were produced by The Genome Center at Washington University School of Medicine in St. Louis (genome.wustl.edu).
Financial Disclosures
This work was funded in part by grants from the NIH (DP2 OD006515 to J.L.S), NIDDK (R01 DK085025 to J.L.S.), from the University of California Discovery Grant Program (to J.B.G.), the California Dairy Research Foundation (to J.B.G.), NIH-NICHD award IR01HD061923 (to C.L.B.) and NIH-NIDDK084214 (to E.C.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Pædiatrica. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J Bacteriol. 2009;191:3629–3638. doi: 10.1128/JB.00811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucl Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M, German JB, Lebrilla CB, Mills DA. The influence of milk oligosaccharides on the microbiota of infants: Opportunities for formulas. Annu Rev Food Sci Tech. 2011;3:331–351. doi: 10.1146/annurev-food-022510-133743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppa GV, Zampini L, Galeazzi T, Gabrielli O. Prebiotics in human milk: a review. Dig Liver Dis. 2006;38 Suppl 2:S291–S294. doi: 10.1016/S1590-8658(07)60013-9. [DOI] [PubMed] [Google Scholar]
- Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- Ferraris RP. Dietary and developental regulation of intestinal sugar transport. Biochemical Journal. 2001;121:265–276. doi: 10.1042/0264-6021:3600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2006;72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Hurd EA, Holman JM, Hansson GC, Domino SE. Gastrointestinal mucins of Fut2-null mice lack terminal fucosylation without affecting colonization by Candida albicans. Glycobiology. 2005;15:1002–1007. doi: 10.1093/glycob/cwi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson NG, Nordman H, Karlsson H, Carlstedt I, Hansson GC. Glycosylation differences between pig gastric mucin populations: A comparative study of the neutral oligosaccharides using mass spectrometry. Biochemical Journal. 1997;326:911–917. doi: 10.1042/bj3260911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koening JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg DS. Oligosaccharides and glycoconjugates in human milk: their role in host defense. J Mammary Gland Biol Neoplasia. 1996;1:271–283. doi: 10.1007/BF02018080. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:1556–1573. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of 0-glycans in normal human mucins along the intestinal tract. Biochemical Journal. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Flahaut C, Michalski JC. Microscale analysis of mucin-type O-glycans by a coordinated fluorophore-assisted carbohydrate electrophoresis and mass spectrometry approach. Electrophoresis. 2003a;24:611–621. doi: 10.1002/elps.200390071. [DOI] [PubMed] [Google Scholar]
- Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta J-P, Michalski JC. Evidence of regio-specific glycosylation in human intestinal mucins. J Biol Chem. 2003b;278:46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- Rotimi VO, Duerden BI. Bacteroides species in the normal neonatal faecal flora. J Hyg Camb. 1981;87:299–304. doi: 10.1017/s0022172400069515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends in Microbiology. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng HJ, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–U1256. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fuc α 1–2 glycosyltransferase. Biochem J. 2002;367:609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. A simple method for assessing sample sizes in microarray experiments. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 108:4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.