Abstract
Purpose
We previously showed that 90% (47 of 52; 95% CI, 0.79 to 0.96) of lung adenocarcinomas from East Asian never-smokers harbored well-known oncogenic mutations in just four genes: EGFR, HER2, ALK, and KRAS. Here, we sought to extend these findings to more samples and identify driver alterations in tumors negative for these mutations.
Experimental Design
We have collected and analyzed 202 resected lung adenocarcinomas from never smokers seen at Fudan University Shanghai Cancer Center. Since mutations were mutually exclusive in the first 52 examined, we determined the status of EGFR, KRAS, HER2, ALK, and BRAF in stepwise fashion as previously described. Samples negative for mutations in these 5 genes were subsequently examined for known ROS1 fusions by RT-PCR and direct sequencing.
Results
152 tumors (75.3%) harbored EGFR mutations, 12 (6%) had HER2 mutations, 10 (5%) had ALK fusions all involving EML4 as the 5′ partner, 4 (2%) had KRAS mutations, and 2 (1%) harbored ROS1 fusions. No BRAF mutation were detected.
Conclusion
The vast majority (176 of 202; 87.1%, 95% CI: 0.82 to 0.91) of lung adenocarcinomas from never smokers harbor mutant kinases sensitive to available TKIs. Interestingly, patients with EGFR mutant patients tend to be older than those without EGFR mutations (58.3 Vs 54.3, P = 0.016) and patient without any known oncogenic driver tend to be diagnosed at a younger age (52.3 Vs 57.9, P = 0.013). Collectively, these data indicate that the majority of never smokers with lung adenocarcinoma could benefit from treatment with a specific tyrosine kinase inhibitor.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide [1], [2]. Although the majority of cases occur in those with a personal history of tobacco smoke exposure, lung cancer also occurs in never smokers, defined as individuals who smoked less than 100 cigarettes in a lifetime. If lung cancer from never smokers was considered as a separate category, the disease would rank among seven to nine most common fatal cancers in the US[3]. While it was reported that up to 30% of lung cancer patients in East Asia were never smokers, it would rank the fifth most common malignancies in China as a separate disease[4], [5].
Lung cancer in never smokers is clinically distinct from other subsets of the disease. First, although lung cancer is comprised of four main histologies including small cell carcinoma, squamous cell carcinoma, large cell carcinoma, and adenocarcinoma, about 70% of never smokers have adenocarcinoma[6]. Second, women have higher risk of developing lung cancer than men among never smokers[3], [7]. Third, never smokers comprise a higher percentage (∼30%) of patients with lung cancer in East Asian countries versus North American and European populations (∼10%)[4], [6]. Reasons for this discrepancy are unknown.
At the molecular level, lung cancer in never smokers is also unique. From genetic analysis of 52 adenocarcinomas samples from patients who never smoked, we previously showed that approximately 90% of samples harbor mutually exclusive driver alterations in just four genes: EGFR, KRAS, HER2, and ALK [8]. By contrast, only ∼40% of cases of non-small cell lung cancer (NSCLC, which includes squamous cell carcinoma, large cell carcinoma, and adenocarcinoma histology) would harbor such mutations.
In this new study, we have further collected another 150 never smoker tumor samples besides the 52 samples previously published for detection of known driver mutations including EGFR, KRAS, HER2, BRAF, and EML4-ALK alterations [8]. Furthermore, we screened ‘pan-negative’ samples for the presence of ROS1 fusions. ROS1 is the human homolog of the avian sarcoma virus UR2 transforming gene v-ros and encodes a receptor tyrosine kinase (RTK) of the insulin receptor family. Activating ROS1 fusions (involving the FIG gene) were previously found in glioblastoma [9] and more recently in lung cancer [10]. We chose to focus first on ROS1 because cell lines harboring ROS1 fusions are sensitive to the tyrosine kinase inhibitor, crizotinib (Bergethon, Pao, Ji, Chen, Iafrate, et al, submitted), making it another potentially targetable mutant kinase in the disease. This study will hopefully provide important insights into molecular defects and identify therapeutic targets in never smoker patients with lung adenocarcinomas.
Materials and Methods
Patients and tissues
Primary tumor samples were obtained from 1103 consecutive patients who underwent potentially curative pulmonary resection at the Fudan University Shanghai Cancer Centre from Oct 2007 through Sep 2010. This study was approved by the Institutional Review Board of the Fudan University Shanghai Cancer Center, Shanghai, China. All participants gave written informed consent. We further collected 150 never smoker tumor samples besides the 52 samples that have been published [8]. A total of 202 patients were enrolled in this specific study based upon the following criteria: they are all never smokers (defined as smoked less than 100 cigarettes in their lifetime), they had a pathologic diagnosis of lung adenocarcinoma, their tumor sample contained a minimum of 50% tumor cells as determined by study pathologists, they did not receive neoadjuvant chemotherapy, and they had sufficient tissue for molecular analysis.
RNA extraction and mutational analysis
All mutational analyses were performed in China. Frozen tissues were grossly dissected into TRIZOL (Invitrogen Inc.) for RNA extraction following standard protocols. Total RNA samples were reverse transcribed into single-stranded cDNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, EU).
EGFR (exons 18–22), HER2 (exons 18 to 21), KRAS (exons 2 to 3), and BRAF (exons 11 to 15) were PCR amplified using cDNA and directly sequenced. For detection of EML4-ALK fusions, primers were designed to amplify all known fusion variants using cDNA. The forward primers were EML4 E2F (5′-TGATGTTTTGAGGCGTCTTG-3′), EML4 E13F (5′-AGATCGCCTGTCAGCTCTTG-3′), and EML4 E18F (5′-TTAGCATTCTTGGGGAATGG-3′), and the reverse primer ALK E20R was (5′-TGCCAGCAAAGCAGTAGTTG-3′). Primers used to detect fusions between ALK and KIF5B or TFG were as previously reported [11], [12]. For detection of CD74-ROS1 and SLC34A2-ROS1 fusions, the forward primers were CD74 E5F (5′-CCTGAGACACCTTAAGAACACCA-3′) and SLC34A2 E4F (5′-TCGGATTTCTCTACTTTTTCGTG-3′). The reverse ROS1 primer was E34R (5′-TGAAACTTGTTTCTGGTATCCAA-3′).
Multiplex PCR analysis was done with KOD plus DNA polymerase (Toyobo, Osaka, Japan). The program to detect ALK fusions was : 94°C 5 minutes; 94°C 30 seconds, 63°C 30 seconds, 68°C 1 minute, 35 cycles; 68°C 10 minutes. The program to detect ROS1 fusions was: 94°C 5 minutes; 98°C 10 seconds, 62°C 30 seconds, 68°C 15 seconds, 35 cycles; 68°C 10 minutes. PCR products were directly sequenced in both forward and reverse directions. All mutations were verified by analysis of an independent PCR isolate.
Real-time PCR quantification
The level of ROS1 mRNA was determined using Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen, CA, USA). The primers for real-time PCR were ROS1-qPCR-F (5′-CAAGAACCCGACCAAAGACCTAC-3′) and ROS1-qPCR-R (5′-CAAATCACATCGCCATCTTCACC-3′). The results were analyzed and expressed as relative mRNA expression of CT (Threshold Cycle) value, which was then converted to fold changes.
Statistical analysis
Associations between mutations and clinical and biological characteristics were analyzed by χ2 or Fisher's exact test. All data were analyzed using the Statistical Package for the Social Sciences Version 16.0 Software (SPSS Inc., Chicago, IL). The two-sided significance level was set at p<0.05.
Results
Patient Characteristics
From Oct 2007 to Sep 2010, we consecutively collected a total of 452 resected lung adenocarcinomas. All patients were Chinese. Interestingly, there were 285 from never smokers while only 167 were from smokers,highlighting a predominant percentage (63.1%) of never smokers in this specific subtype of lung cancer. 202 lung adenocarcinomas from never smokers met eligibility for this study. The median age at diagnosis was 57.3 years ( Table 1 ). The number of patients in stages I-IV was 113, 21, 62 and 6, respectively. 43 tumors were from males and 159 from females. No differences were observed in age, stage, or degree of tumor differentiation between males and females.
Table 1. Clinical Characteristics of Never Smokers With Lung Adenocarcinomas.
| Sex | ||||
| Characteristics | Total | Male | Female | P |
| No. of patients | 202 | 43 | 159 | |
| Age, years | 57.3 | 59.9 | 56.6 | .061 |
| SD | 10.1 | 11 | 9.8 | |
| Clinical Stage | ||||
| I | 113 | 18 | 95 | .058 |
| II | 21 | 7 | 14 | |
| III | 62 | 15 | 47 | |
| IV | 6 | 3 | 3 | |
| Differentiation | ||||
| Well | 38 | 8 | 30 | .160 |
| Moderate | 111 | 19 | 92 | |
| Poor | 53 | 16 | 37 | |
EGFR mutation status
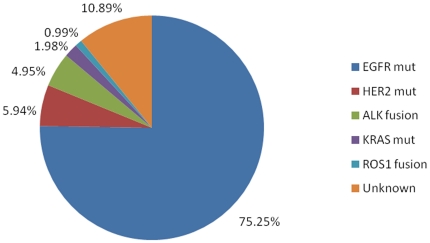
75.3% (152/202) of tumors were found to harbor EGFR kinase domain mutations ( Figure 1 ). Among these, 77 were deletions in exon 19 and 59 were L858R missense changes. Other alterations included 7 exon 20 insertions and 4 exon 18 G719X mutations. 2 samples from patients without previous chemotherapy or TKI treatment harbored concurrent L858R and T790M mutations. Other EGFR mutations included L816Q, I768S, E709K, and K757M.
Figure 1. Spectrum of oncogenic driver mutations in lung adenocarcinomas from never smokers.
From 202 tumors, 75.3% (152/202) harbored EGFR kinase domain mutations, 5.9% (12/202) HER2 mutations, 5.0% (10/202) ALK fusions, and 2% (4/202) KRAS mutations, 1% (2/202) of tumors harbor ROS1 fusion. There are 10.9% (22/202) with unknown oncogenic driver mutations.
Tumors from 76.7% (122/159) of female never smokers harbored EGFR kinase domain mutations. A comparable EGFR mutation rate (69.8%, 30/43) was found in male never smokers (P = 0.348). This confirmed our previous study [13] showing no significant difference in the frequency of EGFR mutations between men and women with lung adenocarcinoma who never smoked. The average age at diagnosis between patients with EGFR mutant and wild-type tumors was 58.3 and 54.3 years, respectively (P = 0.016), showing that patients with EGFR mutant patients tend to be older than those without EGFR mutations. The EGFR mutation rate in well, moderate and poorly differentiated tumors was 86.8%, 75.7% and 66% respectively (P = 0.075) ( Table 2 ). There were no statistically significant differences among subtypes of EGFR mutation and the analyzed clinicopathological features ( Table 3 ).
Table 2. Association between EGFR tyrosine kinase mutations and clinical pathological features.
| Characteristics | Total | Mutant | Wild type | P |
| Age, years | 57.3 | 58.3 | 54.3 | .016 |
| SD | 10.1 | 9.8 | 10.4 | |
| Gender | ||||
| Male | 43 | 30 | 13 | .348 |
| Female | 159 | 122 | 37 | |
| Clinical Stage | ||||
| I | 113 | 87 | 26 | .831 |
| II | 21 | 16 | 5 | |
| III | 62 | 44 | 18 | |
| IV | 6 | 5 | 1 | |
| Differentiation | ||||
| Well | 38 | 33 | 5 | .075 |
| Moderate | 111 | 84 | 27 | |
| Poor | 53 | 35 | 18 |
Table 3. Correlation Between EGFR Mutation Subtypes and Clinical Pathological Features.
| Subtypes of EGFR mutation | |||||
| Characteristics | Total | Exon 19 deletion | L858R | Others | P |
| Age, years | 58.3 | 57.1 | 60.6 | 55.4 | 0.056 |
| SD | 9.8 | 10.2 | 8.9 | 10.4 | |
| Gender | |||||
| Male | 13 | 13 | 4 | 0.608 | |
| Female | 64 | 46 | 12 | ||
| Clinical Stage | |||||
| I | 42 | 36 | 9 | 0.899 | |
| II | 10 | 5 | 1 | ||
| III | 23 | 16 | 5 | ||
| IV | 2 | 2 | 1 | ||
| Differentiation | |||||
| Well | 11 | 15 | 7 | 0.073 | |
| Moderate | 46 | 30 | 8 | ||
| Poor | 20 | 14 | 1 | ||
HER2, KRAS, BRAF, and ALK alterations
5.9% (12/202) of samples had HER2 kinase domain mutations, among which 11 had exon 20 insertions and 1 had an L755P point mutation. EML4-ALK fusions were found in 5% (10/202) of samples, involving EML4 exons 13, 20, and 6 (V1, V2 and V3a/b variants). No KIF5B-ALK or TFG-ALK fusions were detected. Four samples (2%) had a KRAS mutation, including two G12V and two G12D mutations, respectively. No mutations were found in BRAF ( Figure 1 ). Clinical characteristics associated with these different oncogenic driver mutations are shown in Table 4 .
Table 4. Association Between Driver Mutations and Age and Gender in Lung Adenocarcinoma From Never Smokers.
| HER2 | EML4-ALK | KRAS | ROS1 | |||||||||
| Characteristics | Mutant | Wild type | P | Mutant | Wild type | P | Mutant | Wild type | P | Mutant | Wild type | P |
| Age, years | 52.8 | 57.6 | 0.107 | 59.3 | 57.3 | 0.540 | 62.0 | 57.3 | 0.359 | 46.5 | 57.5 | 0.128 |
| SD | 5.4 | 10.3 | 9.8 | 10.2 | 3.4 | 10.2 | 2.1 | 10.2 | ||||
| Gender | ||||||||||||
| Male | 1 | 42 | 0.467 | 4 | 39 | 0.228 | 3 | 40 | 0.032 | 0 | 43 | 1.000 |
| Female | 11 | 148 | 6 | 150 | 1 | 155 | 2 | 154 | ||||
ROS1 fusions
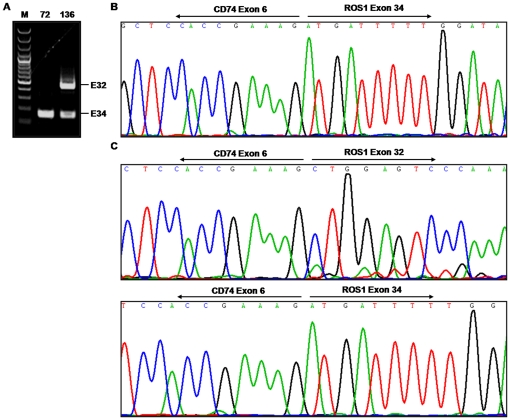
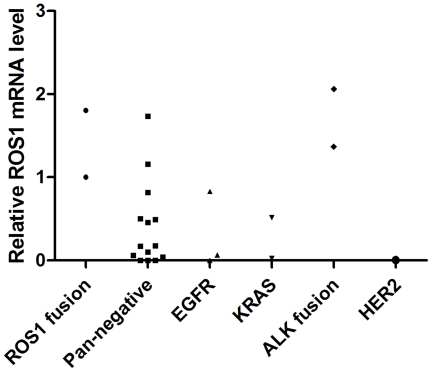
1% (2/202) of samples had CD74-ROS1 fusions ( Figure 2A ). No fusions involving SLC34A2 were detected. Both patients with ROS1 rearrangements were female and diagnosed with stage III disease, and both harbored the previously described CD74-ROS1 fusion involving exon 34 of ROS1 fused to exon 6 of CD74 ( Figure 2B ). Interestingly, one of the samples (No.136) had two types of CD74-ROS1 fusions: exon 32 or exon 34 of ROS1 fused to exon 6 of CD74 (Fig. 2C). Both fusions were in-frame and retained the transmembrane region of ROS1, which may be different splicing products produced from the same translocation event similar to SLC34A2-ROS1 fusions previously found in HCC78 cells [10]. To test if the high ROS1 expression is associated with the ROS1 fusion, we have performed the Q-PCR experiment in 24 samples including the 2 ROS1-fusion positive samples, 3 EGFR mutated samples, 2 KRAS mutated samples, 2 ALK fusion positive samples, 1 HER2 mutated sample and 14 pan-negative samples defined as without above driver mutations. Interestingly, both ROS1-fusion positive samples did show relative high ROS1 expression ( Figure 3 ). However, other samples with either ALK fusion or EGFR mutations or without any known oncogenic drivers also showed comparable expression of ROS1 ( Figure 3 ), indicating that the ROS1 mRNA level may not be as an good indicator for the ROS1 fusion.
Figure 2. Detection of CD74-ROS1 fusions in lung adenocarcinomas from never smokers.
(A) Agarose gel electrophoresis analysis of RT-PCR products for CD74-ROS1 fusions. E32, CD74-ROS1 exon 32 fusion; E34, CD74-ROS1 exon 34 fusion. (B) Sequencing of RT-PCR product from a tumor (No.72) identified a fusion of CD74 exon 6 to ROS1 exon 34. (C) Sequencing of RT-PCR product from a tumor (No.136) identified a fusion of CD74 exon 6 to both ROS1 exon 32 and exon 34.
Figure 3. Detection of ROS1 mRNA levels in lung adenocarcinomas from never smokers either with indicated oncogenic driver mutations or negative for all known oncogenic driver mutations.
Pan-negative samples
Despite identification of ROS1 rearrangements, still 10.9% (22 of 202) samples had no identifiable driver mutation ( Figure 1 ). These data are consistent with our previously published study[8]. Interestingly, further comparative analyses showed that patients without oncogenic driver mutation tend to be diagnosed at a younger age than those with known oncogenic drivers (52.27 Vs 57.93, P = 0.013) ( Table 5 ).
Table 5. Comparison of clinical Characteristics of patients with or without known driver mutations.
| With known driver mutations | Without known driver mutations | P value | |
| Age, years | 57.9 | 52.3 | 0.013 |
| SD | 9.6 | 12.7 | |
| Gender | |||
| Male | 38 | 5 | 0.789 |
| Female | 142 | 17 | |
| Clinical Stage | |||
| I–II | 118 | 16 | 0.502 |
| III–IV | 62 | 6 | |
| Differentiation | |||
| Well | 36 | 2 | 0.441 |
| Moderate | 98 | 13 | |
| Poor | 46 | 7 |
Discussion
During the past decade, a wealth of data from genomic[14], expression[15], mutational[16], and proteomic profiling studies[10] have led to the identification of multiple molecularly distinct subsets of non-small cell lung cancer (NSCLC). Based upon these findings, one of the most promising treatment strategies now involves the subdivision of NSCLC histologies into clinically relevant molecular subsets, using a classification schema based upon specific ‘driver mutations’. Major recurrent mutations in lung adenocarcinoma have been found to occur in EGFR, KRAS, HER2, BRAF, ALK and ROS1.
In our previous study, approximately 90% of lung adenocarcinomas from never smokers harbor known oncogenic mutations in just 4 genes of EGFR, KRAS, HER2, EML4-ALK [8]. Here we comprehensively analyzed all the known oncogenic drivers up to date including all known types of ALK fusions besides EML4-ALK and ROS1 fusions and expanded the sample size. We show that 89.1% of samples harbor known oncogenic driver mutations, consistent with our previous report[8]. In the present study, EGFR kinase domain mutations were found in 75.3% of samples. EGFR kinase domain mutations are more common in elder patients (P = 0.016), consistent with previous studies[17].
The mutation spectrum including EGFR, HER2, KRAS, EML4-ALK in this study shows similar proportions as previously described[8]. In an attempt to provide a more comprehensive map of oncogenic drivers, we have examined other types of ALK fusions, such as KIF5B-ALK and TFG-ALK. However, no tumor positive for these two ALK fusions was found. We have identified 12 samples with HER2 kinase domain mutations, including 11 exon 20 insertions and 1 L755P point mutation. A previous study showed that lung adenocarcinoma patients harboring HER2 exon 20 insertion responded dramatically to Trastuzumab, a monoclonal antibody to HER2 [18]. Pan-ErbB family inhibitors such as Neratinib and BIBW2992[19], [20], under clinical trials to overcome EGFR T790M mutation, may also be used in patients harboring HER2 mutations. Except for KRAS, all the other oncogenic driver mutations have targeted agents being used in clinic or under clinical trials.
ROS1 fusion is recognized as a new oncogenic driver [10]. Here we found about 1% of samples harbored CD74-ROS1 fusion. Interestingly, there is one sample with two forms of ROS1 fusion, which may be derived from the same rearrangement due to alternative splicing mechanism as previously reported[10]. However, it remains unknown why one tumor harbors two forms of fusion and if there is any synergy between them. ROS1 fusion has been shown to contribute to the formation of lung adenocarcinoma [10]. Knockdown of ROS1 in the lung cancer cell line HCC78 which has SLC34A2-ROS1 fusion promotes apoptosis [10], indicating ROS1 fusion is important for cell survival. This validates ROS1 as a good target for clinical treatment. Lung cancer cell line HCC78 positive for ROS1 fusion is sensitive to the tyrosine kinase inhibitor, crizotinib (Bergethon, Pao, Ji, Chen, Iafrate, et al, submitted), indicating that ROS1 fusion is another potentially targetable mutant kinase for lung cancer.
In summary, we have expanded and extended our previous study and examined all known oncogenic driver mutations up to date in lung adenocarcinomas from a large cohort of never smokers. Most of the patients could be classified to a certain type according to oncogenic driver mutations. Except for KRAS, all oncogenic drivers can be effectively targeted in clinic, highlighting the importance of molecular classification of lung adenocarcinoma in never smokers. Interestingly, patients without any oncogenic driver mutation tend to be diagnosed at a younger age. Although we do not have an explanation yet, we are now studying these ‘Pan-negative’ samples using whole exome sequencing in a hope to identify certain novel oncogenic drivers. Our study suggest that most lung adenocarcinoma patients who never smoked could potentially benefit from personalized targeted therapy.
Acknowledgments
The authors thank Qiong Lu, Junhua Zhang, Shihua Yao, Shilei Liu, Rui Wang and Xiaolei Ye for technical support.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts: Dr. William Pao has consulted for AstraZeneca and MolecularMD and has received research funding from Xcovery. Dr. A. John Iafrate has consulted for Pfizer and Abbott Molecular. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported by the National Basic Research Program of China (2010CB912102; 2012CB910800), the National Natural Science Foundation of China (30871284 and 30971461), the Chinese Academy of Sciences (2010KIP504), and the Science and Technology Commission of Shanghai Municipality (09JC1416300). H.J. is a scholar of the Hundred Talents Program of the Chinese Academy of Sciences. W.P. received funding from the National Cancer Institute (USA): R01 CA121210; the Specialized Program of Research Excellence in Lung Cancer Grant (CA90949), and the Vanderbilt-Ingram Cancer Center Core Grant (CA68485). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Thun MJ, Hannan LM, Adams-Campbell LL, Boffetta P, Buring JE, et al. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 2008;5:e185. doi: 10.1371/journal.pmed.0050185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toh CK, Gao F, Lim WT, Leong SS, Fong KW, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol. 2006;24:2245–2251. doi: 10.1200/JCO.2005.04.8033. [DOI] [PubMed] [Google Scholar]
- 5.China. MohotPsRo. Health Statistics of China 2010. 2010.
- 6.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 7.Henschke CI, Yip R, Miettinen OS. Women's susceptibility to tobacco carcinogens and survival after diagnosis of lung cancer. JAMA. 2006;296:180–184. doi: 10.1001/jama.296.2.180. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Ren Y, Fang Z, Li C, Fang R, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28:4616–4620. doi: 10.1200/JCO.2010.29.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charest A, Lane K, McMahon K, Park J, Preisinger E, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 10.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 12.Wong DW, Leung EL, Wong SK, Tin VP, Sihoe AD, et al. A novel KIF5B-ALK variant in nonsmall cell lung cancer. Cancer. 2011. [DOI] [PubMed]
- 13.Sun YH, Fang R, Gao B, Han XK, Zhang JH, et al. Comparable rate of EGFR kinase domain mutation in lung adenocarcinomas from Chinese male and female never-smokers. Acta Pharmacol Sin. 2010;31:647–648. doi: 10.1038/aps.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weir BA, Woo MS, Getz G, Perner S, Ding L, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YH, Lee JK, Kang HJ, Lee TS, Kim HR, et al. Association between age at diagnosis and the presence of EGFR mutations in female patients with resected non-small cell lung cancer. J Thorac Oncol. 2010;5:1949–1952. doi: 10.1097/jto.0b013e3181f38816. [DOI] [PubMed] [Google Scholar]
- 18.Cappuzzo F, Bemis L, Varella-Garcia M. HER2 mutation and response to trastuzumab therapy in non-small-cell lung cancer. N Engl J Med. 2006;354:2619–2621. doi: 10.1056/NEJMc060020. [DOI] [PubMed] [Google Scholar]
- 19.Yap TA, Vidal L, Adam J, Stephens P, Spicer J, et al. Phase I trial of the irreversible EGFR and HER2 kinase inhibitor BIBW 2992 in patients with advanced solid tumors. J Clin Oncol. 2010;28:3965–3972. doi: 10.1200/JCO.2009.26.7278. [DOI] [PubMed] [Google Scholar]
- 20.Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]