Abstract
The molecular mechanism(s) linking tumorigenesis and morphological alterations in the nucleolus are presently coming into focus. The nucleolus is the cellular organelle in which the formation of ribosomal subunits occurs. Ribosomal biogenesis occurs through the transcription of ribosomal RNA (rRNA), rRNA processing and production of ribosomal proteins. An error in any of these processes may lead to deregulated cellular translation, evident in multiple cancers and ‘ribosomopathies’. Deregulated protein synthesis may be achieved through the overexpression of ribosomal proteins as seen in primary leukemic blasts with elevated levels of ribosomal proteins S11 and S14. In this study, we demonstrate that ribosomal protein S6 (RPS6) is highly expressed in primary diffuse large B-cell lymphoma (DLBCL) samples. Genetic modulation of RPS6 protein levels with specifically targeted short hairpin RNA (shRNA) lentiviruses led to a decrease in the actively proliferating population of cells compared with control shRNA. Low-dose rapamycin treatments have been shown to affect the translation of 5′ terminal oligopyrimidine (5′ TOP) tract mRNA, which encodes the translational machinery, implicating RPS6 in 5′ TOP translation. Recently, it was shown that disruption of 40S ribosomal biogenesis through specific small inhibitory RNA knockdown of RPS6 defined RPS6 as a critical regulator of 5′ TOP translation. For the first time, we show that RPS6 associates with multiple mRNAs containing a 5′ TOP tract. These findings expand our understanding of the mechanism(s) involved in ribosomal biogenesis and deregulated protein synthesis in DLBCL.
Keywords: RPS6, diffuse large B-cell lymphoma, 5′ terminal oligopyrimidine, p53
Introduction
Ribosomes in higher eukaryotic cells are large multimeric ribonucleoprotein complexes consisting of four ribosomal RNA (rRNA) molecules and ~80 ribosomal proteins, which make up a large (60S) and a small (40S) subunits (Fatica and Tollervey, 2002; Perry, 2007). In view of the fact that a significant proportion of the cell’s energy is spent in ribosomal biogenesis (Warner, 1999), it is reasonable to expect that the proteins and rRNAs that are the constituents of ribosomes be tightly regulated. Disruption of the processes involved in the regulation of ribosomal biogenesis lead to ‘ribosomopathies’ or ribosomal diseases, including cancer.
Ribosomal biogenesis and assembly is extremely complex, with ribosomal proteins themselves being involved in the regulation of other ribosomal proteins. Of all the essential ribosomal proteins, ribosomal protein S6 (RPS6) has attracted the most attention, as it was the first ribosomal protein shown to be phosphorylated in response to proliferation and stimuli, such as thyrotropin-releasing hormone and epidermal growth factor (Gross et al., 1988; Franco and Rosenfeld, 1990; Pende et al., 2004). The significant body of research conducted on RPS6 over the past few decades has implicated RPS6 in the regulation of protein synthesis (through the regulation of 5′ terminal oligopyrimidine (5′ TOP) RNA synthesis) and cell size. These messages containing an oligopyrimidine tract are important for the ribosomal biogenesis, as they encode most of the translational apparatus, including ribosomal proteins and translation initiation factors (Avni et al., 1997; Jefferies et al., 1997). A selective increase in the translation of 5′ TOP containing messages can be induced in mitogen-stimulated cells (Franco and Rosenfeld, 1990; Pende et al., 2004; Patursky-Polischuk et al., 2009), and can be selectively decreased through treatment with rapamycin or its derivatives (Jefferies et al., 1994), suggesting that this group of mRNAs may have a important role in the control of cellular growth. Therefore, a link between the translation of mRNAs containing 5′ TOP sequences and the phosphorylation state of RPS6 led to a model, suggesting RPS6 regulation of 5′ TOP mRNA translation. However, the mechanism(s) of how RPS6 and its upstream regulatory molecules selectively control 5′ TOP translation has come under intense investigation in recent years, with publications demonstrating that 5′ TOP mRNA are still regulated in mouse embryonic fibroblasts, in which all five possible phosphorylation sites on RPS6 were mutated to alanine (Ruvinsky et al., 2005) or in p70 S6 kinase 1 null mice (Stolovich et al., 2002). Conversely, another report demonstrated that RPS6 phosphorylation was still present in mouse embryonic fibroblast cells (p70 S6 kinase 1−/−) due to a compensatory signal from p70 S6 kinase 2 (Lee-Fruman et al., 1999). To reconcile these discrepancies, a more thorough molecular understanding of the functional role of RPS6 is required.
Experimental evidence has recently revealed that the lack of the ribosomal protein, RPS6, may be catastrophic for the formation of a nascent 40S ribosomal subunit. Owing to the interdependency of ribosomal protein regulation, the loss of RPS6 can lead to an overabundance of other ribosomal proteins and the activation of extraribosomal functions of these proteins secondary to loss of regulation (Fumagalli et al., 2009). Ribosomal proteins have been shown to have extraribosomal functions, including the regulation of the processing and translation of their own mRNA (Fewell and Woolford, 1999; Mitrovich and Anderson, 2000; Badis et al., 2004) or the mRNA of other genes (Takagi et al., 2005; Kapasi et al., 2007; Ofir-Rosenfeld et al., 2008). The presence of RNA-binding domains in some ribosomal proteins RPL26 and RPL13a, suggests that classes of mRNAs may be regulated through extraribosomal functions. A recent report by Fumagalli et al. (2009) demonstrated that modulating RPS6 levels using small inhibitory RNA (siRNA) led to an increase in the translation of 5′ TOP-containing mRNAs, such as RPL11 and RPS16, suggesting that RPS6 is a negative regulator of 5′ TOP message translation. We theorized that RPS6, a protein previously shown to interact with ribosomal-associated RNA (Nygård and Nilsson, 1990), might be involved in the regulation of these 5′ TOP mRNAs by associating with them through the oligopyrimidine tract and preventing their translation in order to preserve proper stoichiometry between rRNA and ribosomal proteins.
In this study, we provide evidence that RPS6 does associate with multiple mRNAs containing a 5′ TOP tract. We also found that RPS6 is highly expressed in primary diffuse large B-cell lymphoma (DLBCL), and targeting this protein using specifically designed short hairpin RNA (shRNA) against RPS6 mRNA decreased proliferation of two DLBCL cell lines compared with control shRNA.
Results
RPS6 regulates the translation of mRNAs containing a 5′ TOP tract
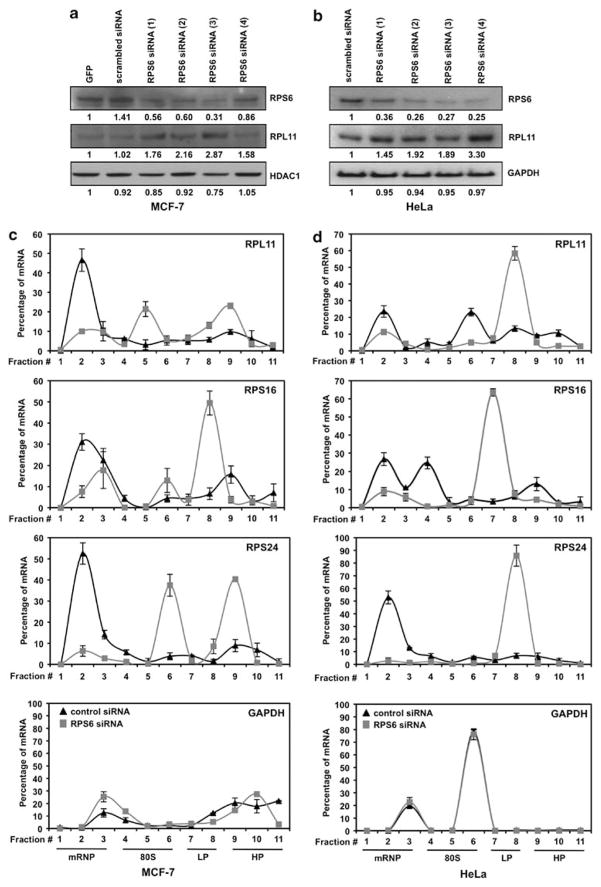
RPS6 has been previously shown to regulate the translation of RPS8 and RPL11, which contain a 5′ TOP tract in the 5′ untranslated region of their mRNA (Fumagalli et al., 2009). We examined whether other ribosomal protein mRNAs containing a 5′ TOP (Table 1) were regulated in a similar manner by modulating RPS6 levels using specific siRNA. To this end, mammary epithelial adenocarcinoma (MCF-7) and cervical carcinoma (HeLa) cells were transfected with multiple siRNA targeting the mRNA of RPS6 (Figures 1a and b). Although the transfection efficiency of siRNA in MCF-7 cells was lower than in HeLa cells, the knockdown of RPS6 led to a specific increase in the protein levels of RPL11 in both cell lines. Knockdown of RPS6 increased the abundance of 5′ TOP mRNA, which encodes RPL11, RPS16 and RPS24, in actively translating polysomal fractions, with no significant changes in glyceraldehyde 3-phosphate dehydrogenase mRNA levels (loading control; Figures 1c and d). The RPS6 siRNA-induced increase in the translation was also shown to be specific for 5′ TOP mRNA, as BCL-2 mRNA (a non-5′ TOP mRNA control) exhibited a decrease in the translational profile (Supplementary Figure 1A). This increase in actively translating mRNA was achieved through a translational mechanism, as no significant changes in total RNA levels were observed between control siRNA and siRPS6 cells (Supplementary Figure 1B).
Table 1.
mRNA encoding ribosomal proteins or translation initiation molecules contain a 5′ terminal oligopyrimidine sequence
| Genes | 5′ Terminal sequences (5′–3′) | No. of pyrimidines | References |
|---|---|---|---|
| RPS6 | cctcttttccgtggcgcctcggaggcgttc | 10 | Patursky-Polischuk et al. (2009) |
| RPS16 | gaaaagcggccagggtggcccctagctttccttttccgg | 12 | Levy et al. (1991) |
| RPL32 | aggggttacgacccatcagcccttgcgcgccaccgtcccttctctcttcctcg | 16 | Levy et al. (1991) |
| eIF3e | gagcacagactcccttttctttgg | 13 | Iadevaia et al. (2008) |
| eIF3h | ctctttcttcctgtctgcttgg | 12 | Iadevaia et al. (2008) |
| RPL21 | tttcctttcggccggaaccgccatcttcca | 6 | Frigerio et al. (1995) |
| RPL34 | gtctgcaggtatggatgttgttctcttttccctgtct | 11 | Rommens et al. (1995) |
| RPL38 | gcccggaaacggaagtctcgttctttttcgtccttttccccgg | 7 | Espinosa et al. (1997) |
| RPS29 | cttttacctcgttgcactgctgagagcaag | 5 | Rommens et al. (1995) |
| RPS27 | ctttccggcggtgacgacctacgcacacgagaa | 6 | Vladimirov et al. (1996) |
| RPS21 | cttgcccgccgatatctctgccgggtgactagctgcttcctttctctctcgcgcgc | 15 | Vladimirov et al. (1996) |
| RPL29 | ttccggcgttgttgaccctatttcccgtgctgcaccgcagcccctttctcttccgg | 14 | Law et al. (1996) |
| RPS24 | aggcatcggcgcggtcagcctcgtggcgcgcccacgcccccacgccggctcttcccgg | 8 | Vladimirov et al. (1996) |
| RPS23 | ggggtccttggctgggcggggcttgctcgcggtggcttgtggctccttcctgcgg | 9 | Vladimirov et al. (1996) |
Examples of ribosomal proteins or translation initiation molecules that contain a 5′ terminal oligopyrimidine sequence (highlighted in bold) in the 5′ untranslated region of their mRNA.
Figure 1.
Specific siRNA knockdown of RPS6 leads to an increased translation of mRNA containing a 5′ TOP motif. (a, b) Representative western blot analysis of MCF-7 or HeLa cells 48 h after transfection with control and RPS6 siRNA. A volume of 30 μg of total protein lysates was loaded and the abundance of RPS6, RPL11, HDAC1 and GAPDH was assessed. (c, d) Cytoplasmic lysates from either scrambled siRNA or RPS6 siRNA cells were fractionated through sucrose gradient centrifugation. Quantitative PCR was performed on the RNA isolated from each fraction using specific primers for RPL11, RPS16, RPS24 and GAPDH. Fold changes were calculated for target mRNA in each fraction and summed together to define a total mRNA population. Signal from each fraction was divided by the total population to calculate the percentage of mRNA present per fraction from either scrambled siRNA or RPS6 siRNA cells. Graphs represent the mean and standard deviation from three independent assays. Cytoplasmic fractions may be grouped together as free messenger ribonucleoprotein (mRNP), 80S ribosomes, and light (LP) or heavy (HP) polysomes. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green florescent protein.
RPS6 associates with mRNA containing a 5′ TOP tract
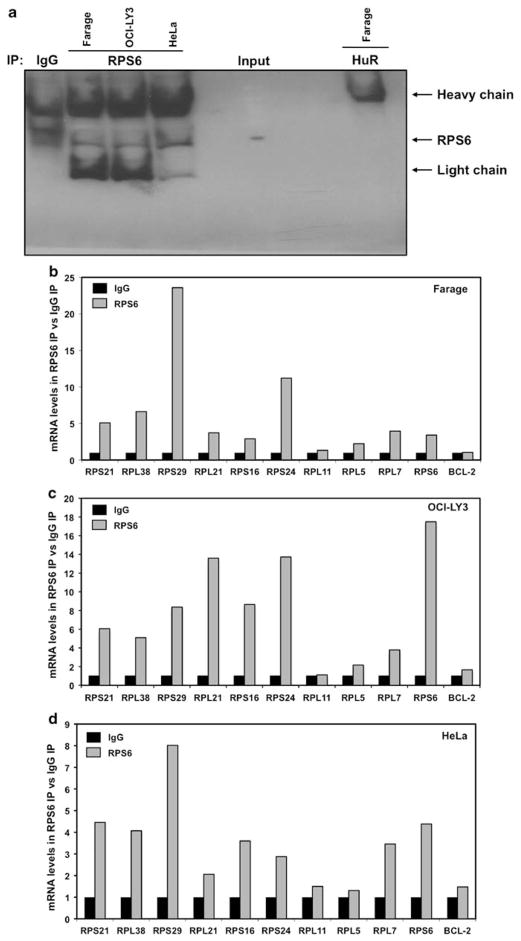
Multiple ribosomal proteins have been shown regulate the translation of mRNA through association with sequences found in the 5′ untranslated region (Takagi et al., 2005; Kapasi et al., 2007; Ofir-Rosenfeld et al., 2008). Given that RPS6 is a known RNA-binding protein and has been shown to be a regulator of 5′ TOP message translation, we wished to study whether RPS6 binds to mRNA containing a 5′ TOP. The ability of endogenous RPS6 to bind mRNA was evaluated by examining immunoprecipitates from an anti-RPS6 mRNP-IP for the presence of 5′ TOP mRNA (Figure 2a). Indeed, real-time quantitative PCR (qPCR) of RNA purified from immunoprecipitates revealed that RPS6 specifically associates with a majority of examined mRNAs containing a 5′ TOP in Farage, OCI-LY3 and HeLa cells (Figures 2b–d).
Figure 2.
RPS6 associates with multiple 5′ TOP messages. (a) Representative immunoprecipitation assays performed as described (Materials and methods) but followed by western blot analysis to assess the abundance of RPS6 protein in the IP material, isotype IgG and non-RPS6-binding protein HuR were included as controls. (b–d) Cytoplasmic lysates from Farage, OCI-LY3 and HeLa cell lines were used for immunoprecipitation assays, using anti-RPS6 antibody. RNA was isolated and reverse transcription followed by qPCR was performed to measure abundance of RNA. Graph represents the mean from three independent assays.
RPS6 associates with both wild-type and mutant 5′ TOP sequences
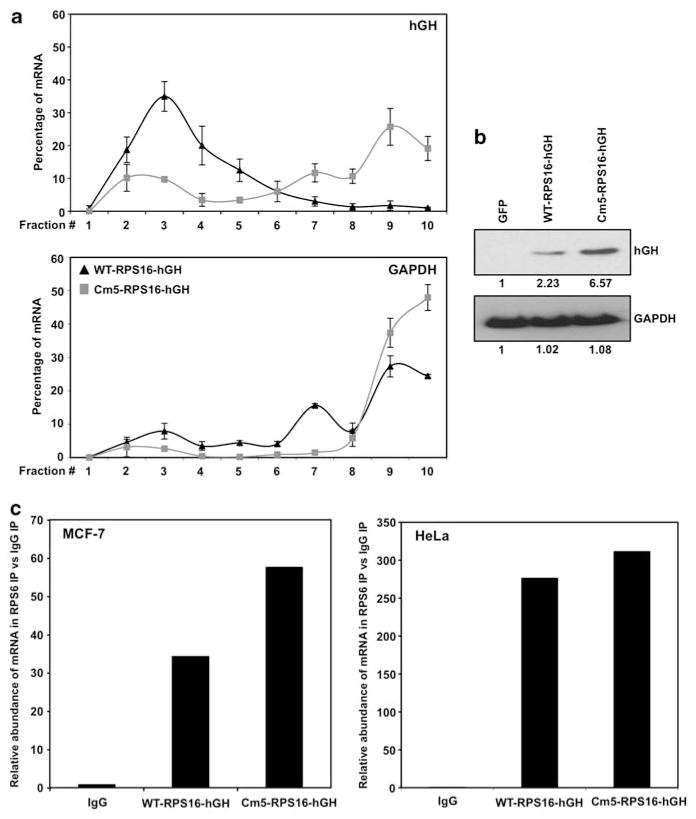
The translation of mRNA encoding ribosomal proteins has been shown to be negatively controlled by the presence of 5′ TOP sequences (Fumagalli and Thomas, 2000; Meyuhas, 2000). The translational regulation of these messages is disrupted when purines are substituted for pyrimidines in the 5′ TOP tract (Levy et al., 1991; Jefferies et al., 1997). To investigate the possibility that RPS6 association to the message may be lost once the 5′ TOP is mutated, we transiently expressed two heterologous reporter constructs. The constructs contain the coding region of human growth hormone (hGH) fused to the first 29 nucleotides of the RPS16 5′ untranslated region (WT-RPS16-hGH) or to the mutant in which five of the eight pyrimidines within the 5′ TOP have been mutated to purines (Cm5) (Levy et al., 1991; Jefferies et al., 1997). This mutation led to a differential recruitment of Cm5-RPS16-hGH mRNA to actively translating polysomes (Figure 3a), resulting in increased protein levels (Figure 3b). Real-time qPCR analysis of purified RNA from RPS6 mRNP-IP demonstrated RPS6 associates with both the wild-type and mutant RPS16 5′ TOP sequences (Figure 3c) in both MCF-7 and HeLa cells, suggesting other mechanism(s) or factors are involved in RPS6-mediated regulation of 5′ TOP mRNA translation.
Figure 3.
RPS6 associates with both wild-type and mutant 5′ TOP messages. (a) Quantitative PCR analysis of the distribution on polysomes of hGH reporter mRNA in HeLa cells transfected with either WT-RPS16-hGH or Cm5-RPS16-hGH plasmids (b) Representative western blot analysis of HeLa cells transfected with either control green florescent protein (GFP), WT-RPS16-hGH or Cm5-RPS16-hGH. A volume of 10 μg of total protein lysates was loaded and the abundance of hGH and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was assessed. (c) Cytoplasmic lysates from MCF-7 and HeLa cell lines were used for immunoprecipitation assays, using anti-RPS6 antibody. RNA was isolated and reverse transcription followed by qPCR was performed to measure abundance of RNA. Graph represents the mean from three independent assays.
Subcellular distribution of RPS6, TIA-1 and DCP2 after rapamycin treatment
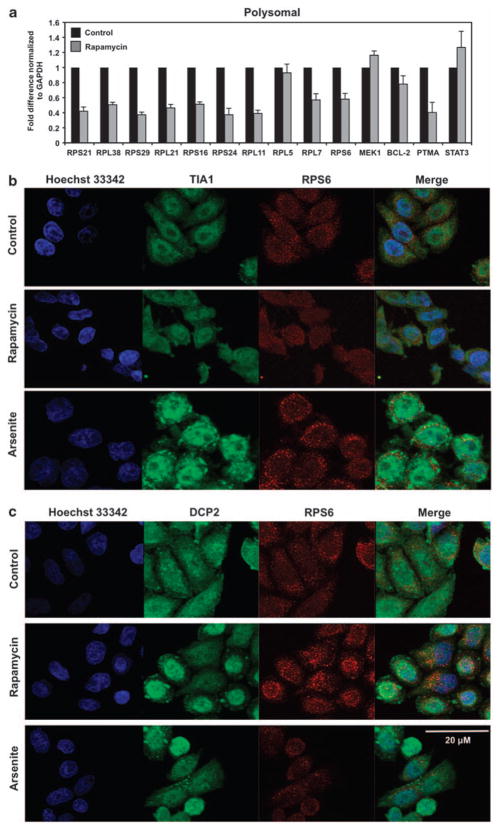
Although the exact molecular mechanism(s) are not fully understood, treatment of mammalian cells with low dose rapamycin has been definitively shown to suppress the translation of 5′ TOP mRNAs (Jefferies et al., 1994, 1997) through inhibition of the mammalian target of rapamycin C1 signaling pathway. To confirm these effects in our system, we monitored actively translating polysomes to visualize the inhibition of 5′ TOP messages in HeLa cells treated with low dose rapamycin (Figure 4a). Following cellular stress, dynamic sites of mRNA storage, such as stress granules or processing bodies, form foci in the cytoplasm; we hypothesized RPS6 may be involved in translocation of mRNA to these storage areas subsequent to treatment with rapamycin. To assess whether rapamycin treatment resulted in any changes in the intracellular localization of RPS6, we examined RPS6, TIA-1 (stress granules marker) and DCP2 (processing bodies marker) through immunoflourescence. We demonstrated that rapamycin treatment does not induce either stress granules or processing bodies in HeLa cells as shown by the lack of TIA-1 (Figure 4b) or DCP2 (Figure 4c) localization to foci in the cytoplasm. HeLa cells treated with arsenite, a known stress granule-inducing agent, were included as positive controls for the formation of TIA-1 aggregates in stress granule foci. Although treatment of cells with arsenite induced stress granules, there was no colocalization of RPS6 with TIA-1, consistent with RPS6 not being involved in trafficking of mRNA to stress granules.
Figure 4.
RPS6 does not associate with either stress granules or processing bodies after rapamycin treatment. (a) Relative association of mRNA with actively translating polysomes was tested by preparing cytoplasmic lysates from HeLa cells treated with either dimethyl sulfoxide or 10 nM rapamycin for 8 h, fractionating them through sucrose gradients and collecting 11 fractions for analysis. mRNA levels from pooled fractions (8–11) comprised of translating RNA were quantified by real-time qPCR and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. At the times indicated following addition of rapamycin or after a 60-min incubation with 0.5 mM sodium arsenite, the subcellular localizations of (b) RPS6 (red) and TIA-1 (green) or (c) RPS6 (red) and DCP2 (green) were monitored by immunofluorescence. Merge: yellow indicates colocalization of RPS6 and TIA-1 or DCP2 signals.
RPS6 is important for the malignant phenotype
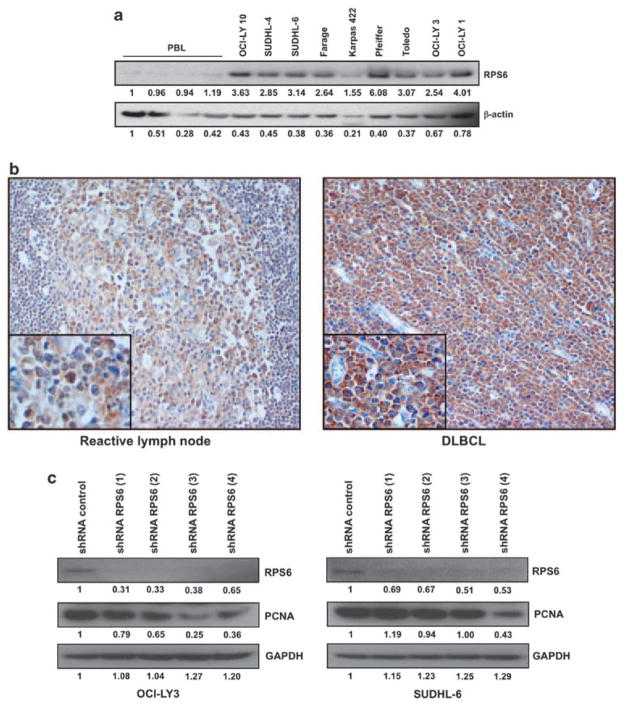
The haploinsufficiency of RPS6 has been linked to multiple different phenotypes including cancer (Nakamura et al., 2008; Ruvinsky et al., 2009), and specifically cancers of the hematopoietic system (Watson et al., 1992). However, as RPS6 may regulate the stoichiometry of other ribosomal proteins (Fumagalli et al., 2009), it is plausible that an overabundance of RPS6 levels in lymphoma would confer an increase in other ribosomal proteins, leading to an increase in the number of ribosomes and a greater capacity to translate oncogenic messages beneficial to the maintenance of the malignant phenotype. Therefore, we examined the levels of RPS6 in a panel of peripheral blood lymphocytes and DLBCL cell lines. We found that DLBCL cell lines have an increased expression of RPS6 when compared with normal peripheral blood lymphocyte samples (Figure 5a). Immunohistochemical analysis of multiple primary patient samples using a RPS6-specific antibody further confirmed that RPS6 is overexpressed in primary DLBCL patients compared with germinal center B cells found in reactive lymph nodes (Figure 5b). Genetic manipulation of ribosomal proteins, including RPS6, was shown to positively regulate the translation of RPL11, a protein that stabilizes p53 protein levels (Figures 1a and b; Takagi et al., 2005; Fumagalli et al., 2009). To investigate how disruption of 40S ribosomal biogenesis may impact cell cycle progression in DLBCL, we transduced p53 wild-type OCI-LY3 and SUDHL-6 DLBCL cell lines with multiple shRNA specifically targeting RPS6 mRNA (Figure 5c). Knockdown of RPS6 resulted in a significant reduction in proliferation, as measured by a decrease in proliferating cell nuclear antigen levels, with the most significant changes in protein levels observed in shRPS6 (3) and (4).
Figure 5.
RPS6 is overexpressed in DLBCL and shRNA knockdown of RPS6 resulted in decreased proliferation. (a) Representative western blot analysis of peripheral blood lymphocytes from multiple patients and several DLBCL cell lines. A volume of 30 μg of total protein lysates was loaded and the abundance of RPS6 and β-actin was assessed. (b) Representative immunohistochemistry staining of paraffin embedded reactive lymph node or primary DLBCL lymph node with anti-RPS6 antibody (original magnifications of × 200 and × 400). (c) Representative western blot analysis of OCI-LY3 and SUDHL-6 cells transduced with multiple shRNA sequences. A volume of 30 μg of total protein lysates was loaded and the abundance of RPS6, PCNA and GAPDH was assessed. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PBL, peripheral blood lymphocyte; PCNA, proliferating cell nuclear antigen.
Discussion
The nucleolus is the subnuclear organelle in which ribosomal subunits are produced and assembled. The relationship between the nucleolus and tumorigenesis was first recognized in 1896 when Pianese observed that irregular morphology of the nucleolus was characteristic of malignant cells (Pianese, 1896). Observed changes in the nucleolar structure were believed to be a consequence of the malignant state and not a causative factor in the tumor cells. However, with the discovery that cancer susceptibility diseases, such as dyskeratosis congenita, cartilage hair hypoplasia and Diamond–Blackfan anemia, were attributed to the dysfunction of factors involved in ribosomal biogenesis (DKC1, RMRP and RPS19, respectively; Draptchinskaia et al., 1999; Ruggero et al., 2003a; Ganapathi and Shimamura, 2008), it became apparent that changes in the nucleolus are actively contributing to the tumorigenic phenotype.
Ribosomal biogenesis and assembly is an extremely complex cellular process and has been shown to be deregulated in cancer. Multiple mechanisms have been discovered to be involved in this deregulation, including increase in rRNA synthesis, alterations in the modification of rRNA and differential levels of ribosomal proteins (Ruggero and Pandolfi, 2003b). The overexpression of multiple ribosomal proteins that belong to both the 40S (small) and 60S (large) has been documented in primary tumors, such as leukemia and hepatocellular carcinoma (Ferrari et al., 1990; Kondoh et al., 2001). Our data illustrated for the first time that the ribosomal protein S6 is highly expressed in primary DLBCL. Genetic manipulation of multiple ribosomal proteins, including RPS6, has been previously shown to positively regulate proteins that stabilize wild-type p53 protein levels (Takagi et al., 2005; Fumagalli et al., 2009). The frequency of p53 mutations in DLBCL ranges from 11 to 25% (Koduru et al., 1997; Sánchez-Beato et al., 2003), which is relatively low when compared with other tumor types. We demonstrated that knockdown of RPS6 using targeted shRNA in two DLBCL cell lines produced a significant decrease in the population of cells undergoing proliferation when compared with control shRNA cells. Interestingly, although shRNA knockdown of RPS6 led to a decreased proliferative index, it did not significantly induce apoptosis as measured by annexin V and propidium iodide staining (Supplementary Figure 2). Therefore, the antiproliferative effect observed through the disruption of RPS6 function in DLBCL with a functional p53 pathway underscores the importance of dysregulated translation and ribosomal biogenesis necessary for maintenance of the malignant phenotype.
Although the prevailing model for mammalian target of rapamycin signaling has implicated RPS6 functional state in the translational control of 5′ TOP mRNAs, several publications have emerged, challenging this notion (Stolovich et al., 2002; Ruvinsky et al., 2005). Using an siRNA approach in our study, we found RPS6 to be a critical regulator of the translation of messages containing a 5′ TOP sequence in multiple malignant human cell lines (Figure 1), which is in agreement with earlier observations (Fumagalli et al., 2009). The exact mechanism(s) of how RPS6 regulates 5′ TOP mRNA translation has yet to be revealed. We hypothesized that RPS6, a known RNA-binding protein (Nygård and Nilsson, 1990), may regulate 5′ TOP mRNA translation through association with these sequences. We found that RPS6 does indeed associate with a number of mRNAs that contains 5′ TOP sequences (Figures 2b–d). The 5′ TOP sequence contained within the 5′ untranslated region of mRNA is a negative regulator of translation (Fumagalli and Thomas, 2000; Meyuhas, 2000). Mutation of pyrimidines to purines found in the 5′ TOP confers constitutive translation to the mRNA (Levy et al., 1991; Jefferies et al., 1997). RPS6 association with both wild-type and mutant 5′ TOP sequences (Figure 3) suggested that its interaction with the mRNA is not the only determinative of translation. On the basis of these data, alternative mechanisms or factors are likely to be involved in the translational regulation of 5′ TOP mRNA. Our data presented allow for the possibility that RPS6 is part of a larger complex of proteins, including other RNA-binding proteins, that assist in 5′ TOP translational regulation.
Our findings presented here further validate that ribosomal biogenesis may be an important factor involved in tumorigenesis, as shown by the elevated levels of RPS6 in DLBCL compared with normal germinal center B cells. The observation that RPS6 associates with 5′ TOP mRNA advances our understanding of the RPS6 regulation of ribosomal biogenesis. A systematic dissection of alternative mechanism(s) or factors involved in RPS6 regulation of 5′ TOP mRNA translation awaits further analysis and is an active area of research in our laboratory.
Materials and methods
Cell culture and transfection
Diffuse large B-cell lymphoma (Farage, SUDHL-6 and OCI-LY3) cells were cultured in RPMI-1640 containing 10% fetal bovine serum and 1% penicillin/streptomycin. Mammary epithelial adenocarcinoma (MCF-7) cells and cervical carcinoma (HeLa) cells were cultured in Dulbecco’s modified essential medium containing 10% of fetal bovine serum and 1% of penicillin/streptomycin (Mazan-Mamczarz et al., 2008). Plasmids used for RPS16 heterologous constructs were previously described (Levy et al., 1991). For RPS6 RNA interference transfections, siRNA targeting the RPS6 coding region (cat. number 140280703) and a control siRNA (Qiagen, Valencia, CA, USA) were used at 10 μM. Cells were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) when plasmids were used and with oligofectamine (Invitrogen) when siRNA oligonucleotides were used. At 48 h after transfection, cells were collected for analysis.
Western blotting
Western blots were carried out as previously described (Reinert et al., 2006). Blots were probed with antibodies recognizing: RPS6, hGH, glyceraldehyde 3-phosphate dehydrogenase, β-actin (Abcam, Cambridge, MA, USA), RPL11 (Abnova, Taipei, Taiwan), proliferating cell nuclear antigen, HuR (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or HDAC1 (Upstate, Lake Placid, NY, USA). Signals were detected with enhanced chemiluminescence according to the manufacturer’s instructions (Pierce, Rockford, IL, USA). Densitometric analysis was performed using Adobe Photoshop CS5 software (Adobe, San Jose, CA, USA).
Immunoprecipitation of mRNP complexes
For immunoprecipitation (IP) of ribonucleoprotein complexes, Farage, OCI-LY 3, MCF-7 and HeLa cells were collected, and cytoplasmic lysates (3 mg) were used for IP for 1 h at 4 °C in the presence of excess (30 μg) IP antibody RPS6 (Abcam) or control immunoglobulin G (IgG; BD Pharmingen, San Jose, CA, USA). Following washes with NT2 buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2 and 0.05% Nonidet P-40), beads were incubated (20 min, 55 °C) in 100 μl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K. RNA from IP material was extracted using phenol and chloroform method in the presence of GlycoBlue (Ambion, Austin, TX, USA). To confirm specificity of immunoprecipitation, an aliquot was resolved by SDS–polyacrylamide gel electrophoresis and immunoblotted for RPS6.
Polysome preparation and qPCR
HeLa and MCF-7 cells were collected by centrifugation and resuspended in a buffer containing 150 m KOAc, 20 m Hepes (pH 7.5), 2.5 m Mg (OAc)2, 2 m dithiothreitol, 1 m phenylmethanesulfonylfluoride and 200 μ/ml RNAsin as described (Lerner et al., 2003). After addition of 100 μg/ml digitonin, lysates were pelleted (10 000 × g, 1 min, 4 °C); the cytoplasmic extracts were then loaded onto 10–50% sucrose gradients and centrifuged (Beckman SW41, Beckman Coulter, Inc., Brea, CA, USA, 35 000 r.p.m., 3 h, 4 °C; Galbán et al., 2003). Subsequently, the material was fractionated into 1 ml aliquots. RNA from polysomal fractions or total RNA was extracted with Trizol reagent (Invitrogen). RNA from total or polysome fractions was reverse transcribed by using iScript complementary DNA synthesis kit (BioRad, Hercules, CA, USA) and the resulting complementary DNA was amplified by quantitative qPCR analysis using gene-specific primer pairs: 5′-cggagt-caacggatttggtcgtat-3′ and 5′-agccttctccatggtggtgaagac-3′ for glyceraldehyde 3-phosphate dehydrogenase, 5′-cgatgaacgcaa acttcgta-3′ and 5′-ttcggaccacataacccttc-3′ for RPS6, 5′-gtcacgt ggcccagatttat-3′ and 5′-tctccttcttggaagcctca-3′ for RPS16, 5′-ga aatgctccgctagcaatc-3′ and 5′-aacctgcctgtgaccttgtc-3′ for RPS21, 5′-aaggacttcctgctcacagc-3′ and 5′-aaaggtatctgctgcatcgaa-3′ for RPL38, 5′-tctcgctcttgtcgtgtctg-3′ and 5′-ccgatatccttcgcgtactg-3′ for RPS29, 5′-tggccacatatatgcgaatc-3′ and 5′-tgccatggtaa-cacttgtgg-3′ for RPL21, 5′-tcgggaaaaactagccaaaa-3′ and 5′-aaatcatgccaaagccagtt-3′ for RPS24, 5′-ccgcaaactctgtctcaaca-3′ and 5′-tgccaaaggatctgacagtg-3′ for RPL11, 5′-attatgctcggaaa cgcttg-3′ and 5′-acgggcataagcaatctgac-3′ for RPL5, 5′-gaggat ggcaagaaaagctg-3′ and 5′-agatttgacgaaggcgaaga-3′ for RPL7, 5′-gggcagatcttcaagcagac-3′ and 5′-ctcgaccttgtccatgtcct-3′ for hGH, 5′-tgttgttcaaacgggattca-3′ and 5′-ggctgggcacatttactgtt-3′ for BCL-2, 5′-atgtcaagccctccaacatc-3′ and 5′-ggcgacatgtaggaccttgt-3′ for MEK1, 5′-cagctttatcgccagagtcc-3′ and 5′-agtccttggtggtgatttcg-3′ for PTMA, or 5′-agtgagtaaggctgggcaga-3′ and 5′-aaggcacccaca gaaacaac-3′ for STAT3 mRNA. A BioRad iCycler instrument and iQSYBR Green Supermix (BioRad) were used to carry out the qPCR analysis.
Immunoflourescence
A total of 5 × 104 HeLa cells were seeded on glass slides and incubated in Dulbecco’s modied Eagle’s medium overnight. Cells were then treated with either dimethyl sulfoxide or 10 nM rapamycin for 8 h or 0.5 mM arsenite for 1 h. Cells were fixed in 3.7% formaldehyde for 10 min at room temperature, permeabilized (0.25% Triton X-100/phosphate-buffered saline, 10 min) and blocked for 1 h (phosphate-buffered saline/5% bovine serum albumin/0.5% NP40). Cells were incubated overnight at 4 °C with anti-DCP2 (Sigma, St Louis, MO, USA; 1:300), anti-RPS6 (Cell Signaling, Danvers, MA, USA; 1:50) and anti-TIA-1 (Santa Cruz Biotechnology; 1:100) in phosphate-buffered saline. Secondary detection was via incubation with anti-goat or anti-rabbit IgG Alexa Fluor 488 or anti-mouse IgG Alexa Fluor 568 secondary antibodies (Molecular Probes, Carlsbad, CA, USA; 1:1000) for 1 hr at room temperature and Hoescht 33342 nuclear dye (Sigma; 1:5000) for 30 s. Imaging was performed on an Olympus Flouview FV1000 (version 1.2.4.0) confocal microscope (Melville, NY, USA) with an Olympus F-view II 12-bit CCD digital camera and Olympus MicroSuiteTM Five acquisition software. Primary dimensions imaged were X and Y with a × 60 objective lens and oil immersion. Images generated were 512 × 512 pixel dimensions and a 16-bit image depth.
Immunohistochemistry
Paraffin sections (3 mm) from four normal reactive and six DLBCL lymph nodes were stained with a Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s protocol. The paraffin section slides were deparaffinized (Dai et al., 2006) with xylenes and rehydrated through graded alcohol washes, followed by antigen retrieval by microwave for 25 min in sodium citrate buffer (10 mmol/l, pH 6.0). Slides were then incubated in 3% hydrogen peroxide to quench endogenous horseradish peroxidase for 10 min. The slides were blocked by incubation in normal goat serum (dilution 1:10) in phosphate-buffered saline (pH 7.4) and subsequently incubated overnight at 4 °C with rabbit anti-human RPS6 antibody (1:150) and rabbit. Slides were then treated with biotinylated anti-rabbit IgG and incubated with preformed avidin-peroxidase complex. The sections were counterstained with hematoxylin, dehydrated and mounted. Negative controls were included.
Flow cytometry
Mission-TRC shRNA-encoding lentiviruses targeting human RPS6 (Sigma), were used for transduction in the OCI-LY3 and SUDHL-6 cell lines according to the manufacturer’s protocol. These shRNA-encoding lentiviruses targeting human RPS6 contained four individual clones: TRCN0000040079 (defined as no. 1), TRCN0000040080 (defined as no. 2), TRCN0000286292 (defined as no. 3) and TRCN0000286294 (defined as no. 4). We also used a control shRNA lentivirus (SHC-002 V) encoding an shRNA targeted against no known mouse or human gene. At 72 h after transduction, cells were collected for western blot analysis or flow cytometric analysis of apoptotic population by measuring Annexin V and Propidium Iodide staining (Southern Biotech ApoScreen Annexin V kit, Southern Biotech, Birmingham, AL, USA).
Statistical analysis
Results were analyzed using Microsoft Excel (Microsoft, Redmond, WA, USA).
Supplementary Material
Acknowledgments
The WT-RPS16-hGH and Cm5-RPS16-hGH constructs that contain the coding region of hGH fused to the first 29 nucleotides of the RPS16 5′ untranslated region (WT-RPS16-hGH) or to the mutant in which five of the eight pyrimidines within the 5′ TOP have been mutated to purines (Cm5) were kindly provided by the laboratories of Dr Oded Meyuhas and Dr George Thomas, This work was funded in part by a Merit Review Award from the Department of Veterans Affairs (RBG) and R01AA017972 from the National Institutes of Health (RBG).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributions: PRH performed the research, analyzed the data and wrote the paper. KMM performed the research, analyzed the data and wrote the paper. EMB, SSM and XFZ analyzed the data. SC performed the research. BD performed the research and analyzed the data. RBG conceived of and designed the research, analyzed the data and wrote the paper.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Avni D, Biberman Y, Meyuhas O. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Dai B, Kim O, Xie Y, Guo Z, Xu K, Wang B, et al. Tyrosine kinase Etk/BMX is up-regulated in human prostate cancer and its overexpression induces prostate intraepithelial neoplasia in mouse. Cancer Res. 2006;66:8058–8064. doi: 10.1158/0008-5472.CAN-06-1364. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Espinosa L, Martín M, Nicolas A, Fabre M, Navarro E. Primary sequence of the human, lysine-rich, ribosomal protein RPL38 and detection of an unusual RPL38 processed pseudogene in the promoter region of the type-1 angiotensin II receptor gene. Biochim Biophys Acta. 1997;1354:58–64. doi: 10.1016/s0167-4781(97)00124-3. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Manfredini R, Tagliafico E, Rossi E, Donelli A, Torelli G, et al. Noncoordinated expression of S6, S11, and S14 ribosomal protein genes in leukemic blast cells. Cancer Res. 1990;50:5825–5828. [PubMed] [Google Scholar]
- Fewell SW, Woolford JL., Jr Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol Cell Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Rosenfeld MG. Hormonally inducible phosphorylation of a nuclear pool of ribosomal protein S6. J Biol Chem. 1990;265:4321–4325. [PubMed] [Google Scholar]
- Frigerio JM, Dagorn JC, Iovanna JL. Cloning, sequencing and expression of the L5, L21, L27a, L28, S5, S9, S10 and S29 human ribosomal protein mRNAs. Biochim Biophys Acta. 1995;1262:64–68. doi: 10.1016/0167-4781(95)00045-i. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G. S6 Phosphorylation and signal transduction. In: Sonenberg N, Hershey JWB, Mathews M, editors. Translational Control of Gene Expression. Vol. 39. Cold Spring Harbor Laboratory Press; 2000. pp. 695–717. [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Martindale JL, Mazan-Mamczarz K, López de Silanes I, Fan J, Wang W, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi KA, Shimamura A. Ribosomal dysfunction and inherited marrow failure. Br J Haematol. 2008;141:376–387. doi: 10.1111/j.1365-2141.2008.07095.x. [DOI] [PubMed] [Google Scholar]
- Gross T, Nischt R, Gatermann K, Swida U, Käufer NF. Primary structure of the ribosomal protein gene S6 from Schizo-saccharomyces pombe. Curr Genet. 1988;13:57–63. doi: 10.1007/BF00365757. [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14:1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the ‘polypyrimidine tract’ mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′ TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, et al. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduru PR, Raju K, Vadmal V, Menezes G, Shah S, Susin M, et al. Correlation between mutation in P53, p53 expression, cytogenetics, histologic type, and survival in patients with B-cell non-Hodgkin’s lymphoma. Blood. 1997;90:4078–4091. [PubMed] [Google Scholar]
- Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 2001;21:2429–2433. [PubMed] [Google Scholar]
- Law PT, Tsui SK, Lam WY, Luk SC, Hwang DM, Liew CC, et al. A novel cDNA encoding a human homologue of ribosomal protein L29. Biochim Biophys Acta. 1996;1305:105–108. doi: 10.1016/0167-4781(95)00224-3. [DOI] [PubMed] [Google Scholar]
- Lee-Fruman KK, Kuo CJ, Lippincott J, Terada N, Blenis J. Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene. 1999;18:5108–5114. doi: 10.1038/sj.onc.1202894. [DOI] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008;27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. Elegans Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura JL, Garcia E, Pieper RO. S6K1 plays a key role in glial transformation. Cancer Res. 2008;68:6516–6523. doi: 10.1158/0008-5472.CAN-07-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O, Nilsson L. Translational dynamics. Interactions between the translational factors, tRNA and ribosomes during eukaryotic protein synthesis. Eur J Biochem. 1990;191:1–17. doi: 10.1111/j.1432-1033.1990.tb19087.x. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol. 2009;29:640–649. doi: 10.1128/MCB.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, et al. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianese G. Beitrag zur histologie und aetiologie der carcinoma. histologische und experimentelle untersuchungen. Beitr Pathol Anat Allgem Pathol. 1896;142:1–193. [Google Scholar]
- Reinert LS, Shi B, Nandi S, Mazan-Mamczarz K, Vitolo M, Bachman KE, et al. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res. 2006;66:8994–9001. doi: 10.1158/0008-5472.CAN-06-1999. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Durocher F, McArthur J, Tonin P, LeBlanc JF, Allen T, et al. Generation of a transcription map at the HSD17B locus centromeric to BRCA1 at 17q21. Genomics. 1995;28:530–542. doi: 10.1006/geno.1995.1185. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003a;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003b;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Katz M, Dreazen A, Gielchinsky Y, Saada A, Freedman N, et al. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4:e5618. doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Beato M, Sánchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101:1220–1235. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, et al. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol. 2002;22:8101–8113. doi: 10.1128/MCB.22.23.8101-8113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Vladimirov SN, Ivanov AV, Karpova GG, Musolyamov AK, Egorov TA, Thiede B, et al. Characterization of the human small-ribosomal-subunit proteins by N-terminal and internal sequencing, and mass spectrometry. Eur J Biochem. 1996;239:144–149. doi: 10.1111/j.1432-1033.1996.0144u.x. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.