Abstract
Little is known about how genetic variations in enhancers influence drug response. In this study, we investigated whether nucleotide variations in enhancers that regulate drug transporters can alter their expression levels. Using comparative genomics and liver-specific transcription factor binding site (TFBS) analyses, we identified evolutionary conserved regions (ECRs) surrounding nine liver membrane transporters that interact with commonly used pharmaceuticals. The top 50 ECRs were screened for enhancer activity in vivo, of which five—located around ABCB11, SLC10A1, SLCO1B1, SLCO1A2, and SLC47A1—exhibited significant enhancer activity. Common variants identified in a large ethnically diverse cohort (n = 272) were assayed for differential enhancer activity, and three variants were found to have significant effects on reporter activity as compared with the reference allele. In addition, one variant was associated with reduced SLCO1A2 mRNA expression levels in human liver tissues, and another was associated with increased methotrexate (MTX) clearance in patients. This work provides a general model for the rapid characterization of liver enhancers and identifies associations between enhancer variants and drug response.
Adverse drug events (ADEs) are a leading cause of morbidity and mortality in the modern world.1 It is estimated that up to 12% of hospitalizations in the United States can be attributed to ADEs.2 Among the many underlying factors, genetic variation has been shown to be an important cause of ADEs.3 Thus far, the majority of identified drug-associated nucleotide variants are protein coding; however, these account for only a fraction of the interindividual pharmacogenetic differences seen in patient populations. With several promoter variants linked to variability in drug response4–9 and association studies linking noncoding single-nucleotide polymorphisms (SNPs) to ADE susceptibility and altered drug response,10 it is likely that nucleotide variants in regulatory elements, such as enhancers, could also contribute to differences in drug response.
Membrane transporters are of great pharmacological importance because they are the targets for many commonly used prescription drugs and are major determinants of the absorption, distribution, metabolism, elimination, and transport of many xenobiotics. More specifically, liver membrane transporters play a critical role in the uptake and elimination of most drugs and xenobiotics through the liver (as reviewed in ref. 11). Numerous pharmacogenetics studies have demonstrated that nucleotide variations in protein-coding regions of liver membrane transporters exert a strong influence on drug response by altering transporter function in the liver.11–14 For example, genetic polymorphisms in OATP1B1 were shown to reduce elimination of the drug pravastatin,15 bosentan-induced liver injury has been observed in individuals with ABCB11 mutations,16 and response to metformin was reduced in carriers of SLC22A1 mutations.17
Haploinsufficiency of liver membrane transporters has also been shown to influence drug response. Removal of SLC22A1 in mice correlated with reduced uptake of metformin in hepatocytes, with heterozygous mice exhibiting intermediate levels,8 pointing to the importance of proper regulation of transporter expression. In addition, nucleotide variations in promoter sequences are thought to influence drug response by altering the transcription of transporters.4–7,18,19 More recently, it has been shown, using allelic imbalance, that differences in gene regulatory elements can lead to tissue-specific expression differences in a drug-associated protein, UGT2B15.20 Taken together, these studies demonstrate that differences in gene expression can alter the pharmacokinetic and pharmacodynamic characteristics of a drug, supporting the possibility that nucleotide variations in enhancers could also lead to similar consequences.
In this study, we identified enhancers that could regulate liver membrane transporters and investigated whether nucleotide variants in these enhancers can alter transporter expression levels. To identify potential enhancers, we used comparative genomics21,22 followed by transcription factor binding site (TFBS) analyses on genomic regions surrounding nine liver membrane transporters (SLC22A1, SLCO1B1, SLCO1B3, ABCC2, ABCB11, SLC22A7, SLCO2B1, SLC10A1, and SLC47A1) that are known to interact with commonly used pharmaceuticals (Table 1). We analyzed these sequences for enhancer activity using the hydrodynamic tail vein injection assay in mice (which involves a rapid intravascular injection of DNA into the mouse tail vein) to obtain specific expression of foreign DNA in the liver.23 Positive liver enhancers were sequenced for genetic variation in a large, ethnically diverse cohort, and common variants were analyzed for differential enhancer activity compared to the reference allele. Our results show that liver enhancer variants/haplotypes can lead to significant differences in reporter gene expression levels, thereby suggesting that regulatory elements can be an important underlying cause for interindividual drug response.
Table 1.
The nine selected liver membrane transporters and their substrates
| Gene | Common name(s) | Chromosomal position (hg18) | Substrates | References |
|---|---|---|---|---|
| ABCB11 | BSEP | chr2: 169,487,695–169,596,079 | Cyclosporin A, pravastatin, taurocholate, troglitazone | 35,36 |
| ABCC2 | MRP2 | chr10: 101,532,453–101,601,652 | Cyclosporin A, etoposide, methotrexate, mitoxantrone, olmesartan, SN-38, valsartan | 11,37,38 |
| SLC10A1 | NTCP | chr14: 69,312,305–69,333,707 | Bile salts, cyclosporine, rosuvastatin | 35,39 |
| SLC22A1 | OCT1 | chr6: 160,462,853–160,499,740 | Cisplatin, metformin | 17,40,41 |
| SLC22A7 | OAT2 | chr6: 43,373,976–43,381,254 | 5-Fluorouracil, allopurinol, bumetanide cGMP, erythromycin, paclitaxel, prostaglandin E2, theophylline | 42,43 |
| SLC47A1 | MATE1 | chr17: 19,377,759–19,422,938 | Cimetidine, metformin, oxaliplatin, paraquat | 44,45 |
| SLCO1B1 | OATPC/OATP1B1 | chr12: 21,175,404–21,283,997 | Bile salts, cerivastatin, cyclosporin A, estrone sulfate, estradiol 17β-D-glucronide, methotrexate, olmesartan, paclitaxel, pravastatin, rifampicin, rosuvastatin, SN-38 | 30,38,46,47 |
| SLCO1B3 | OATP8/OATP1B3 | chr12: 20,906,613–20,946,534 | Bosentan, cyclosporin A, fluvastatin, gemfibrozil, macrolide antibiotics, methotrexate, paclitaxel, pitavastatin, rifampicin, rosuvastatin, sildenafil | 30,38,46 |
| SLCO2B1 | OATPB/OATP2B1 | chr11: 74,548,492–74,594,947 | Atorvastatin, bosentan, fexofenadine, fluvastatin, glibenclamide, pravastatin, rosuvastatin | 11,30 |
Results
Liver membrane transporters have been shown to play an important role in regulating drug absorption, distribution, metabolism, elimination, and transport. For our study, we selected nine liver membrane transporters with well-documented drug substrates (Table 1).
Computational analysis
Using ECRbase (http://ecrbase.dcode.org), we identified human–mouse noncoding evolutionary conserved regions (ECRs) within the genomic regions surrounding the nine liver membrane transporters, up to and including the immediate neighboring genes. We identified 621 ECRs (Supplementary Table S1 online) with at least 70% sequence identity between human and mouse that is at least 100 bp (the commonly used conservation parameters24).
Next, we prioritized our ECRs according to the presence of liver-associated TFBSs, using MATCH,25 a TFBS analysis program. The following TFBSs were chosen, on the basis of their previous implication in liver-specific gene regulation: AP-1, C/ EBPβ, HNF-1, HNF-3β, GATA-3, and NF-1.26–28 Each of the 621 ECRs were analyzed for the prevalence of these TFBSs and then ranked by the total number of liver-specific TFBSs per bp. Topranking ECRs were analyzed manually for repetitive sequences and any coding evidence, using the UCSC Genome Browser database (http://genome.ucsc.edu), and ECRs that contained either of these were removed. The top 50 ECRs (Supplementary Table S2 online) were chosen for enhancer assays.
Enhancer identification screen
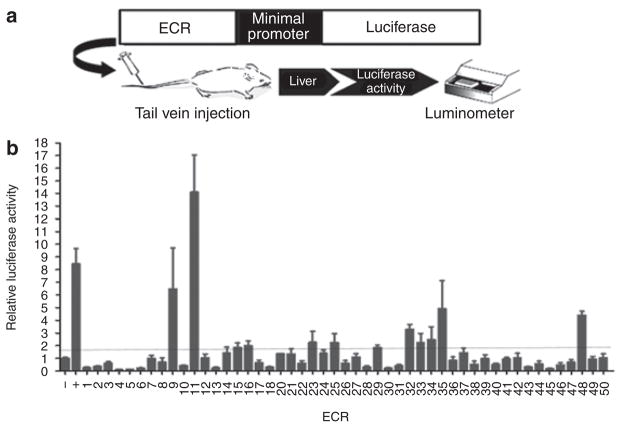
The selected ECRs (Supplementary Table S2 online) were cloned into a luciferase reporter enhancer assay vector (pGL4.23; Promega, Madison, WI) and tested for enhancer activity using the in vivo hydrodynamic mouse tail vein assay (Figure 1a). In addition to the enhancer construct, we injected a Renilla luciferase construct, pGL4.74[hRluc/TK] (Promega), to correct for injection efficiency. On each injection day, we also injected an empty pGL4.23[luc2] vector and the apolipoprotein E (APOE) liver enhancer29 as negative and positive controls, respectively. At least three mice were injected per construct, including our controls. Our initial screen resulted in twelve ECRs exhibiting enhancer activity, as defined by an increase in luciferase activity of at least 1.8-fold relative to the negative control (Figure 1b). Each of these twelve ECRs was followed up by reinjection of the construct in at least nine mice in order to establish statistical significance. Following these reinjections, the enhancer activities of ECRs 11, 29, 32, 35, and 48 were found to be statistically significant (P < 0.05) as compared with the negative control (Table 2). These ECRs were in the vicinity of ABCB11 (ECR11), SLC10A1 (ECR29, ECR32), SLC47A (ECR48), and SLCO1B1 and SLCO1A2 (ECR35). SLCO1A2 was not selected in our initial bioinformatic analyses screen; however, it is located downstream of SLCO1B1 and was therefore a good candidate to consider.
Figure 1.
The hydrodynamic tail vein enhancer assay and results of the initial screen. (a) A schematic illustration of the in vivo hydrodynamic tail vein injection assay. Evolutionary conserved regions (ECRs) were cloned into a luciferase reporter plasmid and injected into the tail vein of the mouse. Relative luciferase activity was assayed 24 hours after the injection. (b) The initial enhancer screen yielded twelve ECRs (9, 11, 15, 16, 23, 25, 29, 32, 33, 34, 35, and 48) that exhibited relative luciferase activity ≥1.8-fold higher than the negative (−) control (pGL4.23). A known liver enhancer of the APOE gene was used as the positive (+) control. Three to five mice were injected for each construct.
Table 2.
Follow-up enhancer assays
| Construct | Transporter | Initial enhancer screen (n = 3–5) | Follow-up screen (n = 6–9) | Combined (n = 9–12) | Combined P value |
|---|---|---|---|---|---|
| ECR9 | SLC10A1 | 6.78 | 0.02 | 2.17 | 0.453 |
| ECR11 | ABCB11 | 14.09 | 4.05 | 8.07 | 0.035 |
| ECR15 | SLC10A1 | 1.85 | 1.19 | 1.39 | 0.604 |
| ECR16 | SLCO1B1 | 2.00 | 1.38 | 1.58 | 0.117 |
| ECR23 | SLC47A1 | 2.26 | 0.94 | 1.30 | 0.483 |
| ECR25 | ABCC2 | 2.23 | 1.38 | 1.62 | 0.222 |
| ECR29 | SLC10A1 | 1.85 | 1.67 | 1.73 | 0.002 |
| ECR32 | SLC10A1 | 3.29 | 3.48 | 3.42 | <0.0001 |
| ECR33 | ABCB11 | 2.23 | 0.04 | 0.70 | 0.533 |
| ECR34 | SLC47A1 | 2.49 | 1.53 | 1.85 | 0.117 |
| ECR35 | SLCO1B1 | 4.93 | 4.92 | 4.92 | 0.008 |
| ECR48 | SLC47A1 | 4.41 | 2.72 | 3.28 | 0.001 |
The follow-up hydrodynamic tail vein experiments for the twelve ECRs that initially exhibited relative luciferase activity ≥1.8-fold higher than the negative control. Results are shown as the relative luciferase activity as compared with the negative control for the initial experiments (n = 3–5 mice), follow-up experiments (n = 6–9 mice), and both combined (n = 9–12 mice). ECRs 11, 29, 32, 35, and 48 were found to have significantly higher relative luciferase activity as compared with the negative control (P values depicted in boldface; Student’s t-test).
ECR, evolutionary conserved region.
Sequencing and functional variant analysis
We sequenced the five positive liver enhancers in 272 subjects from the SOPHIE (Studies of Pharmacogenetics in Ethnically Diverse Populations) cohort (as described in the Methods section). Common variants with a minor allele frequency >1% (Table 3) were selected for differential enhancer activity assays by comparing them to the reference allele (the allele with the largest frequency). Sequencing of the positive enhancers showed that some did not correspond to the reference allele. Consequently, we cloned the reference allele along with the other common alleles into the pGL4.23 enhancer assay vector by using the individuals with the specific haplotype as a template for amplification. Plasmids were verified through sequencing as having the correct haplotype. We injected each of these different haplotypes into at least nine mice. Of the eleven variant/haplotypes tested, 3 exhibited significant differential enhancer activity as compared with their respective reference alleles (Table 3). Two of the three variants were found in ECR35, and the third was found in ECR32.
Table 3.
The various ECR haplotypes and their enhancer activity levels
| ECR | Variant/haplotype | dbSNP ID | PMT SNP ID | Chromosomal location (hg18) | Nucleotide change | Allele frequency by ethnicity | P value (relative to REF) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | CA | AS | ME | RLA | |||||||
| 11 | REF | 0.937 | 0.984 | 0.893 | 0.935 | 3.152 | |||||
| *2*3 | 5973 | Chr2: 169647588 | G→A | 0 | 0 | 0.024 | 0.065 | 4.536 | NS | ||
| rs73026864 | 5972 | Chr2: 169647612 | G→A | ||||||||
| *3*5*6*7 | rs73026864 | 5972 | Chr2: 169647612 | G→A | 0.056 | 0 | 0 | 0 | 1.722 | NS | |
| rs73026864 | 5970 | Chr2: 169647657 | G→A | ||||||||
| 5969 | Chr2: 169647696 | G→A | |||||||||
| rs73026869 | 5968 | Chr2: 169647698 | T→C | ||||||||
| *8 | 5967 | Chr2: 169647904 | A→G | 0.024 | 0.008 | 0 | 0.008 | 1.779 | NS | ||
| 29 | REF | 0.890 | 0.993 | 0.978 | 1.000 | 1.462 | |||||
| *6 | rs729458 | 6669 | Chr14: 69388040 | T→C | 0.081 | 0 | 0 | 0 | 2.086 | NS | |
| 32 | REF | 0.780 | 0.694 | 0.821 | 0.735 | 0.981 | |||||
| *1 | rs10162517 | 6677 | Chr14: 69413537 | A→G | 0.205 | 0.306 | 0.179 | 0.228 | 1.932 | 0.0118 | |
| 35 | REF | 0.455 | 0.676 | 0.662 | 0.787 | 3.727 | |||||
| *1*3 | rs11045981 | 6437 | Chr12: 21378250 | T→G | 0.037 | 0.147 | 0 | 0.074 | 2.577 | NS | |
| rs4148981 | 6439 | Chr12: 21378335 | C→T | ||||||||
| *2*3 | 6438 | Chr12: 21378301 | A→AA | 0.082 | 0.029 | 0 | 0.022 | 0.797 | <0.0001 | ||
| rs4148981 | 6439 | Chr12: 21378335 | C→T | ||||||||
| *3 | rs4148981 | 6439 | Chr12: 21378335 | C→T | 0.425 | 0.147 | 0.338 | 0.118 | 1.081 | <0.0001 | |
| 48 | REF | 0.632 | 0.669 | 0.433 | 0.559 | 2.661 | |||||
| *1 | rs4646800 | 6459 | Chr17: 19502021 | T→G | 0.331 | 0.301 | 0.567 | 0.412 | 2.205 | NS | |
| *3 | 6461 | Chr17: 19502026 | A→T | 0.044 | 0.007 | 0 | 0.007 | 2.759 | NS | ||
Of the eleven variants/haplotypes tested against their reference alleles (REF), three (P values depicted in boldface; Student’s t-test for ECRs 29 and 32; analysis of variance with Dunnett’s test for ECRs 11, 35, and 48) were found to have significant differences in enhancer activity.
AA, African American; AS, Asian; CA, Caucasian; dbSNP, Single-Nucleotide Polymorphism Database (hosted by the National Center for Biotechnology Information); ECR, evolutionary conserved region; ME, Mexican; NS, not significant; PMT, Pharmacogenomics of Membrane Transporters Project; REF, reference allele; RLA, relative luciferase activity; SNP, single-nucleotide polymorphism.
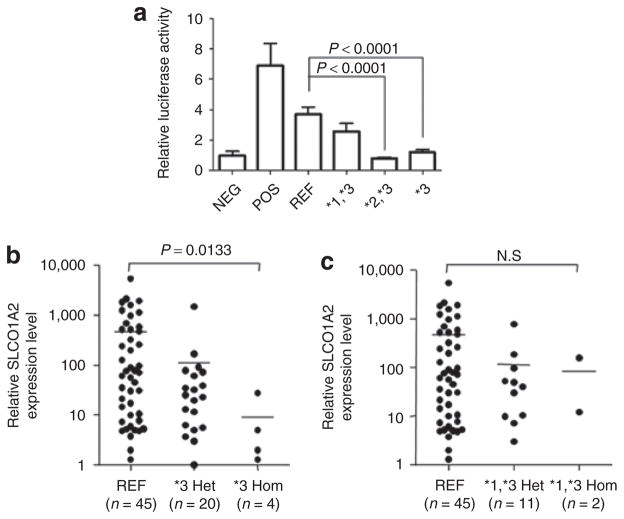
The results for ECR35 and its variants are particularly interesting. ECR35 has three common variants present in the SOPHIE cohort (Table 3). These SNPs, two of which— ECR35*1 (rs11045981) and ECR35*3 (rs4148981)—have been reported, and one novel SNP, ECR35*2, are present in most ethnic groups. ECR35*3 (rs4148981) is found in all four ethnic groups, with an average minor allele frequency of 26%, making it the most prevalent SNP that altered enhancer activity in our study. Of the four common haplotypes, three share ECR35*3; however, only the latter two haplotypes— ECR35*2,*3 and ECR35*3—exhibited significantly reduced enhancer activity (Figure 2a; P < 0.0001; one-way analysis of variance with Dunnett’s multiple-comparison test). The ECR35*1*3 haplotype followed a similar trend but was not found to reduce reporter expression to a statistically significant extent.
Figure 2.
Enhancer activity of the different ECR35 haplotypes and their association with SLCO1A2 mRNA levels. (a) Differential enhancer activity levels of ECR35 haplotypes. ECR35*2,*3 and ECR*3 haplotypes exhibit significantly decreased enhancer activity as compared to the reference allele (one-way analysis of variance with Dunnett’s multiple-comparison test). (b) ECR35*3 haplotype is associated with a significantly lower expression of SLCO1A2 mRNA levels as compared with the reference allele (P = 0.0133, Kruskal–Wallis test followed by Dunn’s test). (c) The ECR35*1,*3 haplotype is not associated with differences in SLCO1A2 mRNA expression. ECR, evolutionary conserved region; Het, heterozygous; Hom, homozygous; REF, reference allele.
Association of ECR35 variants with mRNA transporter expression levels in human liver
Next, we tested for association of mRNA expression levels with our functional enhancer variants. Because of the costs and the limited amount of human tissue samples available (n = 88), we only analyzed the variants for ECR35, which lies in an intron of SLCO1A2, the gene downstream of SLCO1B1. SLCO1A2 is also known to interact with many prescription drugs such as erythromycin, statins, and methotrexate (MTX).30 Using real-time PCR, SLCO1A2 and SLCO1B1 mRNA levels were quantified in the 88 Caucasian human liver tissue samples. Extensive interindividual variation (mean ± SD) was noted for the SLCO1A2 transporter (relative expression 284 ± 722). By contrast, SLCO1B1 mRNA levels were much less variable (relative expression 22.8 ± 24.9).
Sequencing of all 88 samples revealed that the four ECR35 common haplotypes identified in the SOPHIE cohort were present in our tissue samples. However, the ECR35*2,*3 haplotype was found in only one of our samples and was therefore not analyzed further. We examined whether ECR35*1,*3 and ECR35*3 affected mRNA expression of each transporter. We did not observe any significant SLCO1B1 mRNA expression level differences associating with either of these haplotypes (data not shown). However, we observed lower SLCO1A2 mRNA levels associated with increasing numbers of the respective T allele for the ECR35*3 SNP (Figure 2b; P = 0.0133; Kruskal–Wallis followed by Dunn’s test), as in the enhancer assay results. Comparison of ECR35*3 carriers and noncarriers yielded similar findings (P = 0.0128, Mann–Whitney test; data not shown). For the ECR35*1*3 haplotype, we did not observe a significant association with mRNA expression of SLCO1A2 (Figure 2c; P = 0.05, Kruskal–Wallis followed by Dunn’s test); this finding was analogous to our observations in the enhancer assays. Similar results were found when comparing ECR35*1*3 carriers and noncarriers (P > 0.05, Mann–Whitney test; data not shown).
ECR35 SNPs and MTX clearance
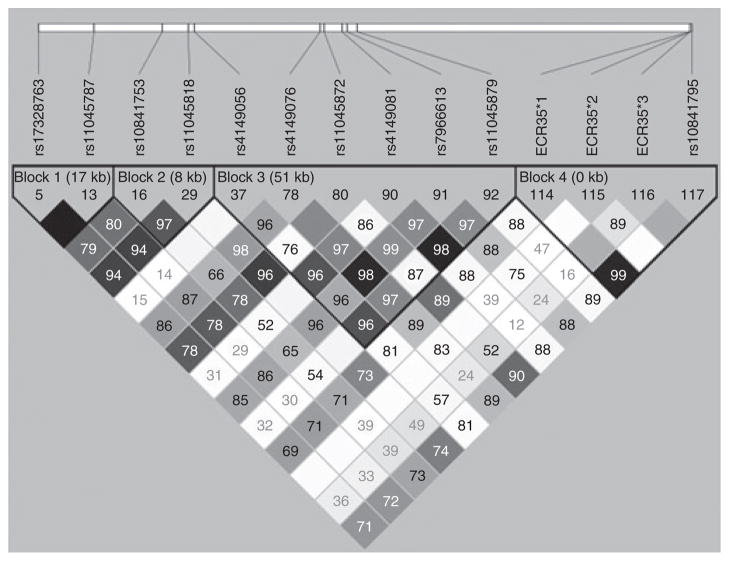
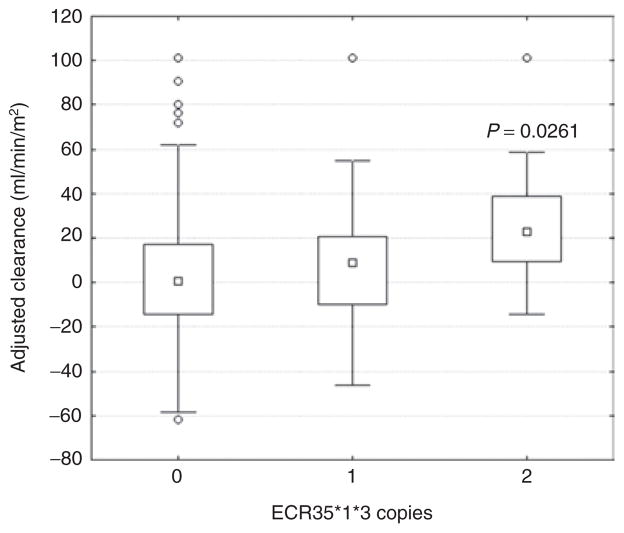
OATP1A2 (the protein coded from SLCO1A2) has been shown to be important for the clearance of MTX, a drug widely used in the treatment of malignant and autoimmune diseases. In addition, through a genome-wide association study (GWAS), the SLCO1B1–SLCO1A2 genomic region was recently associated with MTX clearance and gastrointestinal toxicity.31 We examined the ECR35 variants for their involvement in interindividual differences in MTX clearance. We genotyped ECR35 in the same population from the GWAS study,31 consisting of 639 children, all of whom received multiple courses of MTX at 2–5 g/m2. In this population, ECR35 SNPs were not found to be in strong linkage disequilibrium (LD) with the previously discovered GWAS-associated SNPs (Figure 3; Supplementary Table S3 online). We analyzed these SNPs for their association with MTX clearance. Only haplotype ECR35*1*3 showed a significant association with MTX clearance, and the homozygous allele correlated with increased clearance as compared with the reference allele (P = 0.045; Kruskal–Wallis test; data not shown). The significance (P = 0.0354; Kruskal–Wallis test; data not shown) was also maintained after removing individuals carrying the nonsynonymous SNP, rs4149056 (SLCO1B1 T521C), the most strongly associated SNP with MTX clearance.31 This suggests that the ECR35*1*3 haplotype may influence MTX clearance independent of the GWAS-detected SNPs. Analysis of the ECR35*1,*3 haplotype demonstrated that increasing copies of the ECR35*1,*3 allele significantly associated with increased MTX clearance (Figure 4; P = 0.0261; Kruskal– Wallis). Interestingly, in this population, ECR35 SNP*1 is also in strong LD (Figure 3; D′ = 0.991; r2 = 0.939) with rs10841795 (SLC01A2 T38C), a SNP which has been previously shown to increase MTX transport.32,33
Figure 3.
Linkage disequilibrium (LD) plot of ECR35 SNPs with SLCO1B1 and SLCO1A2 SNPs that are associated with methotrexate clearance. ECR35 SNPs are not in LD with any of the previously described SLCO1B1 genome-wide association study SNPs.31 However, there is strong linkage (D′ = 0.99; r2 = 0.939) between ECR35*1 and the SLCO1A2 SNP rs10841795 (refs. 32,33).
Figure 4.
Haplotype analysis shows an association between the ECR35*1*3 haplotype and increased methotrexate (MTX) clearance. MTX clearance was adjusted for age, sex, race, and treatment arm. Regression analysis of the number of copies of the ECR35*1,*3 haplotype showed a significant association with MTX clearance (P = 0.0261), with the homozygous allele (2) correlating with increased clearance as compared with the reference allele (0). Individuals who are carriers of: *1*1*3*3 = 2; *1*3, *1*2*3*3, and *1*3*3 = 1; *2, *3, *2*3, and wild type = 0. ECR, evolutionary conserved region.
Discussion
Using comparative genomics, we identified noncoding ECRs in the vicinity of nine liver membrane transporters (SLC22A1, SLCO1B1, SLCO1B3, ABCC2, ABCB11, SLC22A7, SLCO2B1, SLC10A1, and SLC47A1) that have well-characterized drug interactions. These sequences were then ranked for the prevalence of previously characterized liver TFBSs, and the top 50 sequences were assayed for liver enhancer activity using the mouse hydrodynamic tail vein injection technique. The initial screening resulted in twelve ECRs that exhibited luciferase activity at or above the cutoff value (1.8-fold). This value could be more stringent in future screenings because we saw that the majority of ECRs with activity between 1.8 and 2.0-fold decreased in reporter gene activity when followed up and subsequently lost statistical significance.
Five sequences that exhibited significant enhancer activity were further characterized for sequence variation in an ethnically diverse cohort. These were in the vicinity of ABCB11, SLC10A1, SLCO1B1, SLCO1A2, and SLC47A1. We found eleven common variants within these five ECRs that we assayed for differential enhancer expression. Of these, three showed significant enhancer activity differences as compared with their reference allele. These variants showed significant minor allele frequency differences between the ethnic groups, suggesting that enhancer variants can also contribute to ethnic-specific differences in drug response, similar to what has been observed for coding SNPs. We performed a more extensive analysis of one enhancer, ECR35. This enhancer is located within an intron of SLCO1A2, a transporter that interacts with a variety of statins, anticancer and antibacterial drugs.30 We identified four different common haplotypes within this ECR, two of which led to significantly reduced enhancer expression levels.
Using human liver tissue samples, we were able to show that ECR35*3 by itself led to reduced SLCO1A2 mRNA levels. However, this SNP in combination with ECR35*1 did not cause a significant reduction of SLCO1A2 mRNA levels despite demonstrating a trend to reduced expression. This suggests that ECR35*1 may mitigate the ability of ECR35*3 to reduce expression of SLCO1A2. Analysis of ECR35 variants for the six liver-specific TFBSs used to rank the ECRs found no changes in binding sites for either ECR35*1 or ECR35*3.
We observed a correlation for the ECR35 variants between the enhancer assay results and SLCO1A2 mRNA expression levels. Our studies were limited due to our small human liver tissue sample size (n = 88) and the large variation in SLCO1A2 mRNA expression levels (22.8 ± 24.9). Additional studies would have to be done to explicitly link ECR35 SNP*3 as the causative SNP that leads to reduced SLCO1A2 expression. We also found another variant that exhibited increased enhancer activity in another enhancer, ECR32, which is near SLC10A1, an uptake transporter for bile salts and cyclosporin A.3 Performing drug-associated studies with this variant would also be of interest because the ECR32*1 variant led to increased reporter gene activity relative to the reference allele.
With ECR35 variants leading to significantly reduced reporter gene and mRNA expression levels, we analyzed their potential effects on OATP1A2 and its associated drug interactions. Coding mutations in OATP1A2 have been shown to affect the transport of MTX32 and other substances,33 and the SLCO1B1–SLCO1A2 genomic region has been found to be associated with reduced MTX clearance and gastrointestinal toxicity.31 Potentially, the reduced MTX clearance could be due to lower SLCO1A2 mRNA expression, attributable to the ECR35 variants. Although we did not observe ECR35 SNPs to be in strong LD with the MTX GWAS-associated SNPs in the SLCO1B1–SLCO1A2 genomic region, we did observe haplotype ECR35*1*3 to be associated with increased MTX clearance. This haplotype led to slightly reduced reporter gene expression in the enhancer assay but did not meet the criteria for significance (Table 3). However, in this population, SNP*1 was found to be in strong LD (D′ = 0.991; r2 = 0.939) with the nonsynonymous SLCO1A2 rs10841795 SNP (T38C; Ile13Thr), which was previously shown to lead to increased MTX uptake when expressed in Xenopus laevis oocytes.32 In the kidney, the predominant organ involved in MTX elimination, OATP1A2 is localized to the apical domain of distal nephrons.33 Higher MTX uptake, as observed in association with rs10841795, should lead to increased reabsorption and lower MTX clearance, which is contrary to our observations. The differences between our in vivo observations and these functional studies might be due to the assay having been carried out in vitro and/or the differences in pH concentrations in the kidney. The pH of the distal tubule is variable, and MTX transport by the reference OATP1A2 allele has been shown to be highly dependent on pH.32
Further studies need to be performed to determine the actual in vivo functional outcomes of rs10841795 and SNP*1, considering their effect on OATP1A2 in the kidney, liver, intestine, and brain. As for ECR35*3, all of our assays were carried out on liver tissues; therefore, its functional role in the kidney—the major organ associated with MTX clearance—is unknown. It would be interesting to investigate whether ECR35 is important for SLCO1A2 expression in the kidney and whether the enhancer affects interindividual drug interactions of other OATP1A2- transported drugs in general.
This study is the first to link nucleotide variations in enhancers with drug-associated genes, providing further evidence that regulatory sequences could be important determinants of differential drug response and ADEs. With the advancement of sequencing technologies, individual genome sequences will soon be available at an affordable price to the general public. The ability to obtain the genetic blueprint of an individual will greatly advance the quality of pharmacological treatment by tailoring drugs to fit that blueprint and ultimately reduce ADEs and improve drug efficacy overall. However, the major hurdle will be the development of high-throughput functional assays that can rapidly interpret the pharmacological nature of the nucleotide change(s) in a given individual, both in coding and in regulatory sequences.
Methods
Identification and ranking of ECRs
Using ECRbase (http://ecrbase.dcode.org), we selected intronic and intergenic ECRs for each transporter that were at least 100 bp long and with at least 70% sequence identity between human and mouse. We parsed conservation data from ECRbase using in-house software (available upon request). ECRs were chosen in proximity to the gene all the way to the next neighboring gene including the neighboring gene’s entire genomic sequence. In the case of ABCB11, given the close proximity of the 3′ neighboring gene, G6PC2 (~13 kb), we searched for ECRs two genes away from the transporter, including the genomic region of SPC25. This analysis generated 621 unique ECRs (Supplementary Table S1 online). To prioritize ECRs for functional assays, we scanned these ECRs for liverspecific TFBSs: AP-1, C/EBPβ, HNF-1, HNF-3β, GATA-3, and NF-1, using MATCH.25 The ECRs were ranked based on the total number of liver-specific TFBS per bp of each ECR.
Cloning of ECRs
The top 50 ECRs ranked by the number of TFBSs per bp of ECR were tested for enhancer activity. Primers (Supplementary Table S2 online) were designed to have up to 200 bp of sequence flanking each side of the human–mouse conserved sequence (previous experiments have shown this addition to be a reliable method for obtaining positive enhancer activity22). ECRs were PCR-amplified using TopTaq (Qiagen, Germantown, MD) from human genomic DNA (Roche, Indianapolis, IN), purified by gel extraction (QIAquick gel extraction kit; Qiagen) and cloned into the pENTR-dTOPO vector (Invitrogen, Carlsbad, CA). After plasmid DNA purification using the QIAprep spin miniprep kit (Qiagen), proper orientation was confirmed by restriction enzyme digest. Clones in correct orientation were subsequently subcloned into the pGL4.23[luc2] vector (Promega) containing the Gateway reading frame A (Invitrogen). Orientation of the insert was re-verified by restriction enzyme digest, and endotoxin-free plasmid DNA was isolated using the EndoFree Plasmid Midi prep (Qiagen). ECR19 was not followed up with injections because of cloning difficulties.
In vivo hydrodynamic tail vein enhancer assay
For the hydrodynamic tail vein assay, each assayed sequence, cloned in the pGL4.23[luc2], was initially injected (10 μg) alongside 2 μg of pGL4.74[hRluc/TK] vector to correct for injection efficiency, into at least three CD1 mice (Charles River Laboratories, Wilmington, MA) using the TransIT EE hydrodynamic gene delivery system (Mirus Bio, Madison, WI) according to the manufacturer’s protocol. Negative (empty pGL4.23[luc2]) and positive (ApoE liver enhancer)29 controls (n = 3–5) were also injected in parallel at each injection date/experiment. After 24 h, the mice were euthanized, and the livers were harvested and homogenized in passive lysis buffer (Promega), followed by centrifugation at 4 °C for 30 min at 14,000 rpm. Firefly luciferase and Renilla luciferase activity in the supernatant (diluted 1:20) were measured on a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT) in replicates of six for each liver, using the Dual-Luciferase reporter assay system (Promega). The ratios for firefly luciferase:Renilla luciferase were determined and expressed as relative luciferase activity. Experiments for each construct were repeated in at least nine mice if analysis of the initial screen had an average increase in activity that was at least 1.8-fold higher than the negative control. All mouse work was approved by the UCSF Institutional Animal Care and Use Committee.
Nucleotide variant sequencing and cloning
Selected positive liver enhancers were sequenced in a panel of 272 ethnically diverse subjects (African Americans (41 female, 27 male), Caucasians (34 female, 34 male), Chinese (44 female, 24 male), and Mexicans (49 female, 19 male)) from the SOPHIE cohort.19 In addition, human liver tissues and the MTX cohort were also sequenced as previously described.31 Enhancers with common variants (minor allele frequency >1%) were PCR-amplified from human genomic DNA from SOPHIE cohort individuals encompassing this variant/haplotype as listed in Table 2. The amplified DNA was inserted into the pENTR-dTOPO vector as previously described and directly sequenced to verify the variant(s) before being subcloned into the pGL4.23[luc2] vector. All enhancer sequencing results can be found on the PMT database (http://pharmacogenetics.ucsf.edu).
Human liver samples
Liver tissue samples were purchased from Asterand (Detroit, MI) and donated by SRI International (Menlo Park, CA). All tissues were flash frozen immediately after collection and stored at −80 °C. Isolation of total RNA and DNA from the tissues was performed using Qiagen Allprep DNA/RNA kits according to the manufacturer’s protocol. Equal amounts of total RNA were reverse transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCRs for SLCO1A2, SLCO1B1, and β-actin were carried out with ready-to-use TaqMan gene expression assays (Applied Biosystems). The thermal cycling conditions used were as follows: 2 min at 50 °C, 10 min at 95 °C followed by 15 s at 95 °C, and 1 min at 60 °C, for 40 cycles. The real-time PCR efficiencies were evaluated with twofold serial dilutions in a pool of all starting RNAs in order to confirm accuracy and linearity. Efficiency was calculated from the slopes of the calibration curve according to the equation E = 10 [−1/slope], as previously described.34 Real-time PCR efficiencies of SLCO1A2, SLCO1B1, and β-actin were 1.90 ± 0.05, 1.91 ± 0.08, and 2.02 ± 0.10, respectively, which were consistently similar to a static efficiency of 2. Therefore, expression values of both target genes were normalized to β-actin, and the 2−ΔCt standard method of relative quantification was used. Genotyping was performed by direct sequencing as described above. This study was approved by the UCSF Committee on Human Research.
Pharmacokinetic data for patients receiving MTX
A total of 639 children with acute lymphoblastic leukemia who were receiving treatment according to St Jude Children’s Research Hospital’s Total XIIIB and Total XV protocols were genotyped for ECR35 by Sanger sequencing.30 Germline DNA was extracted from the blood after remission was achieved. MTX clearance was estimated as described previously.30
Statistical analysis
Data are expressed as the mean ratio firefly luciferase:Renilla luciferase activity as compared with the negative control (+ SEM). Statistical analysis for enhancer identification was carried out using the unpaired Student’s t-test, assuming equal variance compared to the negative control. The activity levels of the variants were also analyzed using the unpaired t-test and compared with the enhancer’s reference luciferase activity. Statistical comparison of luciferase activity for ECR11, ECR35, and ECR48 variants with their respective reference alleles was performed using one-way analysis of variance followed by Dunnett’s multiple- comparison test. Statistical comparisons between different genotypes and haplotypes from the human liver tissue samples were made using the Mann–Whitney and Kruskal–Wallis tests followed by Dunn’s multiple-comparison test. Data were analyzed using GraphPad Prism, version 5.0 (GraphPad, La Jolla, CA). Associations between MTX clearance and ECR haplotypes were evaluated using a general linear model, treating genotype as a numerical variable (0 = AA, 1 = AB, and 2 = BB) and including age, race, sex, and treatment regimen as covariates.30 LD was calculated using Haploview (http://www.broadinstitute.org/haploview). A P value ≤0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This research was supported by NIH NIGMS grant GM61390 and by NIH/ NCRR UCSF-CTSI grant UL1 RR024131. N.A. is also supported by award R01HD059862 from the NICHD and award R01HG005058 from the NHGRI. Grant support for L.B.R. and M.V.R. came from NIH R37 CA36401, U01 GM92666, P30 CA21765, and ALSAC (American Lebanese Syrian Associated Charities). We thank the Ahituv lab for helpful comments on the manuscript. We also thank the clinical and research staff at St Jude Children’s Research Hospital, particularly Wenjian Yang, Pamela McGill, and Jeremy Hunt, as well as the patients and their families for their participation. We thank the staff of the St Jude Hartwell Center for Bioinformatics and Biotechnology. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, the NIGMS, the NICHD, or the NHGRI.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Conflict of Interest
The authors declared no conflict of interest.
References
- 1.Murphy SL. Deaths: final data for 1998. Natl Vital Stat Rep. 2000;48:1–105. [PubMed] [Google Scholar]
- 2.Burt CW, McCaig LF, Rechtsteiner EA. Ambulatory medical care utilization estimates for 2005. Adv Data. 2007;388:1–15. [PubMed] [Google Scholar]
- 3.Sadée W, Dai Z. Pharmacogenetics/genomics and personalized medicine. Hum Mol Genet. 2005;14 (spec 2):R207–R214. doi: 10.1093/hmg/ddi261. [DOI] [PubMed] [Google Scholar]
- 4.Schillani G, et al. 5-HTTLPR polymorphism of serotonin transporter and effects of sertraline in terminally ill cancer patients: report of eleven cases. Tumori. 2008;94:563–567. doi: 10.1177/030089160809400419. [DOI] [PubMed] [Google Scholar]
- 5.Yee SW, et al. Identification and characterization of proximal promoter polymorphisms in the human concentrative nucleoside transporter 2 (SLC28A2) J Pharmacol Exp Ther. 2009;328:699–707. doi: 10.1124/jpet.108.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JH, et al. Identification and characterization of novel polymorphisms in the basal promoter of the human transporter, MATE1. Pharmacogenet Genomics. 2009;19:770–780. doi: 10.1097/FPC.0b013e328330eeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva H, et al. Serotonin transporter polymorphism and fluoxetine effect on impulsiveness and aggression in borderline personality disorder. Actas Esp Psiquiatr. 2007;35:387–392. [PubMed] [Google Scholar]
- 8.Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 2007;12:247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- 9.Lankisch TO, et al. Identification and characterization of a functional TATA box polymorphism of the UDP glucuronosyltransferase 1A7 gene. Mol Pharmacol. 2005;67:1732–1739. doi: 10.1124/mol.104.007146. [DOI] [PubMed] [Google Scholar]
- 10.Sadee W. Regulatory polymorphisms in key candidate genes for disease susceptibility and drug response: a mandate for valid genetic biomarkers. Expert Rev Mol Diagn. 2010;10:9–11. doi: 10.1586/erm.09.72. [DOI] [PubMed] [Google Scholar]
- 11.Giacomini KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ieiri I, Higuchi S. Pharmacogenomics: inter-ethnic and intra-ethnic differences in pharmacokinetic and pharmacodynamic profiles of clinically relevant drugs. Yakugaku Zasshi. 2009;129:231–235. doi: 10.1248/yakushi.129.231. [DOI] [PubMed] [Google Scholar]
- 13.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234:4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 14.Gerloff T. Impact of genetic polymorphisms in transmembrane carriersystems on drug and xenobiotic distribution. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:69–77. doi: 10.1007/s00210-003-0813-5. [DOI] [PubMed] [Google Scholar]
- 15.Kivistö KT, Niemi M. Influence of drug transporter polymorphisms on pravastatin pharmacokinetics in humans. Pharm Res. 2007;24:239–247. doi: 10.1007/s11095-006-9159-2. [DOI] [PubMed] [Google Scholar]
- 16.Lang C, et al. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- 17.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahara H, et al. Functional genetic variation in the basal promoter of the organic cation/carnitine transporters OCTN1 (SLC22A4) and OCTN2 (SLC22A5) J Pharmacol Exp Ther. 2009;329:262–271. doi: 10.1124/jpet.108.146449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesselson SE, et al. Genetic variation in the proximal promoter of ABC and SLC superfamilies: liver and kidney specific expression and promoter activity predict variation. PLoS ONE. 2009;4:e6942. doi: 10.1371/journal.pone.0006942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C, Southard C, Witonsky DB, Olopade OI, Di Rienzo A. Allelic imbalance (AI) identifies novel tissue-specific cis-regulatory variation for human UGT2B15. Hum Mutat. 2010;31:99–107. doi: 10.1002/humu.21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:116–130. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pennacchio LA, et al. In vivo enhancer analysis of human conserved noncoding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 24.Nóbrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- 25.Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krivan W, Wasserman WW. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandenbon A, Nakai K. Modeling tissue-specific structural patterns in human and mouse promoters. Nucleic Acids Res. 2010;38:17–25. doi: 10.1093/nar/gkp866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonet WS, Bucay N, Lauer SJ, Taylor JM. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J Biol Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- 30.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treviño LR, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badagnani I, et al. Interaction of methotrexate with organic-anion transporting polypeptide 1A2 and its genetic variants. J Pharmacol Exp Ther. 2006;318:521–529. doi: 10.1124/jpet.106.104364. [DOI] [PubMed] [Google Scholar]
- 33.Lee W, et al. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280:9610–9617. doi: 10.1074/jbc.M411092200. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen R. Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR: Methods and Applications. Springer Press; Heidelberg, Germany: 2001. pp. 21–34. [Google Scholar]
- 35.Mita S, et al. Inhibition of bile acid transport across Na+/taurocholate cotransporting polypeptide (SLC10A1) and bile salt export pump (ABCB 11)-coexpressing LLC-PK1 cells by cholestasis-inducing drugs. Drug Metab Dispos. 2006;34:1575–1581. doi: 10.1124/dmd.105.008748. [DOI] [PubMed] [Google Scholar]
- 36.Ho RH, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics. 2007;17:647–656. doi: 10.1097/FPC.0b013e3280ef698f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arimori K, et al. Effect of P-glycoprotein modulator, cyclosporin A, on the gastrointestinal excretion of irinotecan and its metabolite SN-38 in rats. Pharm Res. 2003;20:910–917. doi: 10.1023/a:1023847521767. [DOI] [PubMed] [Google Scholar]
- 38.Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 39.Ho RH, et al. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 2006;130:1793–1806. doi: 10.1053/j.gastro.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 40.Jonker JW, et al. Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol Cell Biol. 2001;21:5471–5477. doi: 10.1128/MCB.21.16.5471-5477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, et al. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res. 2006;66:8847–8857. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi Y, Sakai R, Ohshiro N, Ohbayashi M, Kohyama N, Yamamoto T. Possible involvement of organic anion transporter 2 on the interaction of theophylline with erythromycin in the human liver. Drug Metab Dispos. 2005;33:619–622. doi: 10.1124/dmd.104.003301. [DOI] [PubMed] [Google Scholar]
- 43.Cropp CD, et al. Organic anion transporter 2 (SLC22A7) is a facilitative transporter of cGMP. Mol Pharmacol. 2008;73:1151–1158. doi: 10.1124/mol.107.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, et al. Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. Pharmacogenomics J. 2009;9:127–136. doi: 10.1038/tpj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/ H(+)-organic cation antiporters. Biochem Pharmacol. 2007;74:359–371. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Treiber A, Schneiter R, Häusler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug Metab Dispos. 2007;35:1400–1407. doi: 10.1124/dmd.106.013615. [DOI] [PubMed] [Google Scholar]
- 47.Gui C, et al. Effect of pregnane X receptor ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J Pharmacol. 2008;584:57–65. doi: 10.1016/j.ejphar.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.