Abstract
Fibroblast growth factor receptor 3 (FGFR3) mutations are frequently involved in human developmental disorders and cancer. Activation of FGFR3, through mutation or ligand stimulation, results in autophosphorylation of multiple tyrosine residues within the intracellular domain. To assess the importance of the six conserved tyrosine residues within the intracellular domain of FGFR3 for signaling, derivatives were constructed containing an N-terminal myristylation signal for plasma membrane localization and a point mutation (K650E) that confers constitutive kinase activation. A derivative containing all conserved tyrosine residues stimulates cellular transformation and activation of several FGFR3 signaling pathways. Substitution of all nonactivation loop tyrosine residues with phenylalanine rendered this FGFR3 construct inactive, despite the presence of the activating K650E mutation. Addition of a single tyrosine residue, Y724, restored its ability to stimulate cellular transformation, phosphatidylinositol 3-kinase activation, and phosphorylation of Shp2, MAPK, Stat1, and Stat3. These results demonstrate a critical role for Y724 in the activation of multiple signaling pathways by constitutively activated mutants of FGFR3.
INTRODUCTION
The fibroblast growth factor receptor (FGFR) family of receptor tyrosine kinases (RTKs) mediates growth, differentiation, and cellular migration in a diverse range of cell types. These receptors function as high-affinity binding proteins for the FGF family of ligands, of which there are nearly 20 members (Szebenyi and Fallon, 1999). Ligand binding to the extracellular domain of FGFRs results in receptor dimerization and transphosphorylation of tyrosine residues in the intracellular domain.
Many human disorders have been linked to mutations in FGFR3 (reviewed in Webster and Donoghue, 1997b; Burke et al., 1998). For example, mutations in the extracellular domain of FGFR3 such as R248C and G370C result in thanatophoric dysplasia type I (TDI), a lethal skeletal disorder. Mutations in the FGFR3 transmembrane domain can also lead to constitutive receptor activation, such as the G380R mutation responsible for most cases of achondroplasia, or skeletal dwarfism. Mutations in the FGFR3 kinase domain are responsible for developmental disorders that range in severity from the relatively mild hypochondroplasia (N540K) to the neonatal lethal disorder TDII (K650E). A different mutation at this same position, K650M, leads to a second syndrome, SADDAN (severe achondroplasia with developmental delay and acanthosis nigricans), which involves skeletal malformations, CNS disturbances, and skin dysplasia (Tavormina et al., 1999).
An exciting link between FGFR3 and cancer has recently been uncovered. The G375C and K650E mutations involved in skeletal malformation syndromes have also been identified in some patients with multiple myeloma, a proliferative B cell disorder (Chesi et al., 1997; Richelda et al., 1997). The K650E kinase domain mutation, as well as the R248C, S249C, and G370C extracellular domain mutations originally characterized in TDI patients, have also been shown to occur frequently in human bladder and cervical carcinomas (Cappellen et al., 1999).
Although elevated FGFR3 activity is implicated as a causative factor in human disorders, the resulting activation of signaling pathways is much less clear. Previous work has shown that PLC-γ binds to the Y760 autophosphorylation site of FGFR3 (corresponding to Y766 in FGFR1) and represents the only well characterized SH2 domain-containing binding partner for FGFRs (Mohammadi et al., 1991); however, the mutation Y766F, although eliminating phosphatidylinositol (PI) hydrolysis and receptor endocytosis, does not affect FGFR1-mediated mitogenesis (Mohammadi et al., 1992). The adaptor protein FRS2 binds to the FGFR1 juxtamembrane region in a phosphotyrosine-independent manner (Ong et al., 1997, 2000; Xu et al., 1998). FRS2 contains at least five binding sites for the Grb2 adaptor protein, which is constitutively bound to the guanine nucleotide exchange factor Sos. Sos stimulates ras activation, coupling FGFRs to activation of the MAPK cascade (Kouhara et al., 1997). FRS2 also binds to the protein tyrosine phosphatase Shp2 (Hadari et al., 1998), the function of which in FGFR signaling is still undefined. Despite these advances in our understanding of signal transduction through FGFRs, the function of specific autophosphorylation sites remains unresolved.
Seven tyrosine residues have been mapped as autophosphorylation sites in FGFR1 (Mohammadi et al., 1991, 1996a); several of these residues are conserved in all FGFR family members (see Figure 1A). Five of the seven phosphorylated tyrosines in FGFR1 are conserved in FGFR3. Additionally, a C-terminal tyrosine (Y770 in FGFR3; Y776 in FGFR1) is conserved in all four family members (see Figure 1A). The two tyrosine residues in the “YYKK” motif of the activation loop of FGFRs are required for kinase activity of FGFR1 (Mohammadi et al., 1996a) and FGFR3 (Webster et al., 1996). The other autophosphorylation sites have been examined in detail only in FGFR1, and mutation of all nonactivation loop tyrosines does not affect FGFR1-mediated activation of MAPK or mitogenesis in L6 myoblasts, or stimulation of neurite outgrowth in PC12 cells (Mohammadi et al., 1996a); however, Y463 and Y730 are important for induction of urokinase-type plasminogen activator in L6 cells (Dell'Era et al., 1999).
Figure 1.
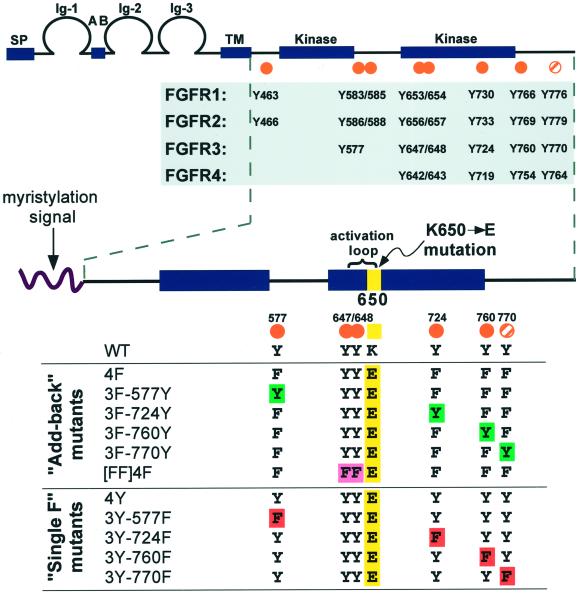
FGFR conserved tyrosine residues and constructs designed for this study. (Top) General FGFR structure and conserved tyrosine residues. The seven autophosphorylation sites (Mohammadi et al., 1996a) mapped in FGFR1 are indicated by orange dots, and an eighth tyrosine (Y776) that is conserved in all four FGFRs is marked by an orange striped dot. (Bottom) Constructs generated for this study. A myristylation signal targets the intracellular domain of FGFR3 to the plasma membrane. All derivatives, with the exception of WT, contain the K650E point mutation (yellow). The [FF]4F construct contains mutations of the Y647 and Y648 activation loop residues to phenylalanine, as indicated by [FF] (pink). The 4F construct was used to derive the “Add-back” mutants, where each tyrosine was individually added back to the phenylalanine mutants (green). The 4Y derivative served as the parental constructs for the “Single F” derivatives, which have each nonactivation loop tyrosine individually removed by mutation to phenylalanine (red).
Several elegant studies have exploited derivatives of PDGF receptor (PDGFR) that either lack specific tyrosine residues or contain only one or two “add-back” phosphorylation sites to study RTK-mediated signaling. Such studies have provided valuable information regarding the role of specific tyrosine phosphorylation sites in regulating PDGFR-dependent signal transduction (Fantl et al., 1992; Kashishian et al., 1992; Kazlauskas et al., 1992; Ronnstrand et al., 1992; Kashishian and Cooper, 1993; Valius and Kazlauskas, 1993; Valius et al., 1993). We used this technique to determine the importance of individual tyrosine phosphorylation sites in FGFR3. Plasma membrane-targeted derivatives of the FGFR3 intracellular domain were designed that are constitutively activated by the K650E mutation. Systematic mutation of all conserved tyrosine residues created a derivative lacking all nonactivation loop autophosphorylation sites. Individual tyrosine residues were then added back to assess their contribution to FGFR3 signaling. We found that adding back Y724 restores nearly 100% of activity when assayed for transformation, MAPK and Shp2 phosphorylation, Stat activation, and PI 3-kinase activation. Results presented here thus provide a detailed model of the role of individual tyrosine residues in mediating downstream signaling by constitutively activated mutants of FGFR3.
MATERIALS AND METHODS
Construction of Add-back and “Single F” Mutants
To facilitate mutagenesis of FGFR3, additional silent restriction sites were engineered into the myristylated wild-type (WT) FGFR3 construct PM-Kin(WT) described previously (Webster and Donoghue, 1997a). The basic organization of the WT construct, with location of restriction sites relative to the relevant tyrosine codons, is as follows: HindIII (5′ MCS) – XhoI (nt 251) – Y577 – BsrDI (nt 744) – YYKK – AccI (nt 890) – Y724 – BsrGI (nt 1050) – HpaI (nt 1135) – Y760 – Eco47III (nt 1172) – Y770 – BsmBI (nt 1233) – BamHI (nt 1289) – STOP – XbaI (3′ MCS). This is the WT clone described in Figure 1.
To insert the K650E activation loop mutation, the XhoI–BsrGI fragments of the YYKE and FFKE FGFR3 constructs (described in Webster et al., 1996) were swapped into the WT clone, generating the 4Y (pKH134) and [FF]4Y (pKH138) constructs. Oligonucleotide pairs were inserted into the Eco47III–BamHI or HpaI–BamHI sites of 4Y (pKH134) and [FF]4Y (pKH138), mutating either Y760 or Y770 to F.
To construct the 4F and [FF]4F mutants, the Y760F and Y770F mutations were constructed simultaneously using D2027/D2028 oligonucleotides inserted into the HpaI–BsmBI sites of 4Y (pKH134) and [FF]4Y (pKH138), making the intermediates 2Y-760/770F (pKH137) and [FF]2Y-760/770F (pKH141) (not examined in this study). These two clones were used for PCR-mediated mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The Y577F mutation was substituted into pKH137, creating 3F-724Y (pKH143), whereas the Y724F mutation was engineered into pKH141, creating [FF]3F-577Y (pKH142) (mutagenic primer sequences available on request).
To generate the Y724F mutation in the full-length WT or TDII FGFR3 receptors, the AccI–XbaI fragment of 3Y-724F was ligated with the Xho–AccI fragments of full-length WT or TDII FGFR3 (described in Webster and Donoghue, 1996) and the Xho–XbaI vector fragment from full-length WT FGFR3. The remaining constructs were generated by swapping different regions between the clones using the restriction sites listed above. Sequences of all regions incorporating oligonucleotides or subjected to PCR were confirmed by dideoxy nucleotide sequencing. Fragments containing PCR-generated mutations were subcloned back into the parental vector. Additionally, all clones were verified by multiple diagnostic restriction enzyme digests.
Cell Culture, Focus Assays, and FGF stimulation
NIH3T3 cells were maintained in DME plus 10% bovine calf serum in a 10% CO2 humidified incubator. 293T cells were grown in DME plus 10% FBS, 10% CO2. COS-1 cells were grown in DME plus 10% FBS, 5% CO2. Chinese hamster ovary (CHO)-K1 cells were grown in Ham's F-12 plus 10% FBS, proline, penicillin/streptomycin, and fungizone, at 5% CO2. NIH3T3, COS-1, CHO-K1, and 293T cells were transfected using a modified calcium phosphate transfection protocol (Chen and Okayama, 1987). Transformation assays were performed using NIH3T3 cells as described previously (Webster and Donoghue, 1997a). Transfection efficiencies were determined by G418-resistant colonies on parallel plates. Data represent the mean and SE of at least three independent experiments. For CHO-K1 cell experiments, cells were starved overnight after transfection, then stimulated for 30 min with 200 ng/ml aFGF (recombinant human; R & D Systems, Minneapolis, MN) plus 20 μg/ml heparin. Cell lysates were TCA-precipitated, and one-fourth of the total sample was analyzed by immunoblotting, as described below.
Immunoprecipitation and Immunoblotting
Transfected 293T cells were lysed in RFR buffer (20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 25 mM β-glycerophosphate, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2.25 μg/ml aprotinin, 1 mM sodium vanadate). Immune complexes were bound to protein A-Sepharose beads and washed four times in lysis buffer. Whole-cell lysates or immunoprecipitations were resolved by SDS-PAGE, and proteins were transferred to Immobilon. Proteins were detected by enhanced chemiluminescence (Amersham Pharmacia Biotech, Arlington Heights, IL). Antisera used for immunoblotting and immunoprecipitation were as follows: FGFR3 (C-15), FGFR4 (C-16), SH-PTP2 (C-18; Shp2), Stat1 (C-136) from Santa Cruz Biotechnology (Santa Cruz, CA); phospho-MAPK (T202/Y204), phospho-Stat1 (Y701), phospho-Stat3 (Y705) from New England Biolabs (Beverly, MA); mouse anti-MAPK (ERK1 + ERK2) from Zymed (San Francisco, CA); 4G10 (antiphosphotyrosine) from Upstate Biotechnology (Lake Placid, NY); Stat3 from Transduction Labs (Lexington, KY); and HRP-anti-mouse and HRP-anti-rabbit secondary antibodies from Amersham.
Transcription Assays
NIH3T3 cells were transfected with 2 μg of the 4xStRE-luciferase reporter (Bromberg et al., 1999) together with 8 μg of each FGFR3 construct. Cells were re-fed the following day with 0.2% calf serum and starved for 20–40 h. Lysates were generated using the protocol from the Luciferase Assay System (Promega, Madison, WI). The mean and SE were determined from three independent experiments.
In Vitro Kinase Assays
FGFR3 proteins were recovered from transfected 293T cells lysed in RFR buffer by immunoprecipitation with C-15 FGFR3 antiserum. In vitro kinase assays were performed as described previously (Webster and Donoghue, 1996). Equivalent amounts of receptor protein were detected by immunoblotting of lysates before immunoprecipitation.
PI 3-Kinase Assays
PI-3 kinase assays were performed as described previously (Fridell et al., 1996; Hart et al., 2000b). Equivalent amounts of lysate were immunoprecipitated with 4G10 phosphotyrosine antiserum. The control was Vps34p-purified PI 3-kinase from yeast, a kind gift from Andrew Wurmser and Scott Emr (University of California, San Diego, CA). Equivalent FGFR3 expression in samples was confirmed by immunoblotting of lysates.
Immunofluorescence
Cells were fixed with 3% paraformaldehyde/PBS for 20 min and permeabilized with 0.5% Triton X-100/PBS for 10 min. Coverslips were mounted on glass slides with 90% glycerol in 0.1 M Tris, pH 8.5, plus phenylenediamine to prevent fading, and photographed using a Nikon Microphot-FXA microscope with a Hamamatsu C5810 camera.
For Stat3 nuclear translocation experiments, COS-1 cells were cotransfected with 3 μg each of myc-tagged Stat3 and FGFR3 plasmid. After 20 h starvation in 0.2% FBS, cells were fixed and permeabilized as described above. FGFR3 derivatives were detected with rabbit C-15 FGFR3 antiserum and Texas red-conjugated anti-rabbit secondary antiserum. Stat3 protein was detected with 9E10 anti-myc mAb and fluorescein-conjugated anti-mouse secondary.
RESULTS
Description of FGFR3 Derivatives
The structure of FGFRs is shown in Figure 1. Seven autophosphorylation sites have been mapped in the FGFR1 intracellular domain (Y463, Y583, Y585, Y653, Y654, Y730, Y766) (Mohammadi et al., 1991, 1996a), and there is an eighth tyrosine (Y776) that is conserved in all four FGFRs (Figure 1). Y653 and Y654 are in the activation loop of the kinase domain and are required for FGFR1 kinase activity (Mohammadi et al., 1996a).
To facilitate the study of specific FGFR3 tyrosine residues in mediating FGFR3 signaling, we used a novel set of derivatives exhibiting two unique features that were based on a previous study of the K650E mutant (Webster and Donoghue, 1997a). First, a myristylation signal was used to target the intracellular domain of FGFR3 to the inner surface of the plasma membrane (Figure 1). Second, derivatives were rendered constitutively active by incorporating the K650E activation loop mutation. These derivatives are unique in that they are completely independent of ligand and thus provide a simplified system in which to study FGFR3 signaling.
All derivatives contain the sequence “YYKE” in the activation loop (residues 647–650) region of FGFR3, except for “[FF]” constructs, which contain the sequence “FFKE” in this region (Figure 1). These latter derivatives lack the activation loop tyrosine residues but contain the activating K650E mutation, and they allowed us to assess the importance of these two tyrosines in the presence of the K650E mutation. Mutation of all other conserved tyrosines in FGFR3 (Y577, Y724, Y760, Y770) to phenylalanine generated the constructs 4F and [FF]4F. Each tyrosine was then added back to examine its individual contribution to FGFR3 signaling (Figure 1, “Add-back mutants”). For example, the 3F-577Y derivative contains the activation loop tyrosines, plus Y577, with the three remaining conserved tyrosine residues mutated to phenylalanine (3F).
Alternatively, each tyrosine residue was individually mutated to phenylalanine (Single F mutants) using the positive control 4Y derivative, which contains all conserved phosphorylation sites and the K650E mutation (Figure 1). As a negative control, we also used the WT derivative, which does not contain the K650E point mutation.
Transforming Activity of Add-back and Single F Mutants
To analyze the ability of FGFR3 derivatives to confer contact- and anchorage-independent growth, NIH3T3 cells were transfected with the indicated constructs (Figure 2). Removal of all four nonactivation loop tyrosine residues (4F) resulted in a significantly decreased transforming activity (∼20% activity) when compared with the 4Y positive control; however, the Add-back construct 3F-724Y restored transforming activity to nearly 100% of 4Y (Figure 2). The presence of Y577 (3F-577Y), Y760 (3F-760Y), or Y770 (3F-770Y) had only a modest effect on the transforming activity of the 4F derivative (Figure 2). The 3F-770Y construct exhibited the lowest level of transforming activity of the Add-back derivatives (∼18% of 4Y) (Figure 2). Removal of Y724 from the 4Y derivative generated the 3Y-724F Single F mutant, which caused a significant reduction in transforming activity to 40% of the control 4Y (Figure 2), again suggesting the importance of Y724 for transformation mediated by constitutively activated FGFR3 mutants. The absence of Y577 (3Y-577F) or Y760 (3Y-760F) resulted in a modest decrease in activity. In contrast, mutation of Y770 (3Y-770F) increased the transforming activity of the FGFR3 construct to ∼130% of the control 4Y (Figure 2).
Figure 2.
Transformation activity of FGFR3 Add-back and Single F mutants. NIH3T3 cells were transfected with the indicated constructs and assayed for focus formation. Foci were counted after 12–14 d. Results are presented as percentage of the 4Y transforming activity and represent the mean and SD of three independent experiments.
A similar pattern of activity was also seen with the FFKE activation loop derivatives, lacking the activation loop tyrosines (Figure 2). Although the [FF]4Y derivative exhibits only ∼6% of the transformation activity of the 4Y positive control (Figure 2), it is still surprising that this derivative is active at all, because it lacks the activation loop tyrosine residues. Removal of Y724 from the [FF]4Y derivative (creating [FF]3Y-724F) reduced transformation activity to ∼1%, whereas mutation of Y770 to F ([FF]3Y-770F) increased the activity to 12%, approximately twofold over [FF]4Y (Figure 2). The [FF]3F-724Y Add-back derivative was just as active as [FF]4Y, suggesting the importance of Y724 as a regulatory site, in addition to the activation loop tyrosines, required for maximal transforming activity. The data also suggest that Y770 may represent a negative regulatory site for signaling pathways involved in transformation.
In Vitro and In Vivo Kinase Activity
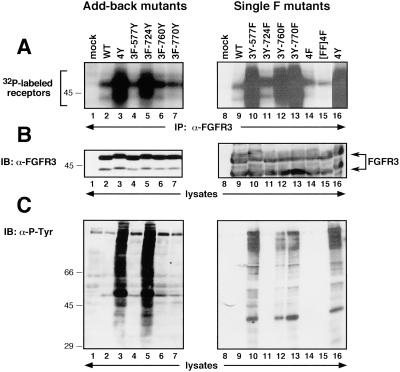
To determine whether the in vitro autophosphorylation activity was altered in any of the FGFR3 derivatives, the Add-back and Single F constructs were immunoprecipitated from transfected cells with FGFR3 antiserum and subjected to an in vitro kinase assay. Significant autophosphorylation was detected in the 4Y derivative (Figure 3A, lanes 3 and 16). Similar kinase activation was exhibited by derivatives containing the Y724 site (Figure 3A, 3F-724Y, lane 5) or lacking residues Y577, Y760, or Y770 (Figure 3A, Single F mutants, lanes 10, 12, and 13). All derivatives were expressed at equivalent levels (Figure 3B).
Figure 3.
In vitro and in vivo kinase activity of FGFR3 Add-back and Single F derivatives. (A) In vitro kinase assay of FGFR3 derivatives. (B) FGFR3 expression levels were similar in each sample, as determined by immunoblotting of whole-cell lysates. (C) Tyrosine-phosphorylated proteins in cells expressing FGFR3 constructs. Molecular weight markers are indicated in kilodaltons.
We next examined whether any of the tyrosine mutations affected the in vivo kinase activity of the receptor derivatives. The Add-back and Single F mutants were transfected into 293T cells, and lysates were analyzed by immunoblotting using 4G10 phosphotyrosine antiserum. Figure 3C shows that both the 4Y and 3F-724Y constructs led to a significant increase in cellular tyrosine phosphorylation (Figure 3C, lanes 3 and 5), when compared with mock-transfected cells or cells expressing the wild-type FGFR3 derivative (Figure 3C, lanes 1 and 8, 2 and 9, respectively). The Single F mutant 3Y-724F abolished all cellular tyrosine phosphorylation (Figure 3C, lane 11). Addition of Y577, Y760, or Y770 to the 4F mutant (Figure 3C, lanes 4, 6, and 7) did not appreciably affect phosphorylation of cellular proteins. Removal of these residues in the Single F mutants (Figure 3C, lanes 10, 12, and 13) did not reduce cellular tyrosine phosphorylation. Taken together, these results suggest that Y724 is a major regulatory site for signaling by constitutively activated mutants of FGFR3.
Activation of MAPK
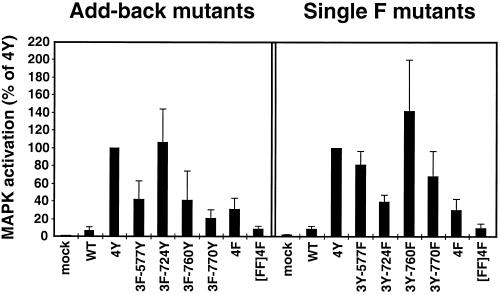
FGF stimulation leads to MAPK activation in multiple cell types (Kouhara et al., 1997; Szebenyi and Fallon, 1999). To determine which tyrosine residues are important for this response, whole-cell lysates of 293T cells transfected with the indicated samples were immunoblotted with Phospho-p44/42 MAP Kinase (Thr202/Tyr204) antiserum, which detects the phosphorylated, activated forms of p42MAPK and p44MAPK. The same membrane was reprobed with ERK1/2 antiserum to confirm equivalent levels of p42MAPK and p44MAPK in each sample. The resulting ECL exposures were scanned and quantitated using NIH Image software. The bar graph, shown in Figure 4, represents the levels of activated MAPK (p42/p44), calculated as a ratio of P-MAPK/MAPK levels, over that exhibited by 4Y.
Figure 4.
Activation of the MAPK cascade. The bar graph represents the fold activation of MAPK (p42/p44), calculated as a ratio of P-MAPK/MAPK levels detected on immunoblots, as a percentage of the maximum activity exhibited by the 4Y derivative. Results represent the mean and SE for three independent experiments.
The 3F-724Y Add-back derivative stimulates activation of MAPK to the same level as 4Y (Figure 4, left panel). Addition of Y577, Y760, or Y770 to 4F did not significantly increase activation of MAPK over the levels stimulated by 4F (Figure 4, left panel). Removal of Y724 from the 4Y construct (3Y-724F Single F mutant) reduces the level of MAPK activation to 40% of the control (Figure 4, right panel); however, removal of Y577 and Y770 only mildly decreased the MAPK activation (Figure 4, right panel). These results indicate that Y724 is required for maximal activation of the MAPK pathway by the panel of mutants examined here.
Phosphorylation of Shp2
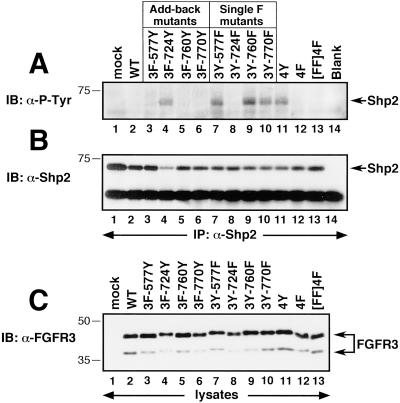
The tyrosine phosphatase Shp2 is tyrosine phosphorylated in response to FGFs (Hadari et al., 1998). To determine the importance of Y724 for Shp2 phosphorylation, lysates of transfected 293T cells were immunoprecipitated with Shp2 antiserum and analyzed by immunoblotting with 4G10 phosphotyrosine antiserum. Although expression of the 4F mutant did not lead to Shp2 phosphorylation (Figure 5A, lane 12), the Add-back mutant containing Y724 (3F-724Y) resulted in significant tyrosine phosphorylation of Shp2 (Figure 5A, lane 4). In the Single F derivatives, removal of Y577 (lane 7), Y760 (lane 9), or Y770 (lane 10) from the 4Y derivative (Figure 5A, lane 11) did not affect the phosphorylation of Shp2; however, mutation of just Y724 (3Y-724F) completely abolished this effect (Figure 5A, lane 8). Expression of Shp2 and FGFR3 were approximately equal in each sample (Figure 5, B and C, respectively).
Figure 5.
Tyrosine phosphorylation of the FGF-responsive effector protein Shp2. (A) Detection of tyrosine-phosphorylated Shp2 in FGFR3-expressing cells. (B) The same membrane in A was stripped and reprobed with Shp2 antiserum, to confirm approximately equal recovery of Shp2 protein in each sample. (C) Whole-cell lysates were analyzed by immunoblotting with FGFR3 antiserum to confirm equivalent expression levels of each construct.
Activation of Stat1 and Stat3
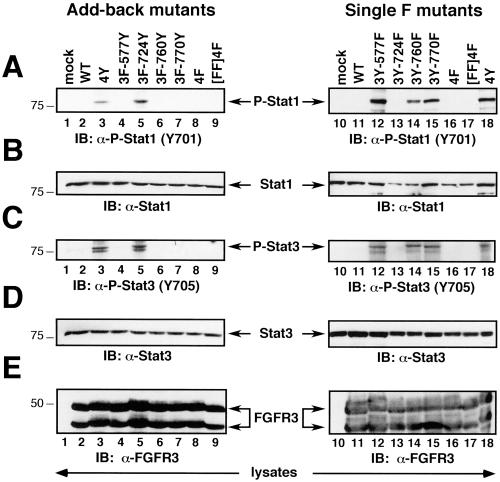
Stat1 and Stat3 proteins are activated by FGFRs, particularly by FGFR3 containing the K650E mutation (Su et al., 1997; Legeai-Mallet et al., 1998; Li et al., 1999; Sahni et al., 1999; Hart et al., 2000a). To examine the importance of each FGFR3 tyrosine residue in mediating this response, lysates from 293T cells transfected with the indicated constructs were immunoblotted with the indicated Stat- and phospho-Stat-specific antisera (Figure 6). Expression of either 4Y or the 3F-724Y Add-back construct led to phosphorylation of Stat1 and Stat3 proteins in 293T cells (Figure 6, A and C, lanes 3 and 5). Removal of Y724 in the Single F mutant 3Y-724F abrogated the ability of this FGFR3 derivative to mediate phosphorylation of Stat1 or Stat3 (Figure 6, A and C, lane 13). The Single F derivatives 3Y-577F, 3Y-760F, and 3Y-770F, retained the ability to stimulate Stat1 and Stat3 phosphorylation (Figure 6, A and C, lanes 12, 14, and 15). Levels of Stat1 and Stat3 were equivalent in each sample (Figure 6, B and D). Expression of the various FGFR3 constructs was equivalent (Figure 6E).
Figure 6.
Phosphorylation of Stat1 and Stat3. (A) Phosphorylation of Stat1 in cells expressing FGFR3 derivatives. (B) Immunoblotting of Stat1 protein indicated equivalent expression in each sample. (C) Phosphorylation of Stat3 in response to FGFR3 constructs. (D) Immunoblotting of Stat3 protein indicated equal expression in each lane. (E) Levels of the FGFR3 Add-back and Single F mutants were examined by immunoblotting of whole-cell lysates with FGFR3 antisera.
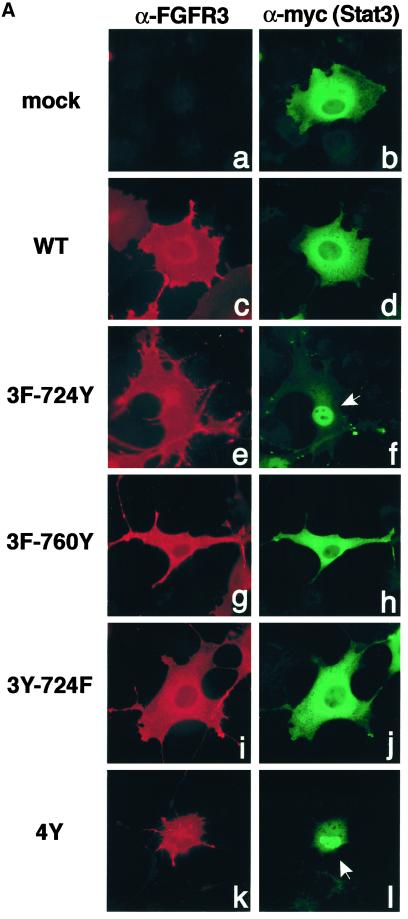
Stat activation results in relocalization of the dimeric Stat complexes to the nucleus (Darnell, 1997). Thus, we next examined whether FGFR3-dependent Stat phosphorylation resulted in nuclear translocation and activation of transcription. COS-1 cells were cotransfected with FGFR3 derivatives and myc-tagged Stat3, and Stat3 localization was examined by indirect immunofluorescence (Figure 7A). The myc-tagged Stat3 is present in the cytoplasm of cells cotransfected with empty vector (Figure 7A, a and b) or in cells coexpressing the wild-type FGFR3 derivative (Figure 7A, c and d). Coexpression of the Add-back derivative 3F-724Y led to relocalization of Stat3 protein to the nucleus (Figure 7A, f). Similar results were seen with the 4Y derivative (Figure 7A, l). Removal of just Y724 from this derivative, in the Single F mutant 3Y-724F, abolished the translocation of Stat3 to the nucleus (Figure 7A, j). Other Add-back derivatives were unable to cause Stat3 nuclear accumulation (Figure 7A, h) (and our unpublished results).
Figure 7.
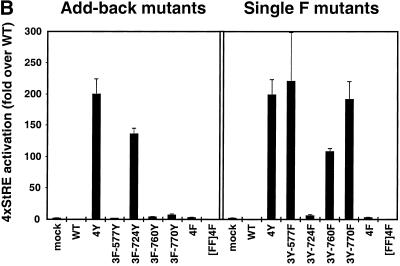
FGFR3-activated Stat proteins translocate to the nucleus and activate transcription. (A) COS-1 cells were transfected with FGFR3 derivatives and myc-tagged Stat3, and proteins were detected by double-label immunofluorescence. The left column shows expression of FGFR3 constructs detected by FGFR3 antiserum (a, c, e, g, i, k). The right column shows localization of myc-tagged Stat3, detected with myc antiserum (b, d, f, h, j, l). (B) NIH3T3 cells were transfected with FGFR3 derivatives and the Stat-responsive reporter construct 4xStRE-luciferase. Lysates were analyzed for luciferase activity, and results are shown as the fold activation over WT and represent the mean and SE for three independent experiments.
We used a Stat1/Stat3-specific luciferase reporter construct (4xStRE-luciferase) (Bromberg et al., 1999) to examine directly Stat1/Stat3 transcriptional activation. These results, shown in Figure 7B, parallel the phospho-Stat results in Figure 6. Addition of Y724 in the Add-back mutant 3F-724Y caused an ∼150-fold stimulation of the reporter construct, when compared with WT (Figure 7B, left panel). None of the other Add-back mutants significantly activated the reporter construct. Removal of Y577 or Y770 in the Single F mutants did not inhibit activation of the reporter and led to ∼200-fold activation over WT (Figure 7B); however, removal of Y724 in the Single F mutant 3Y-724F completely abrogated the activation of the reporter (3Y-724F, right panel). Interestingly, the absence of Y760 in the 3Y-760F Single F mutant decreased activation to ∼50% of the positive control (Figure 7B). Taken together, these results clearly implicate Y724 in the activation of Stat proteins by constitutively activated mutants of FGFR3 and suggest that both Y724 and Y760 are required for maximal Stat activation.
Activation of PI 3-Kinase by FGFR3 Derivatives
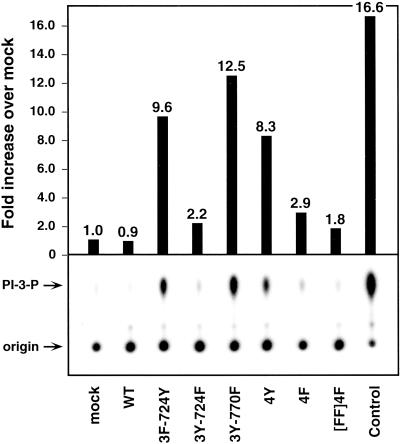
The sequence surrounding Y724 resembles a consensus binding site for the p85 regulatory subunit of PI 3-kinase. To examine PI 3-kinase activation, transfected cell lysates were immunoprecipitated with 4G10 phosphotyrosine antiserum and subjected to an in vitro lipid kinase assay using PI as substrate. The 4Y and 3F-724Y derivatives led to an 8.3- and 9.6-fold increase, respectively, in PI 3-kinase activity over mock-transfected cells (Figure 8). The 3Y-724F construct resulted in marginal PI 3-kinase activation. Interestingly, removal of Y770 from the 4Y construct (3Y-724F) increased the PI 3-kinase activation ∼1.5-fold over the 4Y activity. These results suggest that Y770 may negatively regulate the activation of PI 3-kinase by constitutively activated FGFR3; moreover, the presence of Y724 appears to be important for maximal activation of this pathway.
Figure 8.
PI 3-kinase activation in response to FGFR3 derivatives. Immunoprecipitated tyrosine-phosphorylated proteins were subjected to an in vitro lipid kinase assay using PI and [32P]-γ-ATP as substrates. The control was Vps34p-purified PI 3-kinase from yeast. Arrows indicate the position of the origin and the 32P-labeled PIP product detected by autoradiography after TLC. Fold increase in PIP production over mock-transfected cells was quantitated using NIH Image and is indicated in the bar graph.
Role of Y724 in Full-length FGFR3 Derivatives
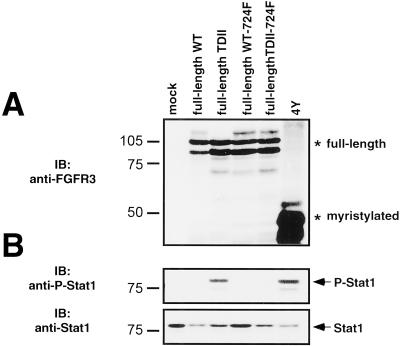
The Y724F mutation was swapped into the full-length WT and TDII derivatives of FGFR3 to determine whether this residue was important for full-length receptor or ligand-stimulated signaling. First, 293T cells were transfected with full-length derivatives, then starved overnight, and lysates were examined for activation of Stat1 (Figure 9). The full-length TDII FGFR3 derivative, as well as the 4Y construct, which are both constitutively activated, led to activation of Stat1 in the absence of ligand; however, removal of Y724 from the full-length TDII construct rendered it unable to active Stat1 (Figure 9B). Similar results were seen when phosphorylation of MAPK was examined (our unpublished results). Similar levels of full-length FGFR3 constructs were expressed in the cells (Figure 9A).
Figure 9.
Role of Y724 in full-length FGFR3. 293T cells transfected with full-length derivatives of FGFR3, with or without the Y724F mutation, were lysed and analyzed by immunoblotting. (A) Immunoblotting with antisera against FGFR3 shows equivalent expression levels of full-length FGFR3 derivatives. (B) Stat1 is phosphorylated in response to full-length constitutively active FGFR3 but not if the Y724F mutation is present. Lysates were examined by immunoblotting with anti-phospho-Stat1 (Y701) sera. Equal amounts of Stat1 were present in each sample.
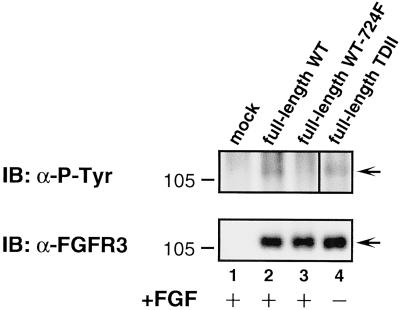
Next we examined whether ligand-stimulated signaling was affected by the Y724F mutation. CHO-K1 cells, which are known to lack endogenous FGFRs, were transfected with full-length FGFR3 derivatives, starved overnight, and then stimulated as indicated with aFGF plus heparin (Figure 10). Ligand stimulation leads to autophosphorylation of FGFR3, as seen in Figure 10 (lane 2). Constitutively active full-length FGFR3 is also autophosphorylated in these cells in the absence of ligand (Figure 10, lane 4); however, when the Y724F mutation is incorporated into WT FGFR3, it becomes insensitive to ligand-stimulated autophosphorylation (Figure 10, lane 3). Taken together, these results suggest that Y724 is also an important residue for signaling through full-length FGFR3.
Figure 10.
Ligand-stimulated FGFR3 activation. CHO-K1 cells, lacking endogenous FGFRs, were transfected with full-length FGFR3 derivatives, with or without the Y724F mutation. Top panel, examination of the lysates with anti-phosphotyrosine sera 4G10 shows that stimulation for 30 min with aFGF (200 ng/ml) plus heparin (20 μg/ml) leads to autophosphorylation of WT FGFR3, but not when the Y724F mutation is present. Bottom panel, equal amounts of each receptor were expressed in the samples. In the top panel, lanes 1–3 are from a 20 s ECL exposure of the immunoblot. In contrast, lane 4 is from a 5 s ECL exposure, because the full-length TDII sample appeared overexposed on the 20 s ECL.
DISCUSSION
Members of the FGFR family mediate a number of important cellular processes and are mutated or overexpressed in several human developmental syndromes and also in various cancers. The activating mutation Lys650→Glu in the activation loop of the FGFR3 kinase domain underlies the lethal human skeletal disorder TDII (reviewed in Webster and Donoghue, 1996) and is also found in patients with multiple myeloma and bladder and cervical carcinomas (Chesi et al., 1997; Richelda et al., 1997; Cappellen et al., 1999). We recently reported a comparative analysis of the signaling activity of FGFR family members, in which the analogous mutation was introduced into FGFR1, FGFR3, and FGFR4 (Hart et al., 2000a). In this previous study, we showed that the kinase domains of FGFR1, FGFR3, and FGFR4 containing the activation loop mutation, when targeted to the plasma membrane by a myristylation signal, can transform NIH3T3 cells and induce neurite outgrowth in PC12 cells. Phosphorylation of Shp2, PLC-γ, and MAPK was also stimulated by all three “TDII-like” FGFR derivatives. Additionally, activation of Stat1 and Stat3, as well as activation of PI-3 kinase activity, was observed in cells expressing the activated FGFR derivatives. This study demonstrated that these activated receptor derivatives exhibit significant overlap in the panel of effector proteins used to mediate downstream signals and suggested that Stat activation by FGFRs is important in their ability to act as oncogenes.
To build on this earlier work, and to further analyze signaling by members of the FGFR family, we chose FGFR3 for further study of the importance of individual tyrosine residues for downstream signaling. The results presented here demonstrate the importance of Y724 for signaling by constitutively activated mutants of FGFR3. Addition of Y724 to the 4F derivative that lacks all nonactivation loop tyrosine residues restored the ability of this mutant to cause morphological transformation, to a level nearly 100% of the 4Y construct. Examination of individual FGFR3 signaling effectors showed that phosphorylation of Shp2, MAPK, Stat1, and Stat3 was also stimulated by both 4Y and the 3F-724Y Add-back mutant. These results are summarized in Figure 11. Mutation of Y724 to F abolished both FGFR3-dependent transformation and activation of signaling pathways, further demonstrating that Y724 functions as the critical regulatory tyrosine residue for the panel of mutants examined here. Examination of full-length FGFR3, both ligand-stimulated and constitutively active derivatives, also reveals an important role for Y724 in FGFR3 signaling. We also found that Y770 may function as a negative regulatory site for FGFR3-stimulated transformation and PI 3-kinase activation. Although this region does not resemble any known tyrosine phosphorylation motifs, it may serve as a novel binding site for a negative regulator of FGFR3 signaling. Additionally, Y760, the PLC-γ binding site, although not required for transformation, MAPK activation, or PI 3-kinase activity, may be important for full Stat activation.
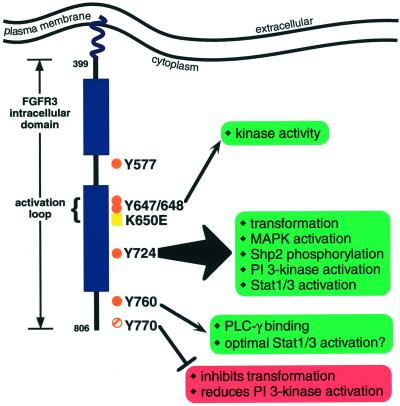
Figure 11.
Model of FGFR3 tyrosine residues and their role in signal transduction. The intracellular domain of FGFR3 is depicted, with the approximate locations of tyrosine residues examined in this study indicated by orange dots (solid dots indicate autophosphorylation sites; striped dot indicates a conserved tyrosine residue). The arrows pointing to green boxes indicate the FGFR3 responses that require those tyrosine residues. The red box associated with Y770 indicates those pathways that are negatively influenced by Y770.
Comparisons with Other RTKs
On the basis of the crystallographic structure of FGFR1, the residue Y730, which corresponds to Y724 in FGFR3, is predicted to lie in α-helix H at the bottom of the large lobe of the kinase domain (Mohammadi et al., 1996b). In the FGFR1 structure, this helix is surface accessible, making this tyrosine available to bind cellular proteins. Interestingly, this tyrosine residue is conserved in many RTKs, including EGFR and PDGFR, but is notably absent in the insulin receptor. Although this residue is phosphorylated in FGFR1 (Mohammadi et al., 1996a), the corresponding residue has not been identified as a phosphorylation site in EGFR or PDGFR, nor has it been previously shown to bind to any cellular proteins.
Our results demonstrate that FGFR3 differs significantly from RTKs such as EGFR, Neu/ErbB2, and PDGFR, which use multiple tyrosine residues to mediate binding to different effector proteins (Fantl et al., 1992; Kashishian et al., 1992; Kazlauskas et al., 1992; Ronnstrand et al., 1992; Decker, 1993; Kashishian and Cooper, 1993; Valius and Kazlauskas, 1993; Valius et al., 1993; Dankort et al., 1997). Binding of multiple effector proteins to a single tyrosine motif has been observed for other RTKs, notably the MET receptor (hepatocyte growth factor receptor). In the MET human receptor, Shc, Gab1, src, PLC-γ, Shp2, and PI 3-kinase all bind to Y1349 and Y1356, whereas Grb2 binds to Y1356 alone. Mutation of these tyrosine residues reduces MET-induced epithelial cell morphogenesis but does not affect MET-mediated activation of the MAPK pathway or the resulting cell scattering (Nguyen et al., 1997; Tulasne et al., 1999). A multiple substrate binding site has also been reported for Y1100 of the Tek/Tie2 receptor (Jones et al., 1999). The ability of one tyrosine or tyrosine-containing motif to mediate binding to several effector proteins may indicate a similar role for Y724 in FGFR3. Future studies will be directed toward identifying cellular proteins that may interact with this site in a phosphotyrosine-dependent manner.
Role of PI 3-Kinase Activation in FGFR3 Signaling
The sequence around Y724 contains a YMXM motif, suggestive of a binding site for the p85 subunit of PI 3-kinase. We found that PI 3-kinase activity is significantly enhanced in cells expressing the 4Y or 3F-724Y derivatives. These derivatives also exhibited high levels of transforming activity. PI 3-kinase has been implicated in transformation as well as apoptosis through activation of the kinase Akt (Sellers and Fisher, 1999; reviewed in Kraslinikov, 2000). Suppression of apoptosis in dysplastic cells could lead to an extended life-span and contribute to FGFR3-mediated transformation. Activation of PI 3-kinase has also been shown to regulate cellular cytoskeletal organization. Abnormal activation of these pathways in cancer cells may facilitate invasion and metastasis by stimulating cellular migration or altering cellular shape and size (Sellers and Fisher, 1999; Kraslinikov, 2000). Thus, demonstration of FGFR3-mediated PI 3-kinase activation may provide at least a partial mechanism for how FGFR3 induces transformation.
Role of Stat Proteins in Signaling by Constitutively Activated FGFR3
Several laboratories have recently shown that FGFRs can activate members of the Stat family of transcription factors (Su et al., 1997; Legeai-Mallet et al., 1998; Li et al., 1999; Sahni et al., 1999; Hart et al., 2000a). A number of human cancers exhibit Stat3 activation, including breast carcinoma, head and neck squamous cell carcinoma, and multiple myeloma (Garcia and Jove, 1998; Grandis et al., 1998; Catlett-Falcone et al., 1999). Recent studies suggest that overactivation of Stat3 protects multiple myeloma cells from apoptosis, thereby contributing to disease progression (Catlett-Falcone et al., 1999). It is fascinating that FGFR3 mutations have also recently been linked to multiple myeloma, as well as bladder and cervical carcinomas (Chesi et al., 1997; Richelda et al., 1997; Cappellen et al., 1999). Further examination of the Y724-dependent Stat activation demonstrated here may suggest additional links between Stats, FGFR3 activation, and human cancer.
FGFR Signaling and Human Disease
FGFR3 mutations are involved in a multitude of developmental syndromes and human cancers. A large number of FGFR1 and FGFR2 mutations are also associated with similar clinical phenotypes (reviewed in Webster and Donoghue, 1997b; Burke et al., 1998). In addition, FGFR1 and FGFR2 chromosomal translocations have been detected in some T-lymphocytic and myeloproliferative disorders (Hattori et al., 1992; Popovici et al., 1998, 1999; Reiter et al., 1998; Smedley et al., 1998; Xiao et al., 1998). Our findings suggest that FGFR3 may differ from FGFR1 in the requirement of a single tyrosine residue (Y724), which may reflect some fundamental difference in the regulation of signaling pathways that are activated by FGFR3 versus FGFR1. Similar add-back mutational studies in other FGFR family members may also facilitate comparison of signaling pathways activated by FGFR1, FGFR2, and FGFR4. Last, because our results reveal a requirement for Y724 in FGFR3 activation of cellular transformation, MAPK, Stat proteins, and PI 3-kinase, Y724 may be a useful therapeutic target for FGFR3-dependent disorders.
ACKNOWLEDGMENTS
We thank April Meyer for generating the WT FGFR3 construct; Ching Wang for helpful information; Kerri Mowan and members of the David laboratory for advice and reagents; Andrew Wurmser and Scott Emr for the PI 3-kinase positive control; members of the Donoghue lab for advice and critical reading of this manuscript; Mark Barsoum for assistance with the Stat localization experiments; and Laura Castrejon for editorial assistance. This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research grant R01-DE12581.
Abbreviations used:
- FGFR
fibroblast growth factor receptor
- PDGFR
PDGF receptor
- RTK
receptor tyrosine kinase
- TD
thanatophoric dysplasia
REFERENCES
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Burke D, Wilkes D, Blundell TL, Malcolm S. Fibroblast growth factor receptors: lessons from the genes. Trends Biochem Sci. 1998;23:59–62. doi: 10.1016/s0968-0004(97)01170-5. [DOI] [PubMed] [Google Scholar]
- Cappellen D, De Oliverira C, Ricol D, de Medina SGD, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, Dalton WS, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4:14)(p16–3;q32–3) in multiple myeloma: association with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort DL, Wang Z, Blackmore V, Moran MF, Muller WJ. Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol Cell Biol. 1997;17:5410–5425. doi: 10.1128/mcb.17.9.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Decker SJ. Transmembrane signaling by epidermal growth factor receptors lacking autophosphorylation sites. J Biol Chem. 1993;268:9176–9179. [PubMed] [Google Scholar]
- Dell'Era P, Mohammadi M, Presta M. Different tyrosine autophosphorylation requirements in fibroblast growth factor receptor-1 mediate urokinase-type plasminogen activator induction and mitogenesis. Mol Biol Cell. 1999;10:23–33. doi: 10.1091/mbc.10.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl WJ, Escobedo JA, Martin GA, Turck CW, del Rosario M, McCormick F, Williams LT. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- Fridell YW, Jin Y, Quilliam LA, Burchert A, McCloskey P, Spizz G, Varnum B, Der C, Liu ET. Differential activation of the Ras/extracellular-signal-regulated protein kinase pathway is responsible for the biological consequences induced by the Axl receptor tyrosine kinase. Mol Cell Biol. 1996;16:135–145. doi: 10.1128/mcb.16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R, Jove R. Activation of STAT transcription factors in oncogenic tyrosine kinase signaling. J Biomed Sci. 1998;5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart KC, Kanemitsu MY, Robertson SC, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000a;19:3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- Hart KC, Robertson SC, Donoghue DJ. Activation of H-ras61L-specific signaling pathways does not require post-translational processing of H-ras. Exp Cell Res. 2000b;257:89–100. doi: 10.1006/excr.2000.4874. [DOI] [PubMed] [Google Scholar]
- Hattori Y, Odagiri H, Katoh O, Sakamoto H, Morita T, Shimotohno K, Tobinai K, Sugimura T, Terada M. K-sam-related gene, N-sam, encodes fibroblast growth factor receptor and is expressed in T-lymphocytic tumors. Cancer Res. 1992;52:3367–3371. [PubMed] [Google Scholar]
- Jones N, Master Z, Jones J, Bouchard D, Gunji Y, Sasaki H, Daly R, Alitalo K, Dumont DJ. Identification of Tek/Tie2 binding partners. J Biol Chem. 1999;274:30896–30905. doi: 10.1074/jbc.274.43.30896. [DOI] [PubMed] [Google Scholar]
- Kashishian A, Cooper JA. Phosphorylation sites at the C-terminus of the platelet-derived growth factor receptor bind phospholipase C gamma 1. Mol Biol Cell. 1993;4:49–57. doi: 10.1091/mbc.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashishian A, Kazlauskas A, Cooper JA. Phosphorylation sites in the PDGF receptor with different specificities for binding GAP and PI3 kinase in vivo. EMBO J. 1992;11:1373–1382. doi: 10.1002/j.1460-2075.1992.tb05182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas A, Kashishian A, Cooper JA, Valius M. GTPase-activating protein and phosphatidylinositol 3-kinase bind to distinct regions of the platelet-derived growth factor receptor beta subunit. Mol Cell Biol. 1992;12:2534–2544. doi: 10.1128/mcb.12.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- Kraslinikov MA. Phosphatidylinositol-3 kinase dependent pathways: the role in control of cell growth, survival, and malignant transformation. Biochemistry Moscow. 2000;65:59–67. [PubMed] [Google Scholar]
- Legeai-Mallet L, Benoist-Lasselin C, Delezoide AL, Munnich A, Bonaventure J. Fibroblast growth factor receptor 3 mutations promote apoptosis but do not alter chondrocyte proliferation in thanatophoric dysplasia. J Biol Chem. 1998;273:13007–13014. doi: 10.1074/jbc.273.21.13007. [DOI] [PubMed] [Google Scholar]
- Li C, Chen L, Iwata T, Kitagawa M, Fu X-Y, Deng C-X. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996a;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Dionne CA, Li W, Li N, Spivak T, Honegger AM, Jaye M, Schlessinger J. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358:681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, Dionne CA, Jaye M, Rubinstein M, Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger M, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996b;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Holgado-Madruga M, Maroun C, Fixman ED, Kamikura D, Fournier T, Charest A, Tremblay ML, Wong AJ, Park M. Association of the multisubstrate docking protein Gab1 with the hepatocyte growth factor receptor requires a functional Grb2 binding site involving tyrosine 1356. J Biol Chem. 1997;272:20811–20819. doi: 10.1074/jbc.272.33.20811. [DOI] [PubMed] [Google Scholar]
- Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J, Lax I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SH, Lim YP, Low BC, Guy GR. SHP2 associates directly with tyrosine phosphorylated p90 (SNT) protein in FGF-stimulated cells. Biochem Biophys Res Commun. 1997;238:261–266. doi: 10.1006/bbrc.1997.7272. [DOI] [PubMed] [Google Scholar]
- Popovici C, Adelaide J, Ollendorff V, Chaffanet C, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is fused to FIM in stem-cell myeloproliferative disorder with t(8;13) Proc Natl Acad Sci USA. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici C, Zhang B, Gregoire MJ, Jonveaux P, Lafage-Pochitaloff M, Birnbaum D, Pebusque MJ. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood. 1999;93:1381–1389. [PubMed] [Google Scholar]
- Reiter A, Sohal J, Kulkarni S, Chase A, Macdonald DH, Aguiar RC, Goncalves C, Hernandez JM, Jennings BA, Goldman JM, Cross NCP. Consistent fusion of ZNF198 to the fibroblast growth factor receptor-1 in the t(8;13)(p11;q12) myeloproliferative syndrome. Blood. 1998;92:1735–1742. [PubMed] [Google Scholar]
- Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, Rocchi M, Otsuki T, Lombardi L, Maiolo AT, Neri A. A novel chromosomal translocation t(4:14)(p16–3;q32) in multiple myeloma involves the fibroblast growth factor receptor 3 gene. Blood. 1997;90:4062–4070. [PubMed] [Google Scholar]
- Ronnstrand L, Mori S, Arridsson A-K, Eriksson A, Wernstedt C, Hellman U, Claesson-Welsh L, Heldin C-H. Identification of two C-terminal autophorphorylation sites in the PDGF β-receptor: involvement in the interaction with phospholipase C-γ. EMBO J. 1992;11:3911–3919. doi: 10.1002/j.1460-2075.1992.tb05484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Fisher DE. Apoptosis and cancer drug targeting. J Clin Invest. 1999;104:1655–1661. doi: 10.1172/JCI9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Hamoudi R, Clark J, Warren W, Abdul-Rauf M, Somers G, Venter D, Fagan K, Cooper C, Shipley J. The t(8;13)(p11;q11–12) rearrangement associated with an atypical myeloproliferative disorder fuses the fibroblast growth factor receptor 1 gene to a novel gene RAMP. Hum Mol Genet. 1998;7:637–642. doi: 10.1093/hmg/7.4.637. [DOI] [PubMed] [Google Scholar]
- Su W, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Fallon JF. Fibroblast growth factors as multifunctional signaling factors. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- Tavormina PL, Bellus GA, Webster MK, Bamshad MJ, Fraley AE, McIntosh I, Szabo J, Jiang W, Jabs EW, Wilcox WR, Wasmuth JJ, Donoghue DJ, Thompson LM, Francomano CA. A novel skeletal dysplasia with developmental delay and acanthosis nigricans is caused by a Lys650Met mutation in the fibroblast growth factor receptor 3 (FGFR3) gene. Am J Hum Genet. 1999;64:722–731. doi: 10.1086/302275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulasne D, Paumelle R, Weidner KM, Vandenbunder B, Fafeur V. The multisubstrate docking site of the MET receptor is dispensable for MET-mediated RAS signaling and cell scattering. Mol Biol Cell. 1999;10:551–565. doi: 10.1091/mbc.10.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valius M, Bazenet C, Kazlauskas A. Tyrosines 1021 and 1009 are phosphorylation sites in the carboxy terminus of the platelet-derived growth factor receptor beta subunit and are required for binding of phospholipase C gamma and a 64-kilodalton protein, respectively. Mol Cell Biol. 1993;13:133–143. doi: 10.1128/mcb.13.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valius M, Kazlauskas A. Phospholipase C-gamma 1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor's mitogenic signal. Cell. 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- Webster MK, D'Avis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for the lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- Webster MK, Donoghue DJ. Enhanced signaling and morphological transformation by a membrane-localized derivative of the FGFR3 kinase domain. Mol Cell Biol. 1997a;17:5739–5747. doi: 10.1128/mcb.17.10.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997b;13:178–182. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman M, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee KW, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]