Abstract
The periplasmic Cu+/Ag+ chaperone CusF features a novel cation-π interaction between a Cu+/Ag+ ion and Trp44 at the metal binding site. The nature and strength of the Cu+/Ag+-Trp44 interactions are investigated using computational methodologies. Quantum mechanical (QM) calculations show that Cu+ and Ag+ interactions with Trp44 are both of similar strength (~14 kcal/mol) and bond order. Quantum-mechanical/molecular mechanical (QM/MM) calculations show that Cu+ binds in a distorted tetrahedral coordination environment in the cation-π interaction-lacking Trp44Met mutant. Molecular dynamics (MD) simulations of CusF in the apo and Cu+ bound states emphasize the importance of the Cu+-Trp44 interactions in protecting Cu+ from water oxidation. The protein structure does not change over the time-scale of hundreds of nanoseconds in the metal bound state. The metal recognition site exhibits small motions in the apo state, but remains largely preorganized towards metal binding. Trp44 remains oriented to form the cation-π interaction in the apo state, and faces an energetic penalty to move away from the metal ion. Cu+ binding quenches the protein’s internal motions in regions linked to binding CusB, suggesting protein motions play an essential role in Cu+ transfer to CusB.
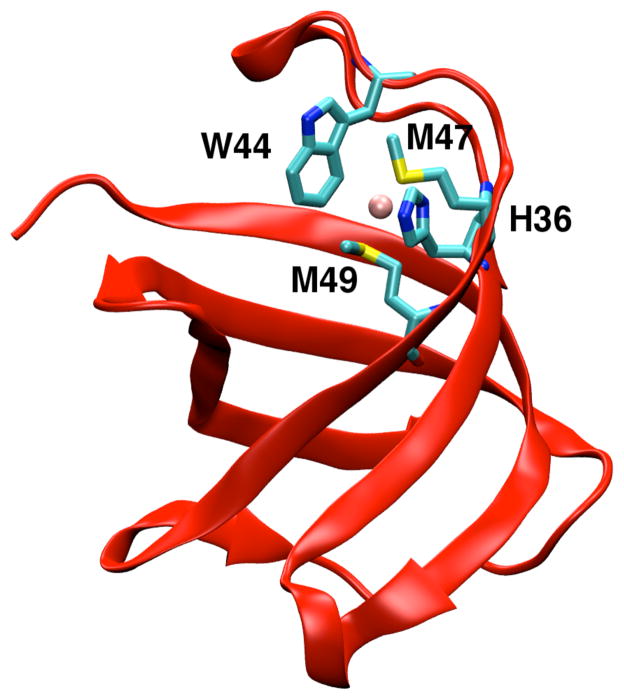
Heavy metal homeostasis regulates the concentration of metal ions in cells and is essential for the survival of organisms.1 A strong coupling between heavy metal tolerance and antibiotic resistance in Gram-negative bacteria makes it imperative to understand the nature of Cu+/Ag+ efflux in these organisms. In Escherichia coli, the cus determinant consisting of the cusCFBA operon provides for Cu+ and Ag+ resistance.2,3 Recent crystal structures suggest that the CusCBA proteins combine to form a tripartite cation efflux pump that expels metal ions from within the cell to the outer membrane.4 CusF is a small β-barrel protein (Fig. 1 and Fig. S.I. 1) that functions as a periplasmic Cu+/Ag+ metallochaperone.5 It binds to CusB and provides Cu+/Ag+ efflux directly from the periplasm by transferring metal ions to the tripartite cation efflux pump.3 CusF is not essential for Cu+/Ag+ ion tolerance and its exact role in heavy metal homeostasis remains a subject for further investigation.6
Figure 1.
Cartoon representation of Cu(I)•CusF (PDB code: 2VB2). Cu+ is shown as a pink sphere and the metal binding residues are represented in stick formation.
Crystallographic structures of the apo and holo forms of CusF are similar, indicating that metal ion binding does not cause a large change in the protein structure.5,7,8 The metal recognition site in CusF is located in a hydrophobic environment and contains a novel cation-π interaction between Cu+/Ag+ and the Cε3 and Cζ3 carbon atoms on the aromatic ring of the neighboring Trp44 residue (Fig. 1 and Figs. SI. 1–3). In addition to this interaction, His36, Met47, and Met49 residues bind to the Cu+/Ag+ ion in a distorted trigonal planar arrangement with the metal ion slightly elevated from the plane (Fig. 1). An exact characterization of the unique Cu+/Ag+-W44 interaction has been hampered by a lack of direct spectroscopic evidence. A crystallographic database search of similar compounds by Xue et al. found this interaction to lie at the periphery of non-bonded interactions, while UV resonance Raman spectroscopy identified this interaction as a novel cation-π interaction rarely observed in a Cu+/Ag+ metal binding site.7 Experiments by Loftin et al. have suggested that Trp44 protects Cu+/Ag+ from oxidative stress in the periplasm.9 The role of the cation-π interaction in the function of CusF remains unclear as a W44M mutation exists in 25% of CusF proteins. The W44M mutant CusF is known to have a higher binding affinity for Cu+.7,9 While the nature of metal coordination in the cation-π interaction-lacking metal binding site of the W44M mutant has been probed with extended X-ray absorption fine structure (EXAFS), the exact geometry of the metal binding site remains to be determined.9
In this study, we investigate the nature of the Cu+/Ag+-W44 interaction and evaluate its impact on the chaperone function of CusF using computational methodologies. We estimate the strength of this interaction and provide a QM basis for its characterization using accurate ab initio calculations. We use QM/MM calculations to evaluate the role of W44 in protecting Cu+ and propose a metal binding geometry for Cu+ in the W44M mutant. In addition, we perform MD simulations to investigate changes in the structure and internal motions of the protein related to Cu+ binding. These simulations provide us with a deeper insight into the role of the cation-π interaction and relate them to the protein’s function. In this study, all ab initio calculations were performed using the Gaussian09 suite of programs,10 QM/MM calculations were performed using the Qsite program in the Schrödinger suite of programs,11 and MD simulations were performed using the Amber11 suite of programs.12 MD trajectories were analyzed using the VMD suite of programs13 and the ptraj utility in AmberTools 1.5. The first nine residues of CusF are not included in the analysis of MD trajectories. Complete details for all calculations are included in the “Methods” section that is presented as part of SI.
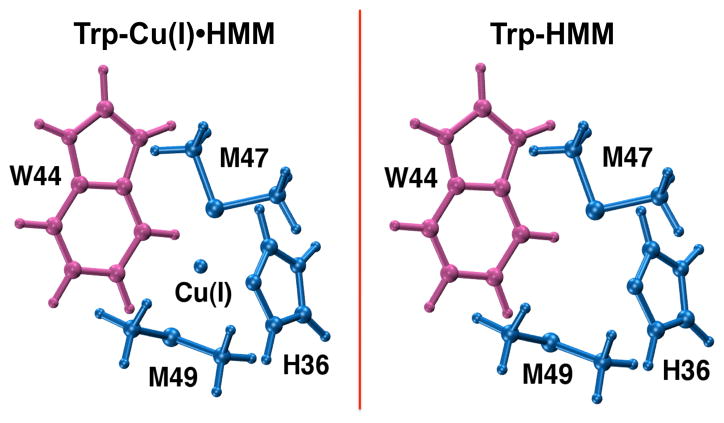
We performed QM calculations at the second order Møller–Plesset perturbation (MP2) level of theory to determine the strength of the Cu+/Ag+-W44 cation-π interaction at the Cu+ and Ag+ bound crystal structure geometries of the protein (Fig. 2). A double ζ-quality LANL2DZ pseudopotential-based basis set14 was used for Ag+ while the augmented Dunning correlation-consistent polarized aug-cc-pVDZ basis set15 was employed for all other atoms in these calculations. We determined the interaction energy between Trp44 and the metal ion bound to H36, M47 and M49 to be on the order of ~14 kcal/mol (Table SI.1). We then estimated the contribution of the W44–(H36•M47•M49) interaction energy to the above energy by calculating the interaction energy between Trp44 and the other metal binding residues in the absence of the metal ion at the same geometry. While this method contains a certain amount of approximation, it yields an estimate for the “apo interaction energy” that is close to the interaction energy calculated from the 1ZEQ crystal structure geometry (−5.43 kcal/mol). From these calculations, we found that the Cu+/Ag+-W44 interactions afforded a stabilization of over 10 kcal/mol to the metal ion complex. A comparison of various functionals within the framework of density functional theory paired with the aug-cc-pVDZ basis set found the M06-L,16 M06-2X17 and wB97XD18 functionals to be most suitable for studying such an interaction (Table SI.2). In addition to calculating these interaction energies, we performed a natural bond orbital analysis19 to characterize the Cu+/Ag+ interaction with the Cε3 and Cζ3 carbon atoms of Trp44 at the MP2 level of theory using the LANL2DZ pseudopotential-based basis set for Ag+ and the aug-cc-pVDZ basis set for all other atoms in these calculations. We found that Cu+ and Ag+ ions interact specifically with both Trp44 atoms (Table SI.3). As expected, these bond orders are lower than those of the Cu+/Ag+–S-Met47 bonds, indicating a relatively longer and weaker interaction. These low bond orders explain the marginal improvements seen by Xue et al.7 and Loftin et al.8 upon including a C/N/O scatterer in their models to fit EXAFS measurements.
Figure 2.
Cu+•CusF metal binding site geometries represented using a model system. Interaction energies were calculated between the pink and blue fragments. The Trp-Cu(I)•HMM interaction energy provided the interaction energy between W44 and bound Cu+ (Cu+•H36•M47•M49), and the Trp-HMM interaction energy gave the interaction energy between W44 and other metal binding residues in the absence of Cu+ (W44–H36•M47• M49) at the same geometry. The stabilization from the cation-π interaction was estimated as Trp-Cu(I)•HMM – Trp-HMM.
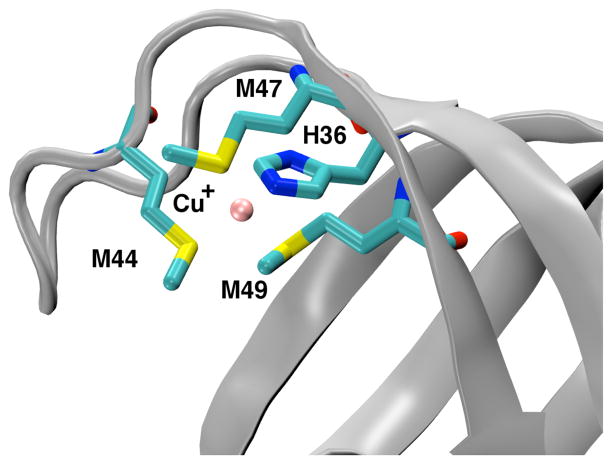
To study the metal ion interactions in the W44M mutant, we investigated its Cu+ binding environment by performing a series of increasingly complex DFT-QM/MM20 calculations employing the M06-L functional. The final geometry optimization was performed using the LACV3P+* basis set. Such methodologies provide powerful tools to elucidate protein function.21–23 In these calculations, the W44M mutation was performed on WT CusF, and Cu+ was allowed to decide its coordination in the solvated protein via energy minimization. Similar to results from NMR experiments performed by Loftin et al., we find that the W44M mutation does not change the protein structure (Figs. SI. 4,5).9 Our calculations found that Cu+ was bound to His36, Met44, Met47 and Met49 residues in a distorted tetrahedral arrangement as shown in Fig. 3. In agreement with EXAFS data, we found the bonds to Cu+ to be longer than the corresponding bonds in WT CusF.9 Interestingly, the Cu+-S(M44) bond was shorter than the other Cu+-S bonds to M47 and M49. The calculated bond lengths and bond angles are listed in Tables SI.4 and SI.5. The interaction energy between the bonded Cu+ ion and Met44 in this binding motif was found to be −20.5 kcal/mol with the Cu+-Met44 interaction accounting for a stabilization energy of −20.7 kcal/mol (Table SI.1). The higher interaction energy and almost two-fold stabilization energy observed in the W44M mutant are consistent with the higher binding affinity for Cu+ in this mutant form.7,9 While these calculations provide new insights about the binding environment of Cu+ in W44M CusF, they are hindered by an inherent lack of conformational sampling that may have introduced errors.
Figure 3.
Cartoon representation of Cu(I)•W44MCusF mutant showing Cu+ as a pink sphere and the metal binding residues represented in stick formation in a distorted tetrahedral coordination environment from QM/MM calculations.
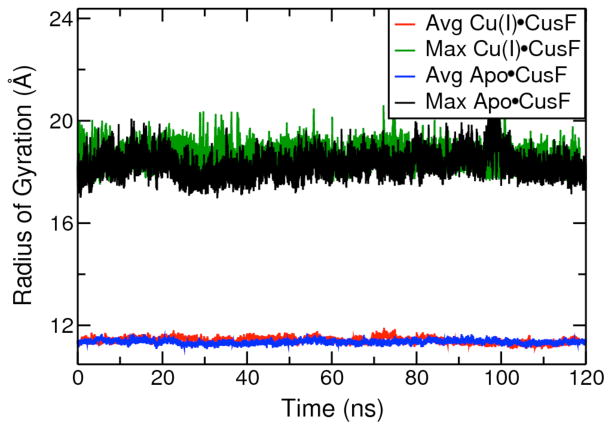
We analyzed changes in the protein’s mobility and structure over 360ns of all atom MD simulations of the apo and Cu+ bound forms of WT CusF in explicit solvent. We developed specific force field parameters to treat Cu+ as a bound ion in simulations of holo CusF. In addition, we modified the Lenard-Jones parameters on W44 to accurately portray the stronger Cu+-W44 interaction obtained from our ab initio calculations. All parameters used in the study are included in SI. A stable protein backbone RMSD was observed over the course of our simulations of apo and Cu+ ion bound CusF. The protein maintained its secondary structure and no large-scale motions were detected in these simulations. The calculated average radius of gyration and maximum radius of gyration for the protein over the course of these simulations of apo CusF and Cu+•CusF were similar (Fig. 4), suggesting that Cu+ binding does not alter the volume and size of the protein. Fig. SI. 6 depicts the calculated Cα backbone carbon atom root mean square fluctuations for the apo and Cu+ bound states of the protein. The first 8 residues in apo CusF remained very mobile. We observed a reduced mobility in most protein residues on Cu+ binding in close agreement with NMR experiments.9 The Cu+ binding residues are less mobile than their neighboring residues in apo CusF and become more rigid on metal ion binding. Trp44 does not actively search for the metal ion in the apo state, and is preorganized to form a cation-π interaction with the metal ion (Figs. SI. 6–8). We found that the loop region defined by residues 72 to 80 moved significantly in our simulation of the apo allosteric form and did not become more rigid as a result of metal ion binding.
Figure 4.
Average and maximum radius of gyration values from simulations of apo CusF and Cu+ bound CusF.
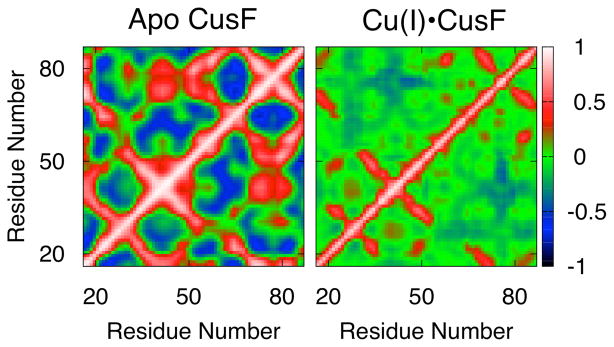
We further examined the impact of metal ion binding on protein residue motions by calculating the cross-correlation matrices for all backbone Cα carbon atoms for the apo state and Cu+-bound state of the protein. These matrices map the correlation between the movements of various residues from +1 for perfect correlation, i.e. they move in the same direction, and −1 for anti-correlation, i.e. they move in opposite directions. The apo state of the protein is characterized by a number of residues involved in correlated and anti-correlated motions (Fig. 5). The metal binding residues move in a correlated manner to each other in this allosteric form. We found that Cu+ binding largely quenches the motions in the protein and causes some previously correlated residues to become anti-correlated and vice versa. The mobile loop region described by residues 40 to 43 and lying adjacent to the metal binding site, now moves in an anti-correlated manner with the mobile loop between residues 72 to 80, and in a correlated manner with the loop between residues 26 to 30.
Figure 5.
Cross correlation plots for apo CusF and Cu+•CusF.
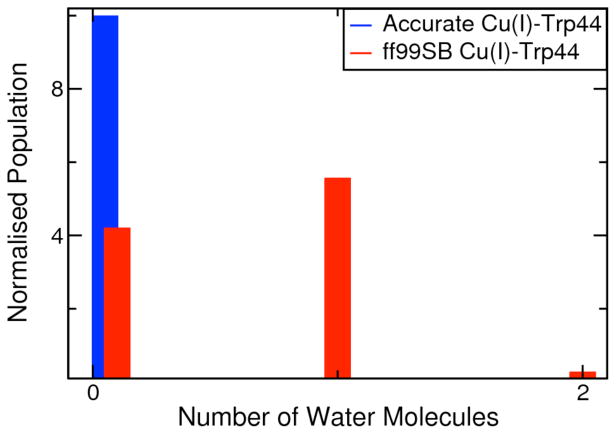
We investigated the function of Trp44 in the chaperone activity of CusF using QM/MM (M06-2X/LACVP*) calculations11 and MD simulations. We calculated the potential of mean force to “swing” Trp44 away from Cu+ along the Cu+-Cβ-Cγ-Cδ1 dihedral coordinate using QM/MM calculations (Fig. SI.7). A scan along the dihedral coordinate at the crystal structure geometry of Cu+ bound CusF revealed a significant energetic penalty associated with swinging Trp44, preventing it from moving away from the metal ion. In addition, we found Trp44 to be locked in its position in our simulations for the apo and holo states of the protein. The C-Cα-Cβ-Cγ dihedral of Trp44 remains steady over the course of our simulation of apo CusF (Fig. SI.8), suggesting that the metal binding site is preorganized for the cation-π interaction between the metal ion and the Trp44 residue. We also examined the protective role of Trp44 in shielding the monovalent metal ion from oxidation by water molecules by performing an additional simulation of holo CusF with a weaker Cu+-W44 interaction by using standard ff99SB24 Lenard-Jones parameters on W44. On comparing our two simulations of holo CusF, we observed no water molecules in the first solvation shell of the metal ion in our simulation with the correct Cu+-W44 interaction energy, while water molecules were present near Cu+ in over 60% of snap-shots from our simulation with a weaker Cu+-W44 interaction energy. These results in conjunction with the low mobility of Trp44 demonstrate that the strong cation-π interaction and the hydrophobic nature of Trp44 successfully shield the Cu+ ion from water molecules.
Our simulations highlight the role of protein motions in the chaperone function of CusF. CusF and CusB form a transient complex only when one of them is in the metal bound state.6, 19 A recent chemical cross-linking and mass spectrometry study of CusF binding to CusB by Mealman et al. suggests that residues 31 to 50 and 51 to 63 of CusF actively participate in interactions with CusB.25 These interactions are centered at Lys31 and Lys58 residues. We find further support for these findings in our simulations. Lys31 and Lys58 remain relatively rigid in simulations of apo and Cu+ bound CusF. Our simulations also find that residues in the CusF-CusB interacting regions become less mobile in the metal bound state. In contrast, other parts of the protein not associated with CusB binding remain mobile in the Cu+ bound states as well. These changes in protein residue mobility are accompanied with changes in the correlated motions as well. Our simulations strongly suggest that metal ion binding alters the internal motions in critical regions of the protein. These changes in the protein’s conformational motions plays an essential role in allowing CusF to form a transient complex with CusB, and transfer Cu+/Ag+ to the cation efflux pump.
In this study, we used multiple computational approaches to provide detailed insights into the function and nature of the cation-π interaction at the metal binding site of the periplasmic protein CusF. We quantified the Cu+/Ag+-W44 cation-π interaction using high-level ab initio calculations and found a number of DFT functionals that accurately predict the energetics of the interaction. The calculated covalencies of the Cu+/Ag+ bonds to the Cε3 and Cζ3 carbon atoms of W44 indicate that they are of a weak and long-range nature. We proposed a distorted tetrahedral coordination geometry for Cu+ in the W44M mutant, and found the Cu+-M44 interaction to be stronger than the cation-π interaction in WT CusF. Our calculations show that Trp44 remains preorganized for metal ion binding and the cation-π interaction helps protect Cu+/Ag+ from oxidation in the periplasm. Our simulations suggest that no large conformational changes take place and the protein maintains its structure in both allosteric forms on the time scale of hundreds of nanoseconds. We found that Cu+ binding quenched the internal dynamics of the protein. This was especially noticeable in residues that participate in the metal ion dependent interaction with the transmembrane periplasmic protein CusB. These results as a whole show that the Cu+/Ag+-W44 cation-π interaction at the metal recognition site in WT CusF maintains a delicate balance between protecting Cu+ from oxidation by water molecules and allowing for the metal ion to transfer to the cation efflux tripartite pump.
Supplementary Material
Figure 6.
Distribution of water molecules in the first solvation shell of Cu+ in simulations with a refined Cu+-W44 interaction energy and standard (weak) Cu+-W44 interaction energy.
Acknowledgments
We thank Benjamin P. Roberts, Sarah Gordon and Naoya Asada for helpful discussions. We thank Thomas O’Halloran for directing our attention towards the W44M mutant. We gratefully acknowledge the National Institutes of Health (GM044974 and GM066859) for funding this project, and high performance computing at the University of Florida for their support.
ABBREVIATIONS
- MD
molecular dynamics
- PMF
potential of mean force
- QM/MM
quantum mechanical molecular mechanical
- DFT
density functional theory
- MP2
second order Møller-Plesset perturbation
- WT
wild-type
- EXAFS
extended X-ray absorption fine structure
Footnotes
Supporting Information. Input files and information on all our calculations, figures, structures, force field parameter files, W44M protein structure, complete citation for references 10 and 12. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ma Z, Jacobsen FE, Giedroc DP. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munson GP, Lam DL, Outten FW, O’Halloran TV. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke S, Grass G, Rensing C, Nies DH. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Nature. 2011;470:558–U153. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. Biochemistry. 2005;44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 6.Kim EH, Rensing C, McEvoy MM. Natural Product Reports. 2010;27:711–719. doi: 10.1039/b906681k. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, Focia P, Spiro TG, Penner-Hahn JE, O’Halloran TV. Nature Chemical Biology. 2008;4:107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Protein Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loftin IR, Blackburn NJ, McEvoy MM. J Biol In-org Chem. 2009;14:905–912. doi: 10.1007/s00775-009-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frisch MJ, et al. Gaussian, Inc. Wallingford CT: 2009. [Google Scholar]
- 11.Qsite. Schrödinger, Inc. 5.7. New York, NY: 2011. [Google Scholar]
- 12.Case DA, et al. University of California. San Francisco: 2010. [Google Scholar]
- 13.Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 14.Hay PJ, Wadt WR. J Chem Phys. 1985;82:299–310. [Google Scholar]
- 15.Woon DE, Dunning TH. J Chem Phys. 1993;98:1358–1371. [Google Scholar]
- 16.Zhao Y, Truhlar DG. J Chem Phys. 2006;125:194101. doi: 10.1063/1.2370993. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Truhlar DG. Theor Chem Acc. 2008;120:215–241. [Google Scholar]
- 18.Chai JD, Head-Gordon M. Phys Chem Chem Phys. 2008;10:6615–6620. doi: 10.1039/b810189b. [DOI] [PubMed] [Google Scholar]
- 19.Foster JP, Weinhold F. J Am Chem Soc. 1980;102:7211–7218. [Google Scholar]
- 20.Warshel A. Computer Modeling of Chemical Reactions in Enzymes and Solutions. John Wiley & Sons, Inc; New York: 1991. [Google Scholar]
- 21.Chakravorty DK, Soudackov AV, Hammes-Schiffer S. Biochemistry. 2009;48:10608–10619. doi: 10.1021/bi901353v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang JK, Warshel A. J Am Chem Soc. 1996;118:11745–11751. [Google Scholar]
- 23.Chakravorty DK, Hammes-Schiffer S. J Am Chem Soc. 2010;132:7549–7555. doi: 10.1021/ja102714u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mealman TD, Bagai I, Singh P, Goodlett DR, Rensing C, Zhou H, Wysocki VH, McEvoy MM. Biochemistry. 2011;50:2559–2566. doi: 10.1021/bi102012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.