Abstract
Streptococcus intermedius is a facultatively anaerobic, opportunistic pathogen that causes purulent infections and abscess formation. The DnaK chaperone system has been characterized in several pathogenic bacteria and seems to have important functions in stress resistance and pathogenicity. However, the role of DnaK in S. intermedius remains unclear. Therefore, we constructed a dnaK knockout mutant that exhibited slow growth, thermosensitivity, accumulation of GroEL in the cell, and reduced cytotoxicity to HepG2 cells. The level of secretion of a major pathogenic factor, intermedilysin, was not affected by dnaK mutation. We further examined the function and property of the S. intermedius DnaK chaperone system by using Escherichia coli ΔdnaK and ΔrpoH mutant strains. S. intermedius DnaK could not complement the thermosensitivity of E. coli ΔdnaK mutant. However, the intact S. intermedius DnaK chaperone system could complement the thermosensitivity and acid sensitivity of E. coli ΔdnaK mutant. The S. intermedius DnaK chaperone system could regulate the activity and stability of the heat shock transcription factor σ32 in E. coli, although S. intermedius does not utilize σ32 for heat shock transcription. The S. intermedius DnaK chaperone system was also able to efficiently eliminate the aggregated proteins from ΔrpoH mutant cells. Overall, our data showed that the S. intermedius DnaK chaperone system has important functions in quality control of cellular proteins but has less participation in the modulation of expression of pathogenic factors.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-011-0284-4) contains supplementary material, which is available to authorized users.
Keywords: Streptococcus intermedius, Escherichia coli, DnaK chaperone system, Thermosensitivity, Intermedilysin
Introduction
Streptococcus intermedius is a facultative anaerobe that belongs to the Anginosus group of streptococci, forming part of the normal flora of the human oral cavity as well as the upper respiratory, gastrointestinal, and female urogenital tracts (Whiley et al. 1990, 1992). S. intermedius is an opportunistic human pathogen and a leading cause of deep-seated purulent infections, including brain and liver abscesses (Whiley et al. 1990, 1992; Jacobs et al. 1995; Jerng et al. 1997; Claridge et al. 2001). This pathogen secretes a human-specific cytolysin, intermedilysin (ILY), which is a member of the cholesterol-dependent cytolysin family (Nagamune et al. 1996). ILY is a major virulence factor in S. intermedius, which is essential for invasion and cytotoxicity to human cells (Nagamune et al. 2000; Sukeno et al. 2005).
Various Gram-negative and Gram-positive intracellular pathogens have been shown to adapt to stress conditions, such as those set by an effective immune system and fever in the host organism, by producing stress proteins (HSPs) such as molecular chaperones (DnaK chaperone system, GroEL and GroES, etc.) and several proteases (Henderson et al. 2006). The ensemble of HSPs constitutes a cellular system for de novo folding and quality control of proteins that relies on the ability of chaperones and proteases to refold or degrade misfolded proteins under stress conditions (Ben-Zvi and Goloubinoff 2001; Dougan et al. 2002). The DnaK chaperone system is composed of DnaK and co-chaperones (DnaJ and GrpE). Co-chaperones are necessary for operation of the DnaK chaperone cycle (Mayer et al. 2000; Ben-Zvi and Goloubinoff 2001; Genevaux et al. 2007); DnaJ binds the substrate (newly synthesized protein, denatured protein, etc.) and interacts with DnaK by transferring its bound substrate to the substrate-binding domain of DnaK and simultaneously stimulating ATP hydrolysis by DnaK. Consequently, DnaK is transformed to the ADP-bound state, which exhibits high affinity and low exchange rate for its substrate. GrpE is a nucleotide exchange factor of DnaK that helps release ADP from DnaK. The nucleotide-free DnaK immediately binds ATP. The ATP-bound state of DnaK exhibits low affinity to the substrate, release of substrate, and resetting of the DnaK chaperone cycle.
DnaK null mutant was originally isolated from the Gram-negative bacterium Escherichia coli (Bukau and Walker 1989a). This mutant has been reported to exhibit pleiotropic defects in growth, cell division, plasmid replication, and thermal and chemical stress resistance (Bukau and Walker 1989a, b; Genevaux et al. 2007). In addition, the DnaK chaperone system negatively controls the activity and stability of the heat shock transcription factor σ32 in E. coli (Arsène et al. 2000; Yura and Nakahigashi 1999). Hence, constitutive induction of heat shock proteins and accumulation of σ32 are observed in the E. coli ΔdnaK mutant. The phenotypes of the DnaK mutants from most Gram-positive bacteria are similar to the phenotypes of the E. coli ΔdnaK mutant in showing sensitivity to several stresses (Koch et al. 1998; Hanawa et al. 1999; Köhler et al. 2002; Singh et al. 2007).
It has been reported that the DnaK chaperone system is required not only for stress tolerance but also for the pathogenicity of bacteria; DnaK of the Gram-negative intracellular pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) positively controls the expression of Salmonella pathogenicity islands 1 and 2 and the dnaK null mutant was non-pathogenic (Takaya et al. 2004). Similar phenotypes were also reported in Gram-positive pathogens: a dnaK null mutant of Staphylococcus aureus showed reduced survival in a mouse host during systemic infection (Singh et al. 2007). Constitutive and lower-level expression of DnaK induced by promoter replacement of the dnaK gene in the intracellular pathogen Brucella suis resulted in non-proliferation of the organism within the macrophage-like U937 cells (Köhler et al. 2002) and in Listeria monocytogenes DnaK, although not largely involved in intracellular growth within macrophages is nevertheless required for efficient phagocytosis by macrophage-like J774.1 cells (Hanawa et al. 1999). In contrast to the examples described above, the exact role of the DnaK chaperone system in stress resistance and pathogenicity control of the facultatively anaerobic opportunistic pathogen S. intermedius remains unclear. In order to investigate this we constructed a dnaK null mutant of S. intermedius. Our data show that S. intermedius DnaK chaperone system has the same chaperone activity in vivo as observed with the E. coli DnaK chaperone system plays an important role in key functions including growth, thermoresistance, and heat shock regulation but has less involvement in the modulation of pathogenic factor expression.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. S. intermedius was cultured at the indicated temperature under anaerobic conditions. Brain–heart infusion (BHI) broth (Becton-Dickinson, Palo Alto, CA, USA) was used for the liquid culture of S. intermedius. E. coli was grown in Luria–Bertani (LB) medium at the indicated temperature under aerobic conditions. Antibiotics were added at the following concentrations: ampicillin (Ap) at 100 μg/mL for culturing E. coli, chloramphenicol (Cm) at 20 μg/mL for E. coli and 2 μg/mL for S. intermedius, erythromycin (Em) at 1 μg/mL for S. intermedius, and kanamycin (Km) at 20 μg/mL for E. coli.
Table 1.
Bacterial strains and plasmids used in this study
| Strains | Relevant characteristics | Reference or source |
|---|---|---|
| S. intermedius | ||
| NCDO2227 | Type strain | Nagamune et al. 2000 |
| UNS38 | High intermedilysin-producing strain from human brain abscess | Sukeno et al. 2005 |
| UNS38 B3 | As UNS38 except for Δily::Em | Sukeno et al. 2005 |
| UNS38 ΔdnaK | As UNS38 except for ΔdnaK::Em | Present study |
| UNS38 ΔdnaK R37 | As UNS38 ΔdnaK, but able to grow to 37°C by an unknown suppressor mutation | Present study |
| E. coli | ||
| DH5αZ1 | F− Φ80Δ(lacZ)M15 Δ(lacZYA-argF)U169 deoR recA1endA1 hsdR17(r−Km+K) phoA supE44 λ−thi-1 gyrA96relA1 tetR lacIqaadA+ | Lutz and Bujard 1997 |
| MC4100 | F−araD139 Δ(argF-lac)U169 deoC1 thiA flbB5301 ptsF25 relA1 rpsL150 tonA21 | Bukau and Walker 1990 |
| BB1554 | As MC4100 except for ΔdnaK52::Cm sidB2 | Bukau and Walker 1990 |
| BB1554 lacIq | BB1554 carrying pDMI, I | Present study |
| BB7224 | As MC4100 except for ΔrpoH::Km suhX401 araD+ | Tomoyasu et al. 2001 |
| CS5262 | BB7224 carrying pBB528 | Tomoyasu et al. 2010b |
| Plasmids | ||
| pDMI, I | Lac repressor–producing plasmid, p15A ori, Kmr | Lanzer and Bujard 1988 |
| pBB528 | Lac repressor–producing plasmid, pSC101 ori, Cmr | Tomoyasu et al. 2001 |
| pSETN1 | Reported as pSET1 Δlac p15A, Cmr | Tomoyasu et al. 2010a, b |
| pSETN1 CP25 | pSETN1-carrying CP25 promoter | Present study |
| pSETN1 EKJ | pSETN1-carrying grpE, dnaK, and dnaJ operon | Present study |
| pZE13 | IPTG-inducible expression vector, pMB1 ori, Apr | Lutz and Bujard 1997 |
| pZE13 EcK | pZE13-carrying E. coli dnaK | Present study |
| pZE13 SiK | pZE13-carrying S. intermedius dnaK | Present study |
| pZE13 SiEK | pZE13-carrying S. intermedius grpE and dnaK | Present study |
| pZE13 SiKJ | pZE13-carrying S. intermedius dnaK and dnaJ | Present study |
| pZE13 SiEKJ | pZE13-carrying S. intermedius grpE, dnaK and dnaJ | Present study |
Databases and multiple sequence alignment
Nucleotide and protein sequences of the DnaK chaperone system from Streptococcus pneumoniae R6 (DnaK: GenBank Acc. No. NP_358049, DnaJ: NP_358050, GrpE: NP_358048), Lactococcus lactis subsp. lactis II1403 (DnaK: NP_267110, DnaJ: NP_268381, GrpE: NP_267109), Bacillus subtilis subsp. subtilis str. (DnaK: NP_390425, DnaJ: NP_390424, GrpE: NP_390426), and E. coli str. K-12 substr. W3110 (DnaK: AP_000678, DnaJ: AP_000679, GrpE: AP_003194) were obtained from GenBank by an Entrez cross-database search at the National Center for Biotechnology Information (NCBI; National Institutes of Health, Bethesda, MD, USA). The nucleotide sequence of the hrcA-grpE-dnaK-dnaJ operon of S. intermedius type strain NCDO2227 has been submitted to the DDBJ database and given a specialized accession number: AB608790. Sequence similarity in the DnaK chaperone systems of these bacteria was analyzed using the NCBI BLAST Needleman–Wunsch Global Sequence Alignment Tool. Multiple sequence alignment was performed using the CLUSTAL W (1.81) program and the sequence motif search was performed using the PROSITE Pattern and PROSITE Profile databases (Kyoto University Bioinformatics Center, Japan; http://www.bic.kyoto-u.ac.jp/).
Generation of dnaK knockout mutants of S. intermedius
dnaK knockout mutants (ΔdnaK) were produced by homologous recombination. DNA fragments of S. intermedius type strain NCDO2227 genomic DNA were amplified by PCR. The 5′ region of the dnaK DNA fragment (1,009 bp) includes a 315-bp coding region of dnaK that was amplified by using dnaK EcoRI F and internal primer Si dnaK BamHI R (Table 2), and then digested with BamHI. The 3′ region of the latter (1,058 bp) DNA fragment includes a 429-bp coding region of dnaK that was amplified using internal primers, Si dnaK SalI F and Si dnaK SphI R (Table 2), and then digested with SalI. The erythromycin-resistance cassette (erm cassette) was amplified from genomic DNA from an ily knockout mutant UNS38 B3 (Sukeno et al. 2005) using the primers, erm (BamHI) F and erm (SalI) R (Table 2) and the erm cassette ligated to the BamHI-digested 5′ region and SalI-digested 3′ region of the dnaK ORF. This ligated fragment was amplified by PCR with primers, Si dnaK EcoRI and Si dnaK SphI R (Table 2). UNS38 ΔdnaK was produced by transformation of competence-stimulating peptide (CSP: DSRIRMGFDFSKLFGK)-treated strain UNS38 with the PCR amplicon. Colonies were selected and isolated on BHI agar-containing erythromycin at 30°C. Disruption of dnaK was confirmed by immunoblotting using anti-Tetragenococcus halophilus DnaK rabbit antiserum (Sugimoto et al. 2008) and by PCR.
Table 2.
Oligonucleotides used in this study
| Purpose | Name | Sequence (5′–3′) |
|---|---|---|
| Disruption of dnaK | ||
| Si dnaK EcoRI F | ACAGAATTCGAACTAGCTAATGAGCGTG | |
| Si dnaK SphI R | TTGCATGCCGCTCAAGGTTTCATAAGCC | |
| Si dnaK SalI F | ATTGTCGACCAATCTAACAGCGGCCTGAC | |
| Si dnaK BamHI R | CTGGATCCCCAAGATATTCTTCTGCATAG | |
| Erythromycin-resistant cassette | ||
| erm (BamHI) F | AATGGATCCCCCGATAGCTTCCGCTATTG | |
| erm (SalI) R | CAGTAGTCGACCTAATAATTTATCTAC | |
| Construction of a constitutive promoter in S. intermedius | ||
| CP25 oligo1 F | CGCCCGGGCTTTGGCAGTTTATT | |
| CP25 oligo1 R | GTCAAGAATAAACTGCCAAAGCCCGGGCG | |
| CP25 oligo2 F | CTTGACATGTAGTGAGGGGGCTGGT | |
| CP25 oligo2 R | GATTATACCAGCCCCCTCACTACAT | |
| CP25 oligo3 F | ATAATCACATAGTACTGTTGAGCTCGC | |
| CP25 oligo3 R | GCGAGCTCAACAGTACTATGT | |
| RBS-F (SacI) | AACGAGCTCAAAGGAGAACGTTGGATCCAC | |
| CP25 PstI | CGCTGCAGCTTTGGCAGTTTATTCTTGAC | |
| CP25 BamHI | GGGGATCCAACGTTCTCCTTTGAGCTCAAC | |
| Complementation of ΔdnaK mutant, conformation of dnaK disruption and complementation | ||
| Si grpE BamHI F | GGAGGATCCTTGTGGCAAAACATAAACAAGAAGAAC | |
| Si dnaK BamHI F | GAAGGATCCATGTCTAAAATTATCGGTATTGAC | |
| Si dnaK R | CTCTCTAGATTACTTTTCCGTAAACTCTCCGTC | |
| Si dnaJ R | CGATCTAGACCCGGGCGTGCAACACATCATTACAAG | |
| Si 3′ hrcA F | GATATCTGAGTAGTAATCACTATGAAGTCC | |
| Si 5′ dnaK R | GCTGAGTTTGTTGTACCTAAGTCAATACCG | |
| 5′ erm R | GCAAACATATAACCGAGGAACAAAGGTATG | |
| Si 3′ dnaK F | GTGATGATGTCGTAGACGGAGAGTTTACGG | |
| pSETN1 R | CCCAGTCACGACGTTGTAAAACGACGGCC | |
Complementation of S. intermedius UNS38 ΔdnaK strain
For complementation of the UNS38 ΔdnaK mutant, the Streptococcus suis–E. coli shuttle vector pSETN1 (previously reported as pSET1 Δlac p15A) was modified (Tomoyasu et al. 2010a). For constitutive expression of the grpE-dnaK-dnaJ operon in the ΔdnaK mutant, a DNA fragment (CGCTGCAGCT TTGGCAGTTT ATTCTTGACA TGTAGTGAGG GGGCTGGTAT AATCACATAG TACTGTTGAG CTCAAAGGAG AACGTTGGAT CCAC) containing synthetic promoter CP25 (Jensen and Hammer 1998) and Shine–Dalgarno sequence was synthesized by PCR, using synthetic primers (Table 2) and multiple cloning steps (data not shown). This fragment was amplified using primers, CP25 PstI and CP25 BamHI, digested with PstI and BamHI, and cloned into the corresponding sites in pSETN1 and transformed into E. coli DH5αZ1 (Lutz and Bujard 1997). The resultant plasmid (pSETN1 CP25) was used for the construction of the plasmid for complementation of the ΔdnaK mutant. The grpE-dnaK-dnaJ operon was amplified from S. intermedius NCDO2227 genomic DNA by PCR using primers, Si grpE BamHI F and Si dnaJ R (Table 2). The amplified fragment was digested with BamHI and SmaI and cloned into the corresponding sites in pSETN1. The resultant plasmid (pSETN1 EKJ) and pSETN1 CP25 were digested with SphI and BamHI. A 9.24-kbp fragment from pSETN1 EKJ and a 0.92-kbp fragment from pSETN1 CP25, which contains the CP25 promoter, were recovered and then ligated. The ligated plasmid was transformed into a CSP-treated UNS38 ΔdnaK mutant (ΔdnaK R37). Transformants were selected and isolated on a BHI agar plate containing 2 μg/mL chloramphenicol. Complementation of ΔdnaK was confirmed by immunoblotting using anti-T. halophilus DnaK rabbit antiserum, recovery of thermosensitive phenotype, and PCR.
PCR analysis
To confirm disruption of the dnaK gene by the erm cassette in the ΔdnaK mutant and to analyze the status of the plasmid in a complemented strain, chromosomal DNA from the parental (UNS38), ΔdnaK mutant, and complemented strains was applied to PCR using the primers listed in Table 2. The primer sets used to amplify each gene fragment were as follows: Si dnaK BamHI F and Si dnaK R to amplify the dnaK gene; Si 3′ hrcA F and Si 5′ dnaK R to amplify a fragment from the 3′ region of hrcA to the 5′ region of dnaK; Si 3′ hrcA F and 5′ erm R to amplify a fragment from the 3′ region of hrcA to the erm cassette: Si 3′ hrcA F and Si dnaK R to amplify a fragment from the 3′ region of hrcA to the 3′ region of dnaK: Si 3′ dnaK F and pSETN1 R to amplify a fragment from the 3′ region of dnaK to pSETN1 CP25; erm (BamHI) F and pSETN1 R to amplify a fragment from the erm cassette to pSETN1 CP25; Si dnaK BamHI F and pSETN1 R to amplify a fragment from the 5′ coding region of dnaK to pSETN1 CP25; and CP25 PstI and Si dnaK R to amplify a region from the CP25 promoter region to the 3′ region of dnaK.
Hemolysis assay
S. intermedius cells were grown in BHI broth at 37°C for 24 h under anaerobic conditions. The culture supernatant was separated by centrifugation (5,000×g). Hemolysis was assayed as described by Nagamune et al. (1996), with minor modifications. Human blood was obtained from healthy Japanese volunteers and stored in sterilized Alsever solution at 4°C. Erythrocytes were washed three times with PBS at 4°C by centrifugation (1,000×g) before use. Chilled PBS containing 5 × 107 erythrocytes per milliliter and standardized amounts of the culture supernatants (optical density at 600 nm; OD600 = 1.0), which was diluted 200- to 6,400-fold with PBS, were mixed in microcentrifuge tubes (total of 0.5 mL). Hemolysis was performed at 37°C for 1 h. After the reaction, non-lysed erythrocytes were removed by centrifugation (1,000×g) at 4°C for 5 min. Absorbance at 540 nm (A540) of 200 μL of supernatant was measured in a Microplate Reader Model 550 (Bio-Rad, Hercules, CA, USA). The percent hemolysis was calculated as follows: hemolysis (%) = [(A540 of supernatant from the sample containing diluted culture supernatant − A540 of supernatant from the sample containing no diluted culture supernatant)/(A540 of supernatant from the sample completely hemolyzed by hypotonic processing − A540 of supernatant from the sample containing no diluted culture supernatant)] × 100.
Construction of isopropyl β-d-1-thiogalactopyranoside-regulated S. intermedius dnaK, grpE-dnaK, dnaK-dnaJ, grpE-dnaK-dnaJ, or E. coli dnaK expression plasmids
To construct S. intermedius DnaK-, GrpE-DnaK-, DnaK-DnaJ-, or GrpE-DnaK-DnaJ producing plasmids (pZE13 SiK, pZE13 SiEK, pZE13 SiKJ, pZE13 SiEKJ), S. intermedius NCDO2227 genomic DNA was used for PCR amplification of (1) the dnaK gene was amplified using primers, Si dnaK BamHI F and Si dnaK R (Table 2); (2) the grpE-dnaK operon amplified using primers, Si grpE BamHI F and Si dnaK R (Table 2); (3) the dnaK-dnaJ operon was amplified using primers, Si dnaK BamHI F and Si dnaJ R (Table 2) and (4) the grpE-dnaK-dnaJ operon amplified using primers, Si grpE BamHI F and Si dnaJ R (Table 2). Each fragment was digested with BamHI and XbaI, and then cloned into the corresponding sites in pZE13 (Lutz and Bujard 1997). E. coli DnaK-producing plasmid (pZE13 EcK) was also created as follows: E. coli dnaK gene was excised by BamHI and HindIII digestion in pBB535, and a 2.17-kbp fragment containing intact E. coli dnaK was cloned into the corresponding sites in pZE13. Then, E. coli ΔdnaK strain BB1554 lacIq (Table 1) was transformed by the plasmids constructed above to analyze the complementation of this mutant.
In vivo chaperone activity of S. intermedius DnaK chaperone system in E. coli ΔrpoH mutant
The chaperone activity of S. intermedius DnaK chaperone system was determined by measuring and comparing the amount of aggregated proteins in both the ΔrpoH mutant (CS5262) and the pZE13 SiEKJ transformed CS5262 (Table 1). Cells were grown in 10 mL of LB medium containing the indicated concentrations of β-d-1-thiogalactopyranoside (IPTG) for 4 h at 30°C and then shifted to 42°C for 1 h. Isolation of total and aggregated proteins was performed as previously described with minor modifications (Tomoyasu et al. 2001, 2010b). After heat treatment, bacterial cultures were rapidly cooled on ice and centrifuged for 10 min at 5,000×g at 4°C to harvest cells. Pellets were resuspended in 80 μL of buffer A (10 mM potassium phosphate buffer [pH 6.5] containing 1 mM EDTA, 20% [w/v] sucrose, 1 mg/mL lysozyme) and incubated for 30 min on ice. Spheroplasts were destroyed by the addition of 720 μL of buffer B (10 mM potassium phosphate buffer [pH 6.5] containing 1 mM EDTA) and sonication with microtip (level 3, 50% duty ratio, for 10 s) in an Astrason Ultrasonic Processor (model XL2020; MISONIX Inc., Farmingdale, NY, USA) while cooling on ice. The insoluble cell fraction from the total proteins was isolated by centrifugation at 17,000×g for 5 min at 4°C. The pellet fractions were frozen, resuspended in 800 μL of buffer B by sonication, and centrifuged (17,000×g, 5 min, 4°C). The washed pellet fractions were again resuspended in 640 μL of buffer B by brief sonication; then, 160 μL of 10% (v/v) Nonidet-P40 (NP40) was added, and the aggregated proteins were isolated by centrifugation (17,000×g, 5 min, 4°C). This washing procedure was repeated for the complete removal of contaminating membrane proteins. NP40-insoluble pellets were washed with 800 μL of buffer B and resuspended in 200 μL of buffer B by brief sonication. Quantification of the amount of total and aggregated proteins was performed using the Bradford assay reagent (Bio-Rad, Hercules, CA, USA) with bovine serum albumin as a standard.
Infection assay
S. intermedius cells were grown in BHI broth at 37°C for 48 h under anaerobic conditions. The infection assay was performed as previously described with minor modifications (Sukeno et al. 2005). HepG2 (1 × 105 cells per well) in 350 μL of DMEM containing 10% fetal bovine serum (FBS) without antibiotics was dispensed into 48-multiwell tissue culture plates and cultured overnight at 37°C in the presence of 5% CO2. For cell infection, bacterial cultures were centrifuged at 13,000×g for 1 min, and cells were suspended to a density of 3 × 105 cells in 350 μL of DMEM containing 10% FBS without antibiotics. The bacterial suspension was added to HepG2 cells, and infection was allowed to proceed for 3 h in the 48-multiwell tissue culture plates. The supernatant was completely removed, and cells were washed three times with PBS. Infected cells were then cultured in 350 μL of fresh medium. A portion of the culture medium (200 μL) was replaced with fresh medium every 12 h to avoid accumulation of ILY. The viability of infected cells was determined using the neutral red (NR) method (Borenfreund and Puerner 1985). After infection, the medium was removed at the indicated time point, and the cells were incubated with 350 μL of NR solution (50 μg/mL) in DMEM for 3 h at 37°C. The cells were subsequently washed three times with PBS and then fixed with 200 μL of formaldehyde (1.0%, v/v) containing 1 mM HEPES-KOH (pH 7.3), 0.85% NaCl, and 1.0% CaCl2. To extract the dye, viable cells were lysed with 1% acetic acid in 50% (v/v) ethanol. The absorbance was then measured at 540 nm (A540). The 0% viability control consisted of cells exposed to 1 M HCl and the 100% viability control consisted of cells incubated in DMEM without bacteria. The level of cytotoxicity was calculated as follows: Viability (%) = (A540 of extract from the infected cells − A540 of extract from the 0% control)/(A540 of extract from the 100% control − A540 of extract from the 0% control) × 100.
Spot tests for thermosensitivity and acid tolerance
The thermosensitivity and acid tolerance of S. intermedius cells were determined as follows: Cells were grown in BHI medium for 48 h at 30°C under anaerobic conditions, and the OD600 of the cultures was measured. Standardized cultures (OD600 adjusted to 1.0) were diluted from 10−1 to 10−5 in BHI medium. To analyze thermosensitivity, 5 μL aliquots were spotted onto BHI agar plates and incubated at 37°C or 42°C for 48 h. For acid tolerance, testing aliquots of the standardized (OD600 = 1.0) cultures were diluted to 10−2 in pH-conditioned glycine-buffered BHI medium (pH 3.5 or pH 7.0) and cultured for 1 h at room temperature. Aliquots of each incubated culture were further diluted from 10−3 to 10−5 in BHI medium and 5 μL spotted onto BHI agar plates. Plates were incubated at 30°C for 72 h.
To analyze thermosensitivity, E. coli cells were grown in LB medium overnight at 30°C. Two microliters of aliquots were spotted onto LB agar plates containing from 0 to 100 μM IPTG, and plates were incubated at 30°C or 42°C for 24 h. To analyze acid tolerance, E. coli cells were grown in LB medium for 3 h at 30°C in the presence of 50 μM IPTG, and the OD600 of the cultures was measured. Standardized amounts (OD600 = 1.0) of cultures were diluted to 10−2 in pH-conditioned glycine-buffered LB medium (pH 3.5 or pH 7.0) and then cultured for 1 h at room temperature. Aliquots of each culture were further diluted from 10−3 to 10−5 in LB medium. Five microliters of aliquots were spotted onto LB agar plates containing 50 μM IPTG and incubated at 30°C for 48 h.
Gel electrophoresis and immunoblotting
S. intermedius cells were grown in BHI broth at 37°C under anaerobic conditions. The culture supernatant and cells were separated by centrifugation (5,000×g). The cells were washed three times with PBS and resuspended in 1 or 0.5 mL of 20 mM Tris–HCl (pH 8.0) containing 100 mM NaCl and 1 mM EDTA. Samples were then added to Lysing Matrix B tubes (Qbiogene Inc., Carlsbad, CA, USA) and lysed in a FastPrep cell disruptor (Savant Instruments, Holbrook, NY, USA). To obtain the soluble protein fraction, samples were centrifuged at 17,400×g for 30 min, and the supernatants were retained. The total protein (10 μg) of the supernatants or standardized amounts of the culture supernatants were analyzed by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Laemmli 1970). E. coli cells were grown in 10 mL of LB medium containing the concentrations of IPTG indicated for 4 h at 30°C. The OD600 of the cultures was measured, and the cells were harvested by centrifugation (17,400×g). The cell pellets were lysed by adding sample buffer (Laemmli 1970), and then standardized amounts (10 μL) of the cell lysate at OD600 = 5.0 were analyzed by 12% SDS–PAGE. For immunoblotting analysis, the resolved proteins were transferred to a poly(vinylidene difluoride) membrane (Millipore, Bedford, MA, USA), incubated with anti-T. halophilus, anti-E. coli GroEL (Kusukawa et al. 1989), or anti-σ32 (Gamer et al. 1992) rabbit antiserum, a 2nd then developed with a 5-bromo-4-chloro-3′-indolyl phosphate/nitro-blue tetrazolium chloride, using alkaline phosphatase-conjugated anti-rabbit immunoglobulin G as the secondary antibody.
Results
Comparison of the amino acid sequence of S. intermedius DnaK chaperone system with those of other DnaK chaperone systems in Gram-positive and Gram-negative bacteria
The genetic organization of the DnaK chaperone system of S. intermedius was similar to that found in other Gram-positive bacteria with genes of this system localized at the hrcA-grpE-dnaK-dnaJ operon. A highly homologous region with the CIRCE operator sequence, which is the binding site for the HrcA heat shock repressor, was detected in the hrcA promoter region (data not shown). We compared the sequence homology of each component of the S. intermedius DnaK chaperone system to the corresponding component of the DnaK chaperone systems of S. pneumoniae, L. lactis, B. subtilis, and E. coli (Table 3). S. intermedius DnaK sequence was well conserved among the Gram-positive bacteria, and >70% amino acid homology was observed. However, S. intermedius DnaK showed weak homology and 54% identity to the DnaK of the Gram-negative bacterium E. coli. S. intermedius GrpE showed less amino acid homology (26%) to E. coli GrpE. We surmised that since a nucleotide exchange factor GrpE for DnaK does not have chaperone activity, it might allow more amino acid substitutions than DnaK and DnaJ. We therefore further characterized a sequence feature of S. intermedius DnaK, DnaJ and GroE by multiple sequence search analysis of these proteins among B. subtilis and E. coli (Fig. 1). This analysis indicated that S. intermedius DnaK has features typical of DnaK of a Gram-positive bacterium, i.e. lacking segments 75–98 and 211–214 in the ATPase domain that are present in almost all proteobacteria (Fig. 1a). The J domain in S. intermedius DnaJ was well conserved (Fig. 1b). However, the glycine- and phenylalanine-rich region (G/F region) was variable among these bacteria. Although S. intermedius GrpE showed only weak amino acid homology to E. coli GrpE, the C-terminal region corresponding to the GrpE protein signature motif was significantly conserved (Fig. 1c)
Table 3.
Sequence identity between the S. intermedius DnaK chaperone system and those of other Gram-positive and Gram-negative bacteria
| GrpE (%) | DnaK (%) | DnaJ (%) | |
|---|---|---|---|
| S. pneumoniae | 79 | 96 | 86 |
| L. lactis | 66 | 85 | 71 |
| B. subtilis | 38 | 75 | 56 |
| E. coli | 26 | 54 | 48 |
Sequence identity was examined by the Needleman-Wunsch alignment.
Fig. 1.
Multiple sequence alignment of DnaK, DnaJ, and GrpE. a Part of the ATPase domains of DnaK from S. intermedius (SI DnaK), B. subtilis (Bs DnaK), and E. coli (Ec DnaK) were aligned with the CLUSTAL W program. b The N-terminal regions of DnaJ including the J domain (underlined) and G/F region (overlined) from S. intermedius (SI DnaJ), B. subtilis (Bs DnaJ), and E. coli (Ec DnaJ) were aligned. c The C-terminal regions of GrpE from S. intermedius (SI GrpE), B. subtilis (Bs GrpE), and E. coli (Ec GrpE) were aligned. The GrpE protein signature motif is underlined. Identical amino acid residues to S. intermedius DnaK, DnaJ, or GrpE are shown in bold
Construction of S. intermedius dnaK null mutants and its complemented strain
To analyze the in vivo function of the DnaK chaperone system in S. intermedius, we disrupted the dnaK gene by insertion of the erm cassette between the dnaK coding sequences. In addition, a dnaK-complemented strain was also constructed by transformation of a ΔdnaK mutant (ΔdnaK R37) with the expression vector pSETN1 CP25 carrying the grpE-dnaK-dnaJ operon to exclude the possibility of dnaK null phenotypes resulting from other mutations in the chromosome. We obtained chloramphenicol (selective marker for pSETN1 CP25)-resistant colonies, although we did not recover the plasmid from the independent colonies. Therefore, the status of the plasmid in a chloramphenicol-resistant strain was analyzed by PCR (Supplementary Fig. 1A and B). When the primers for amplifying the dnaK gene were used, we detected two fragments from the chloramphenicol-resistant strain. The shorter fragment (1.9 kbp) corresponded to the dnaK gene and the longer fragment (2.9 kbp) corresponded to the erm cassette-inserted dnaK gene. These data confirmed that the chloramphenicol-resistant strain obtained actually complemented the dnaK gene disrupted with the erm cassette through the presence of intact dnaK. Subsequently, we analyzed the status of the hrcA-grpE-dnaK-dnaJ operon in the complemented strain. PCR using primers to amplify from the 3′ region of hrcA to the 5′ region of dnaK produced a 0.9-kbp fragment from all strains examined. PCR using primers to amplify from the 3′ region of hrcA to the erm cassette produced a 1.3-kbp fragment from both ΔdnaK R37 and its complemented strain. The primers from the 3′ region of hrcA to the 3′ region of dnaK produced a 2.7-kbp fragment from UNS38 or a 3.7-kbp fragment from both ΔdnaK R37 and the complemented strain. These results suggest that dnaK in the hrcA-grpE-dnaK-dnaJ operon was disrupted by the erm cassette in the complemented strain. We further analyzed the plasmid localization in the S. intermedius chromosome. PCR amplification using primers to amplify from the 3′ region of dnaK, erm cassette, or 5′ coding region of dnaK to pSETN1 CP25 produced 1.7-, 4.3-, or 4.6-kbp fragments respectively only from the complemented strain. These results suggest that pSETN1 CP25 was integrated downstream of the hrcA-grpE-ΔdnaK-dnaJ operon (Supplementary Fig. 1A). In addition, we tried to amplify the fragment between the CP25 promoter and the 3′ region of dnaK and obtained 2.7-kbp fragments only from the complemented strain indicating that grpE and intact dnaK were localized downstream of the CP25 promoter. Overall, our results suggest that the plasmid was recombined with the hrcA-grpE-ΔdnaK-dnaJ operon of ΔdnaK R37 (Supplementary Fig. 1A). Thus far, we have obtained only the plasmid-integrated strain. Since a S. suis-E. coli shuttle vector, pSET1, was not stable in S. intermedius (data not shown), the integrated strain might be preferentially selected.
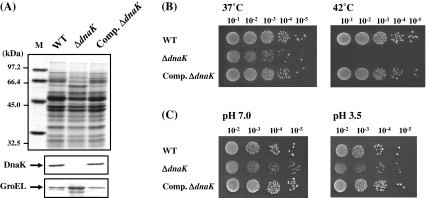
Because the homology between S. intermedius DnaK and E. coli DnaK (Table 3) was weak, we could not detect S. intermedius DnaK by immunoblotting analysis using anti-E. coli DnaK antiserum (data not shown). On the other hand, T. halophilus DnaK (Sugimoto et al. 2008) has high homology with S. intermedius DnaK with 77% amino acid identity. Therefore, immunoblotting analysis was carried out using anti-T. halophilus DnaK antiserum to confirm the amount of DnaK in ΔdnaK R37 and its complemented strain (Fig. 2a). The disappearance of the band corresponding to DnaK was confirmed in the cell extract from ΔdnaK R37, while on the other hand, obvious recovery of DnaK was observed in the complemented strain. In addition, accumulation of a 60-kDa protein was observed in ΔdnaK R37, which cross-reacted with anti-E. coli GroEL antibody. The complemented strain showed a significant reduction of the accumulated protein to the level of the parental strain.
Fig. 2.
Immunoblotting analysis and stress-sensitivity of S. intermedius ΔdnaK mutant. a Immunoblotting analysis of ΔdnaK R37 and its complemented strain. Whole-cell extracts (10 μg) were separated by 12% SDS–PAGE. Immunodetection was carried out with anti-T. halophilus DnaK antiserum or anti-GroEL antiserum. b Spot test for the examination of thermosensitivity. Spotted plates were incubated for 48 h at the indicated temperature. c Spot test for the examination of acid tolerance. Spotted plates were incubated for 48 h at 30°C. M molecular weight marker; WT UNS38; ΔdnaK, ΔdnaK R37; Comp. ΔdnaK the complemented strain of ΔdnaK
Thermosensitivity and acid sensitivity of S. intermedius dnaK null mutants
As observed for other Gram-negative and Gram-positive dnaK null mutants, ΔdnaK R37 showed a thermosensitive phenotype and did not form colonies at 42°C, while UNS38 and the complemented strain formed colonies at both 37°C and 42°C (Fig. 2b). We have isolated four independent dnaK null mutants from UNS38 thus far. Three dnaK null mutants (ΔdnaK S37 1–3) did not grow at 37°C (data not shown), although one mutant (ΔdnaK R37) grew at this temperature. To investigate the cytotoxicity of S. intermedius to human-derived culture cells, the infection assay was carried out at 37°C. Therefore, since the ΔdnaK mutant, which can grow at 37°C, was necessary for the assay, ΔdnaK R37 was used for these experiments. E. coli ∆dnaK mutant is known to have an acid-sensitive phenotype. Then, the acid sensitivity of ΔdnaK R37 was examined (Fig. 2c). The viability of ΔdnaK R37 was the same as UNS38 and the complemented strain under acidic conditions. Unlike E. coli ∆dnaK mutant, S. intermedius ΔdnaK R37 could not show significant acid sensitivity.
Effect of dnaK null mutation on growth and secretion of pathogenic factors
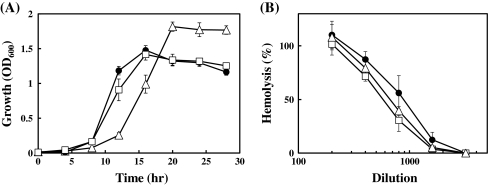
The growth rate of UNS38, ΔdnaK R37, and the complemented strain in BHI medium at 37°C were examined (Fig. 3a). The growth curves of UNS38 and the complemented strain were similar (i.e., both cells enter the stationary phase within 16 h). On the other hand, ΔdnaK R37 exhibited a significantly slower growth rate and entered the stationary phase within 20 h. Although OD600 of ΔdnaK R37 at the stationary phase was slightly higher than UNS38 and the complemented strain.
Fig. 3.
Effect of dnaK null mutation on cell growth and secretion of ILY. a Growth curves of UNS38, ΔdnaK R37, and its complemented strain. Strains were cultured in BHI medium and the OD600 measured at the indicated time points. The graphical data are the mean values ± standard deviation of at least four replicated independent experiments. b Hemolytic activity in the culture supernatant. Strains were cultured in BHI medium 24 h at 37°C, and the culture supernatants collected. Culture supernatant standardized at OD600 was diluted from 200- to 3,200-fold by 2-fold serial dilutions, and the cytolytic activity of ILY in the diluted culture supernatant was estimated by hemolysis assay. Solid circle UNS38; open triangle ΔdnaK R37; open square the complemented strain
We determined the amount of ILY secreted in the culture supernatant at 37°C from UNS38, ΔdnaK R37, and the complemented strain by the hemolysis assay (Fig. 3b). The results showed no large difference in hemolytic activity due to ILY between these strains. The level of ILY in the culture supernatant was estimated by immunoblotting using anti-ILY antibody and showed that ΔdnaK R37 could secrete ILY at levels similar to UNS38 and the complemented strain (data not shown). We further examined the amount of the secreted hyaluronidase in the culture supernatant at 37°C from UNS38, ΔdnaK R37, and the complemented strain by the substrate-agarose plate assay using hyaluronan as substrate. No significant difference in hyaluronidase activity was observed among these strains (data not shown). In addition, although we also examined the hemolytic and hyaluronidase activities in the culture supernatant at 30°C from ΔdnaK S37 1–3 and ΔdnaK R37, they showed the same activities as UNS38 (data not shown).
Cytotoxicity of S. intermedius dnaK null mutant on HepG2 cells
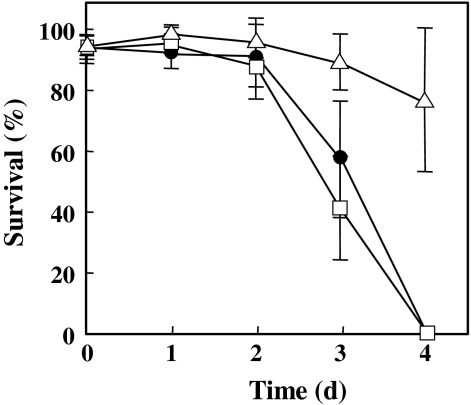
Several studies on pathogenic bacteria have reported that mutation of dnaK causes attenuation of cytotoxicity and pathogenicity. Therefore, we examined the cytotoxicity of ΔdnaK R37 and the complemented strain using HepG2 human cells (Fig. 4). The viability of HepG2 was significantly reduced after 3 days of infection with UNS38 or the complemented strain, and all HepG2 cells were killed at 4 days post-infection. However, the ΔdnaK mutant showed only slight cytotoxicity to HepG2 compared to UNS38 or the complemented strain, with HepG2 cells surviving even after 4 days post-infection. These data show that disruption of the dnaK gene of S. intermedius causes attenuation of cytotoxicity despite ILY secretion being unaffected by this mutation.
Fig. 4.
Cytotoxic effect on HepG2 cells of ΔdnaK R37 and its complemented strain. Cytotoxic effects were observed over 4 days post-bacterial infection. Solid circle UNS38; open triangle ΔdnaK R37; open square the complemented strain
Complementation of E. coli ∆dnaK by S. intermedius DnaK chaperone system
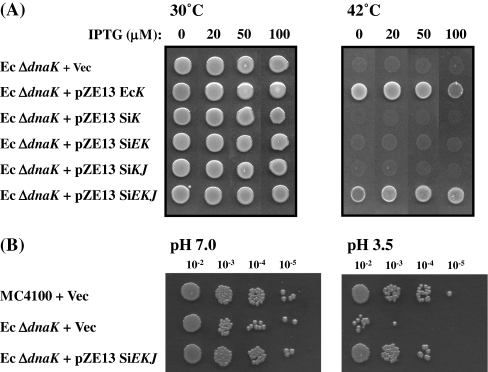
To examine the functions and characteristics of the S. intermedius DnaK chaperone system, S. intermedius dnaK (SiK), grpE-dnaK (SiEK), dnaK-dnaJ (SiKJ), or grpE-dnaK-dnaJ (SiEKJ) was cloned on the E. coli IPTG-inducible vector pZE13 and these plasmids used to transform E. coli ∆dnaK mutant (Ec ∆dnaK). E. coli dnaK (EcK) was also cloned on the same vector as a positive control. The thermosensitivity of these transformants was examined using the spot test (Fig. 5a). These strains were spotted onto LB agar plate containing the concentrations of IPTG indicated and then cultured at 30°C or 42°C. The results showed that all strains grew at 30°C in the presence or absence of IPTG. However, only the pZE13 EcK- and pZE13 SiEKJ-transformed Ec ∆dnaK grew at 42°C. These strains grew well in the presence of 20 or 50 μM IPTG, although a slight growth defect was observed in the absence of IPTG (pZE13 SiEKJ-transformant) or in the presence 100 μM IPTG (pZE13 EcK-transformant). In contrast, pZE13 SiK-, pZE13 SiEK-, or pZE13 SiKJ-transformed Ec ∆dnaK did not grow at 42°C indicating that S. intermedius DnaJ and GrpE were not compatible with E. coli DnaJ and GrpE. However, the intact DnaK chaperone system of S. intermedius could complement the thermosensitivity in E. coli.
Fig. 5.
Complementation of E. coli ΔdnaK by S. intermedius DnaK chaperone system. a Spot test for determining thermosensitivity. Cells were cultured for 24 h at 30°C and then spotted on LB agar plates containing the indicated amounts of IPTG. Spotted plates were incubated 24 h at the indicated temperatures. b Spot test for the detection of acid tolerance. Spotted plates were incubated for 48 h at 30°C. MC4100 + Vec MC4100 transformed with plasmid pZE13; Ec ΔdnaK + Vec E. coli ΔdnaK mutant transformed with plasmid pZE13; Ec ΔdnaK + pZE13 EcK E. coli ΔdnaK mutant transformed with plasmid pZE13 EcK; Ec ΔdnaK + pZE13 SiK E. coli ΔdnaK mutant transformed with plasmid pZE13 SiK; Ec ΔdnaK + pZE13 SiEK E. coli ΔdnaK mutant transformed with plasmid pZE13 SiEK; Ec ΔdnaK + pZE13 SiKJ E. coli ΔdnaK mutant transformed with plasmid pZE13 SiKJ; Ec ΔdnaK + pZE13 SiEKJ E. coli ΔdnaK mutant transformed with plasmid pZE13 SiEKJ
E. coli ∆dnaK mutant is known to be acid sensitive, and we have also confirmed this phenotype. The E. coli ∆dnaK mutant showed lower viability than the wild strain (MC4100) when exposed to low pH conditions (pH 3.5) for 1 h. The S. intermedius DnaK chaperone system could also complement acid sensitivity in the E. coli ∆dnaK mutant, and pZE13 SiEKJ-transformed Ec ∆dnaK showed nearly equivalent acid tolerance compared to the wild strain (Fig. 5b).
Function of S. intermedius DnaK chaperone system in E. coli
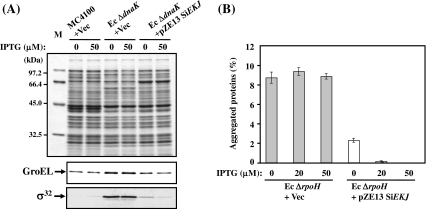
Several studies have reported that negative regulation of the hrcA-grpE-dnaK-dnaJ and groES-groEL operons of many Gram-positive bacteria including streptococci are carried out by the HrcA repressor (Lemos et al. 2001; Woodbury and Haldenwang 2003; Kim et al. 2008), and this activity seemed to be controlled by the DnaK chaperone system and GroESL (Mogk et al. 1997; Koch et al. 1998). On the other hand, heat shock response in many Gram-negative bacteria, including E. coli whose heat shock response is known to be regulated by the heat shock transcriptional factor σ32, differs from that of streptococci. Because the activity and stability of σ32 is controlled by the DnaK chaperone system, constitutive induction of heat shock response and accumulation of σ32 were observed in Ec ∆dnaK (Fig. 6a). Although the control mechanism of heat shock response differs between S. intermedius and E. coli, we examined whether S. intermedius DnaK chaperone system could regulate the E. coli heat shock response by controlling the stability and activity of σ32. Our data showed that S. intermedius DnaK chaperone system could reduce the accumulation of σ32 and GroEL in Ec ∆dnaK up to the level of the wild-type.
Fig. 6.
Function of S. intermedius DnaK chaperone system in E. coli. a Cellular level of GroEL and σ32 in E. coli ΔdnaK mutants in the presence or absence of the S. intermedius DnaK chaperone system. Cells were grown in LB medium containing the indicated concentrations of IPTG for 4 h at 30°C, the OD600 of the cultures measured and standardized amounts of the cell lysates analyzed by 12% SDS–PAGE. Immunodetection was carried out with anti-E. coli GroEL or anti-σ32 antiserum. M molecular weight marker; MC4100 + Vec MC4100 transformed with a plasmid pZE13; Ec ΔdnaK + Vec E. coli ΔdnaK mutant transformed with a plasmid pZE13; Ec ΔdnaK + pZE13 SiEKJ E. coli ΔdnaK mutant transformed with a plasmid pZE13 SiEKJ. b Amounts of aggregated protein in E. coli ΔrpoH mutants in the presence or absence of the S. intermedius DnaK chaperone system. Cells were grown in LB medium containing the indicated concentrations of IPTG for 3 h at 30°C and further cultured at 42°C for 1 h. Aggregated proteins were isolated as described in “Materials and methods.” The amount of aggregated protein was quantified by the Bradford assay reagent and calculated in relation to total protein content (set at 100%). Each value is the average from at least three different experiments. Ec ΔrpoH + Vec E. coli ΔrpoH mutant transformed with a plasmid pZE13; Ec ΔrpoH + pZE13 SiEKJ E. coli ΔrpoH mutant transformed with a plasmid pZE13 SiEKJ
Since the σ32-encoding gene is rpoH, E. coli ∆rpoH mutant (Ec ∆rpoH) is largely devoid of all the major cytosolic chaperones, except for GroEL/GroES, and has lower levels of proteases (Tomoyasu et al. 2001). The lack of chaperones and proteases results in a large accumulation of aggregated proteins by heat treatment, and approximately 9% of the total proteins were aggregated (Fig. 6b). Therefore, Ec ∆rpoH could be used to estimate the in vivo chaperone activity of S. intermedius DnaK chaperone system. When S. intermedius DnaK chaperone system was expressed in Ec ∆rpoH by adding 20 or 50 μM IPTG (Fig. 6b), aggregated proteins dramatically decreased to less than 1% of the total proteins at 42°C. Thus, it was concluded that S. intermedius DnaK chaperone system could efficiently eliminate the aggregated proteins from ∆rpoH mutant cells.
Discussion
DnaK is a major cytosolic chaperone and has an important function in quality control of cellular protein in bacteria, in cooperation with DnaJ and GrpE (Mayer et al. 2000; Ben-Zvi and Goloubinoff 2001; Genevaux et al. 2007). Therefore, the construction and isolation of a dnaK null mutant is believed to be difficult and only a few knockout null mutants have been reported thus far. However, it has been reported in studies on dnaK knockout mutants that the requirement and importance for quality control of the cellular protein DnaK differs among bacterial species. For example, the dnaK null mutants of Gram-negative E. coli and S. Typhimurium showed pleiotropic phenotypes, such as thermosensitivity, cell elongation, and constitutive heat shock induction (Bukau and Walker 1989a, b, 1990; Takaya et al. 2004). In contrast, Gram-positive B. subtilis DnaK mutant showed no apparent phenotype and indicated that DnaK was not essential for normal growth (Schulz et al. 1995). Attempts to isolate a dnaK null mutant of Streptococcus mutans have been unsuccessful thus far and only a dnaK downregulated strain has been reported (Lemos et al. 2007). Therefore, we tried to create dnaK null mutants of S. intermedius to examine the cellular function of the DnaK chaperone system in detail. Since S. intermedius dnaK null mutants showed a thermosensitive phenotype, as observed in the dnaK mutants of other Gram-positive cocci (Koch et al. 1998; Lemos et al. 2007; Singh et al. 2007), the DnaK chaperone system seemed to have important function in the maintenance of cellular proteins at high temperatures among these bacteria.
Moreover, since constitutive heat shock induction and accumulation of heat shock proteins such as GroEL in Gram-positive bacteria were reported in L. lactis dnaK null mutants and S. mutans dnaK downregulated strain, the DnaK chaperone system seemed to be required to maintain the DNA-binding activity of HrcA in these strains (Koch et al. 1998; Lemos et al. 2007). Accumulation of GroEL was also observed in a S. intermedius dnaK null mutant, ΔdnaK R37 (Fig. 2a). The DnaK chaperone system of this bacterium might require the activity of HrcA. However, this does not exclude the possibility that the accumulated misfolded proteins caused by dnaK null mutation could promote depletion of GroEL, which is also known to be required to maintain HrcA activity. On the basis that (1) ΔdnaK R37 did not show obvious acid sensitivity, in contrast to E. coli dnaK mutant and S. Typhimurium dnaK null mutant (Figs. 2c, 5b, and our unpublished result), and (2) a S. mutans dnaK downregulated strain exhibited a slightly higher acid tolerance than the parental strain (Lemos et al. 2007), the activity control or folding of protein(s) which is involved in acid tolerance by the DnaK chaperone system in streptococci might be less important than in E. coli dnaK and S. Typhimurium.
It has been reported that not only the stress-inducible chaperone (DnaK) but also the stress-inducible proteases (ClpXP, Lon) can regulate the pathogenicity of several Gram-negative pathogenic bacteria including S. Typhimurium, enterohemorrhagic E. coli, and Yersinia pestis (Yamamoto et al. 2001; Takaya et al. 2002; Jackson et al. 2004; Tomoyasu et al. 2005). Therefore, these bacteria are believed to control the expression of virulence factors and possess the ability to sense stresses such as those presented by an effective immune system and accompanying fever in the host organism. Therefore, we investigated whether such the stress-inducible proteins participated in the expression control of virulence factors of S. intermedius as well as in the Gram-negative bacteria. ILY is a major virulence factor of S. intermedius, which is essential for the invasion and cytotoxicity to human cells (Nagamune et al. 2000; Sukeno et al. 2005). Compared to the wild strain, S. intermedius dnaK null mutation did not cause reduction in hemolytic (ILY) activity and hyaluronidase activity in the culture medium; the attenuation of cytotoxicity in this mutant might be caused indirectly by slightly reduced viability and the slower growth phenotype at 37°C (Figs. 2b and 3a). We also examined the contribution of stress-inducible ClpP peptidase, which is a catalytic subunit of ClpXP protease, to the expression of virulence factors and cytotoxicity of S. intermedius. For this purpose, a clpP null mutant was constructed, and its phenotype was analyzed. Our result also showed that clpP null mutation did not cause reduction of hemolytic and hyaluronidase activities and cytotoxicity to HepG2 cells (unpublished result).
It has been reported that many streptococcal virulence factors are controlled by the carbon catabolite repression (CCR) sensing the extracellular amount of utilizable carbohydrates (Abranches et al. 2008; Shelburne et al. 2008a, b; Kietzman and Caparon 2010). One of the important factors for CCR is catabolite control protein A (CcpA), which can control the expression not only of carbon catabolite genes but also of streptococcal virulence factors. Recently, we have shown that ily expression is also controlled by CcpA (Tomoyasu et al. 2010b). Thus, the expression of virulence factors and cytotoxicity of S. intermedius seemed to be mainly regulated by the amount of extracellular utilizable carbohydrates rather than extracellular stress conditions.
Previous studies have demonstrated that DnaK of Gram-positive bacteria was unable to complement the viability of E. coli dnaK null mutants (Sussman and Setlow 1987; Tilly et al. 1993; Minder et al. 1997; Mogk et al. 1999; Sugimoto et al. 2008). Multiple sequence alignment by CLUSTAL W shows that DnaK of S. intermedius is typical of Gram-positive bacteria, lacking segment 75–98 in the ATPase domain that is conserved in almost all proteobacteria (Fig. 1a). This segment seems to play a crucial role in the cooperative function with E. coli DnaJ and GrpE co-chaperones as shown in the analysis using a segment deletion mutant of E. coli DnaK (Sugimoto et al. 2007). These data also support our results that the expression of S. intermedius DnaK could not be functionally activated by DnaJ and GrpE in the E. coli ΔdnaK mutant (Fig. 5a). Interestingly, the intact S. intermedius DnaK chaperone system was active in E. coli, was able to complement the thermosensitive phenotype of E. coli ΔdnaK mutant, and could eliminate aggregated proteins in the ΔrpoH mutant (Figs. 5a and 6b). Although the heat shock response of many Gram-positive bacteria including streptococci is regulated by the HrcA repressor, the S. intermedius DnaK chaperone system could also regulate the stability and activity of σ32 (Fig. 6a). It has been reported that binding with E. coli DnaK and DnaJ induces conformational changes of σ32, and consequently, in vivo, could modulate stability and activity (Rodriguez et al. 2008). Similarly, S. intermedius DnaK and DnaJ might bind to σ32 and induce their conformational changes.
Overall, our results show that the DnaK chaperone system of the Gram-positive bacterium, S. intermedius, has the same function as the system in E. coli and plays a fundamental role in vital functions such as growth, thermoresistance, and heat shock regulation, but not in the modulation of expression of pathogenic factors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PPT 2371 kb)
Acknowledgements
The authors would like to thank Mr. S. Ishida, Mr. K. Nagato, and Ms. N. Yamamoto for technical assistance. This work was partially supported by KAKENHI (Grant-in-Aid for Scientific Research (C) 19590449, 23590510) from the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government.
Reference
- Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsène F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi AP, Goloubinoff P. Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J Struct Biol. 2001;135:84–93. doi: 10.1006/jsbi.2001.4352. [DOI] [PubMed] [Google Scholar]
- Borenfreund E, Puerner JA. Toxicity determined in vitro by morphological alterations and neutral red absorption. Toxicol Lett. 1985;24:119–124. doi: 10.1016/0378-4274(85)90046-3. [DOI] [PubMed] [Google Scholar]
- Bukau B, Walker GC. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Walker GC. ΔdnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989;171:6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Walker GC. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge JE, Attorri S, Musher DM, Hebert J, Dunbar S. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus ("Streptococcus milleri group") are of different clinical importance and are not equally associated with abscess. Clin Infect Dis. 2001;15:1511–1515. doi: 10.1086/320163. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Bukau B. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol Life Sci. 2002;59:1607–1616. doi: 10.1007/PL00012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-M. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Hanawa T, Fukuda M, Kawakami H, Hirano H, Kamiya S, Yamamoto T. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones. 1999;4:118–128. [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Allan E, Coates AR. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun. 2006;74:3693–3706. doi: 10.1128/IAI.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MW, Silva-Herzog E, Plano GV. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol Microbiol. 2004;54:1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x. [DOI] [PubMed] [Google Scholar]
- Jacobs JA, Pietersen HG, Stobberingh EE, Soeters PB. Streptococcus anginosus, Streptococcus constellatus and Streptococcus intermedius. Clinical relevance, hemolytic and serologic characteristics. Am J Clin Pathol. 1995;104:547–553. doi: 10.1093/ajcp/104.5.547. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng JS, Hsueh PR, Teng LJ, Lee LN, Yang PC, Luh KT. Empyema thoracis and lung abscess caused by viridans streptococci. Am J Respir Crit Care Med. 1997;156:1508–1514. doi: 10.1164/ajrccm.156.5.97-03006. [DOI] [PubMed] [Google Scholar]
- Kietzman CC, Caparon MG. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect Immun. 2010;78:241–252. doi: 10.1128/IAI.00746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SN, Bae YG, Rhee DK. Dual regulation of dnaK and groE operons by HrcA and Ca++ in Streptococcus pneumoniae. Arch Pharm Res. 2008;31:462–467. doi: 10.1007/s12272-001-1179-4. [DOI] [PubMed] [Google Scholar]
- Koch B, Kilstrup M, Vogensen FK, Hammer K. Induced levels of heat shock proteins in a dnaK mutant of Lactococcus lactis. J Bacteriol. 1998;180:3873–3881. doi: 10.1128/jb.180.15.3873-3881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Ekaza E, Paquet JY, Walravens K, Teyssier J, Godfroid J, Liautard JP. Induction of dnaK through its native heat shock promoter is necessary for intramacrophagic replication of Brucella suis. Infect Immun. 2002;70:1631–1634. doi: 10.1128/IAI.70.3.1631-1634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanzer M, Bujard H. Promoters determine largely the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:6632–6636. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Chen YY, Burne RA. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J Bacteriol. 2001;183:6074–6084. doi: 10.1128/JB.183.20.6074-6084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Luzardo Y, Burne RA. Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol. 2007;189:1582–1588. doi: 10.1128/JB.01655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Rüdiger S, Bukau B. Molecular basis for interactions of the DnaK chaperone with substrates. Biol Chem. 2000;381:877–885. doi: 10.1515/BC.2000.109. [DOI] [PubMed] [Google Scholar]
- Minder AC, Narberhaus F, Babst M, Hennecke H, Fischer HM. The dnaKJ operon belonging to the σ32-dependent class of heat shock genes in Bradyrhizobium japonicum. Mol Gen Genet. 1997;254:195–206. doi: 10.1007/s004380050408. [DOI] [PubMed] [Google Scholar]
- Mogk A, Homuth G, Scholz C, Kim L, Schmid FX, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Bukau B, Lutz R, Schumann W. Construction and analysis of hybrid Escherichia coli–Bacillus subtilis dnaK genes. J Bacteriol. 1999;181:1971–1974. doi: 10.1128/jb.181.6.1971-1974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley RA, Tsuji A, Matsuda Y. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune H, Whiley RA, Goto T, Inai Y, Maeda T, Hardie JM, Kourai H. Distribution of the intermedilysin gene among the anginosus group streptococci and correlation between intermedilysin production and deep-seated infection with Streptococcus intermedius. J Clin Microbiol. 2000;38:220–226. doi: 10.1128/jcm.38.1.220-226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Arsène-Ploetze F, Rist W, Rüdiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol Cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- Sugimoto S, Higashi C, Saruwatari K, Nakayama J, Sonomoto K. A gram-negative characteristic segment in Escherichia coli DnaK is essential for the ATP-dependent cooperative function with the co-chaperones DnaJ and GrpE. FEBS Lett. 2007;581:2993–2999. doi: 10.1016/j.febslet.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Sugimoto S, Saruwatari K, Higashi C, Tsuruno K, Matsumoto S, Nakayama J, Sonomoto K. In vivo and in vitro complementation study comparing the function of DnaK chaperone systems from halophilic lactic acid bacterium Tetragenococcus halophilus and Escherichia coli. Biosci Biotechnol Biochem. 2008;72:811–822. doi: 10.1271/bbb.70691. [DOI] [PubMed] [Google Scholar]
- Sukeno A, Nagamune H, Whiley RA, Jafar SI, Aduse-Opoku J, Ohkura K, Maeda T, Hirota K, Miyake Y, Kourai H. Intermedilysin is essential for the invasion of hepatoma HepG2 cells by Streptococcus intermedius. Microbiol Immunol. 2005;49:681–694. doi: 10.1111/j.1348-0421.2005.tb03647.x. [DOI] [PubMed] [Google Scholar]
- Sussman MD, Setlow P. Nucleotide sequence of Bacillus megaterium gene homologous to the dnaK gene of Escherichia coli. Nucleic Acids Res. 1987;15:3923. doi: 10.1093/nar/15.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J Bacteriol. 2002;184:224–32. doi: 10.1128/JB.184.1.224-232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A, Tomoyasu T, Matsui H, Yamamoto T. The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect Immun. 2004;72:1364–73. doi: 10.1128/IAI.72.3.1364-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Hauser R, Campbell J, Ostheimer GJ. Isolation of dnaJ, dnaK, and grpE homologues from Borrelia burgdorferi and complementation of Escherichia coli mutants. Mol Microbiol. 1993;7:359–369. doi: 10.1111/j.1365-2958.1993.tb01128.x. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Takaya A, Handa Y, Karata K, Yamamoto T. ClpXP controls the expression of LEE genes in enterohaemorrhagic Escherichia coli. FEMS Microbiol Lett. 2005;253:59–66. doi: 10.1016/j.femsle.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Tabata A, Hiroshima R, Imaki H, Masuda S, Whiley RA, Aduse-Opoku J, Kikuchi K, Hiramatsu K, Nagamune H. Role of catabolite control protein A in the regulation of intermedilysin production by Streptococcus intermedius. Infect Immun. 2010;78:4012–4021. doi: 10.1128/IAI.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Tabata A, Nagamune H. Investigation of the chaperone function of the small heat shock protein–AgsA. BMC Biochem. 2010;24:11–27. doi: 10.1186/1471-2091-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Fraser H, Hardie JM, Beighton D. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the "Streptococcus milleri group". J Clin Microbiol. 1990;28:1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Beighton D, Winstanley TG, Fraser HY, Hardie JM. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243–244. doi: 10.1128/jcm.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury R, Haldenwang WG. HrcA is a negative regulator of the dnaK and groESL operons of Streptococcus pyogenes. Biochem Biophys Res Commun. 2003;302:722–727. doi: 10.1016/S0006-291X(03)00254-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Sashinami H, Takaya A, Tomoyasu T, Matsui H, Kikuchi Y, Hanawa T, Kamiya S, Nakane A. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect Immun. 2001;69:3164–3174. doi: 10.1128/IAI.69.5.3164-3174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(PPT 2371 kb)