Abstract
The mitochondrial 70-kDa heat shock protein (mtHsp70), also known in humans as mortalin, is a central component of the mitochondrial protein import motor and plays a key role in the folding of matrix-localized mitochondrial proteins. MtHsp70 is assisted by a member of the 40-kDa heat shock protein co-chaperone family named Tid1 and a nucleotide exchange factor. Whereas, yeast mtHsp70 has been extensively studied in the context of protein import in the mitochondria, and the bacterial 70-kDa heat shock protein was recently shown to act as an ATP-fuelled unfolding enzyme capable of detoxifying stably misfolded polypeptides into harmless natively refolded proteins, little is known about the molecular functions of the human mortalin in protein homeostasis. Here, we developed novel and efficient purification protocols for mortalin and the two spliced versions of Tid1, Tid1-S, and Tid1-L and showed that mortalin can mediate the in vitro ATP-dependent reactivation of stable-preformed heat-denatured model aggregates, with the assistance of Mge1 and either Tid1-L or Tid1-S co-chaperones or yeast Mdj1. Thus, in addition of being a central component of the protein import machinery, human mortalin together with Tid1, may serve as a protein disaggregating machine which, for lack of Hsp100/ClpB disaggregating co-chaperones, may carry alone the scavenging of toxic protein aggregates in stressed, diseased, or aging human mitochondria.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-011-0285-3) contains supplementary material, which is available to authorized users.
Keywords: Mitochondrial Hsp70, Tid1, Hep1, Ssc1, Disaggregation
Introduction
The 70-kDa heat shock proteins (Hsp70s) are ubiquitous molecular chaperones, which are present in all the ATP-containing compartments in the eukaryotic cell, where they perform diverse functions all with a common denominator: they use the energy of ATP hydrolysis to unfold, at least partially, various protein substrates (Goloubinoff and De Los Rios 2007; Sharma et al. 2010). The bacterial chaperone DnaK has long served as a prototype for the study of Hsp70 chaperones and its investigation provided important insights into the structure and function of this large group of proteins (Swain et al. 2007). Three types of chaperone activities have been described for bacterial DnaK: (1) passively preventing the aggregation of artificially unfolded proteins (holding function). (2) ATP-dependent refolding of chemically unfolded proteins to their native forms (Szabo et al. 1994). (3) The ATP-dependent reactivation of stable protein aggregates into native proteins. The last two functions reflect the ability of this chaperone system to act as a polypeptide unfoldase. In this capacity, Hsp70 specifically recognizes misfolded polypeptides, bind them with high affinity, uses the energy of ATP hydrolysis to unfold them, and releases low-affinity unfolded products that in turn can spontaneously refold to the native state (Sharma et al. 2010). In vitro, DnaK can perform this function when assisted by its two co-chaperones, a J-domain-containing protein (either DnaJ, CbpA, or DjlA; Genevaux et al. 2007) and the nucleotide exchange factor GrpE (Sharma et al. 2009). Specificity of the chaperone system is conferred primarily by the DnaJ co-chaperone, which recognizes and binds to a misfolded polypeptide (Hinault et al. 2010), followed by the binding of the polypeptide to a “low-affinity” DnaK, which becomes entrapped upon hydrolyzing ATP. GrpE, acting as an ADP release factor, accelerates discharge of the unfolded polypeptide and spontaneous refolding to the native state (Sharma et al. 2010).
Yeast mitochondria harbor three Hsp70 homologues named Ssc1, Ssq1, and Ecm10 (for sequence alignments see Supplemental Fig. 1). Ssc1 was shown to be a key player in mediating protein import into the mitochondrial matrix. This particular function of the chaperone occurs through association of the ATP-bound state to the TIM23 import channel, by way of the specific protein anchors, Tim44 and PAM16/18. Dissociation of ADP-bound Ssc1 from the anchors takes place as soon as it has locked onto an incoming polypeptide (De Los Rios et al. 2006; D’Silva et al. 2004; Neupert and Herrmann 2007). Ssc1 plays an additional important role in mediating the refolding of matrix-localized proteins, a function which is carried out with the help of two matrix-localized co-chaperones Mdj1 and Mge1, which are homologous to the Escherichia coli DnaJ and GrpE, respectively (Deloche et al. 1997; Horst et al. 1997; Liu et al. 2001; Rowley et al. 1994) (Supplemental Fig. 2). Using purified proteins, it was shown that Ssc1 can mediate ATP-dependent disaggregation and proper refolding of stably misfolded reporter proteins, when supplemented with bacterial DnaJ and GrpE (De Los Rios et al. 2006). Ssq1 is known to play a role in the biogenesis of iron sulfur clusters and the function of Ecm10 is still unclear (Baumann et al. 2000; Dutkiewicz et al. 2003).
The human Hsp70 family consists of eight members that differ in their amino acid sequences, expression levels, and cellular localizations (Daugaard et al. 2007). Six Hsp70 chaperones are located mainly in the cytosol and in the nucleus; one Hsp70 is located in the lumen of the ER and mortalin and is found in the mitochondrial matrix. Thus, in human mitochondria, a single Hsp70, mortalin, must take over all functions that are performed by its three homologues in yeast mitochondria (Bhattacharyya et al. 1995; Daugaard et al. 2007; Mizzen et al. 1989; Wadhwa et al. 1993). Mortalin shares high identity with other members of the Hsp70 family, including E. coli DnaK (51%) and Saccharomyces cerevisiae Ssc1 (65%) (Supplemental Fig. 1). Interestingly, mortalin was also detected in extra-mitochondrial compartments, which reflects the ability of this protein to perform additional functions that are not related to protein folding (reviewed in Kaul et al. 2007).
Homologues for Mdj1 and Mge1 were found in humans and named Tid1 and hMge1, respectively (Supplemental Fig. 2). Human Tid1 encodes for two mitochondrial matrix-localized splice variants, one of 43 (Tid1-L) and one of 40 kDa (Tid1-S). Both variants have an N-terminal mitochondrial signal. However, they differ in their carboxyl terminus tails: Tid1-L has 33 amino acids, whereas Tid1-S has only six amino acids (Syken et al. 1999). Several lines of evidence indicate that both Tid1 isoforms are functional homologues of Mdj1: (1) they contain the characteristic J-domain of the 40-kDa heat shock protein (Hsp40) family; (2) both isoforms were co-immunoprecipitated with mortalin (Syken et al. 1999); and (3) Tid1-L and Tid1-S were both able to compensate for an Mdj1 deletion in yeast. This capacity was dependent on the presence of an intact J-domain and a mitochondrial targeting sequence (Lu et al. 2006). Tid1 isoforms were also found to be involved in extra-mitochondrial protein interactions and functions (see below). These functions include regulation of cell death, proliferation, and signal transduction (Mitra et al. 2009; Syken et al. 1999). It was shown that the two isoforms differ in their non-mitochondrial functions due to differences in their abilities to interact with cytosolic and nuclear proteins (Lu et al. 2006). Moreover, It was demonstrated that expression of the long variant increased apoptosis, while expression of the short variant suppressed apoptosis (Syken et al. 1999). Interestingly, Tid1-L was shown to be more stable in the cytosol as compared with Tid1-S, with a longer cytosolic residency time prior to mitochondrial import (Lu et al. 2006).
Molecular chaperones are key players in maintaining cellular protein homeostasis. Consistent with this role, several studies have shown that mortalin may be implicated in the progression of neurodegenerative and protein aggregation disorders, such as Parkinson and Alzheimer (Burbulla et al. 2010; De Mena et al. 2009; Deocaris et al. 2008; Deocaris et al. 2007; Koren et al. 2009; Shi et al. 2008). Moreover, when mammalian mitochondria are challenged with an imported protein with a strong tendency to form aggregates, the mitochondrial unfolded protein response (mUPR) is activated (Aldridge et al. 2007; Zhao et al. 2002). mUPR upregulated the ClpP protease and the cpn60, cpn10, and Tid1 chaperones but not mortalin.
The strong cyto-protective, even curing effects of the Hsp70–Hsp40 chaperones systems, both in the cytoplasm and in mitochondria can be attributed to their ability to act as polypeptide unfolding enzymes that can target cytotoxic misfolded and aggregated protein conformers (Sharma et al. 2009; Sharma et al. 2010). In organisms and cellular compartments that also express Hsp100/ClpB homologues, the Hsp70–Hsp40 unfoldase chaperone systems benefit from a powerful synergic mechanism of forceful disaggregation that may act both upstream and downstream to the polypeptide unfoldase activity of Hsp70 chaperones (Glover and Lindquist 1998; Goloubinoff et al. 1999; Weibezahn et al. 2005). Thus, in the yeast cytoplasm, a large number of Hsp70–Hsp40 can collaborate with the ClpB-like (Hsp104) disaggregating co-chaperone to prevent the formation of misfolded species and even solubilize-resistant prions. Similarly, yeast mitochondria can use the unfolding abilities of Ssc1, which together with Mdj1 can collaborate with the ClpB homologue, Hsp78, at the active unfolding and disaggregation of potentially toxic misfolded protein conformers (von Janowsky et al. 2006).
Interestingly, mammalian cytoplasm contains no Hsp104 and mammalian mitochondria contain a single Hsp70 (mortalin) and no ClpB-like disaggregating co-chaperones. Notably, up to date it remained unclear whether mortalin in cooperation with Tid1 can reactivate protein aggregates. Here, we used purified components and found that mortalin and both Tid1 variants can mediate the reactivation of stable protein aggregates, implying that mortalin can serve as a unique scavenger of toxic protein conformers in human mitochondrial making it an attractive target for therapies against protein conformational diseases.
Results and discussion
Purification of mortalin and Tid1-L/Tid1-S
Upon expression in bacteria, mitochondrial Hsp70s tend to aggregate and are detected in inclusion bodies (Blamowska et al. 2010; Zhai et al. 2008). In order to obtain soluble mortalin (or Ssc1), each protein was co-expressed in bacteria together with Hep1/Zim17, which has been shown to prevent the aggregation of Ssc1 and of mortalin in mitochondria as well as upon expression in bacteria (Sichting et al. 2005; Zhai et al. 2008). Furthermore, mortalin, Ssc1 and yeast Hep1 were each cloned carrying an octa-histidine tag (His-tag), which in the case of mortalin, and Ssc1, could be cleaved by tobacco etch virus (TEV) protease (Iosefson and Azem 2010). This cloning strategy enabled the efficient removal of Hep1 on a second Ni-Agarose column following cleavage of the hsp70s His-tag by TEV. Using the purification protocol that is detailed under “Materials and methods,” we obtained purified mortalin and Ssc1 (Supplemental Fig. 3) that, in contrast to previously described protocols, are lacking the His-tag (Zhai et al. 2008; Goswami et al. 2010). The absence of Hep1 contamination in the purified protein was further confirmed by western blots with His-tagged antibodies (not shown).
Upon their expression in bacteria, both Tid1-L and Tid1-S were detected in the inclusion bodies. Thus, in the first step, Tid1-L/S were solubilized by treatment with 3 M urea as described previously for Mdj1 (Horst et al. 1997). Final purification was carried out using a second Ni-NTA column followed by a refolding step using dialysis, as detailed under “Materials and methods.” The purity of Tid1-L is presented under Supplemental Fig. 4. Since a previous study showed that both Tid1-L and Tid1-S are equally able to complement deletion of yeast Mdj1, we initially focused on studying Tid1-L (Lu et al. 2006).
Functional cooperation between Hsp70 and DnaJ homologues as examined by single-turnover ATPase assays
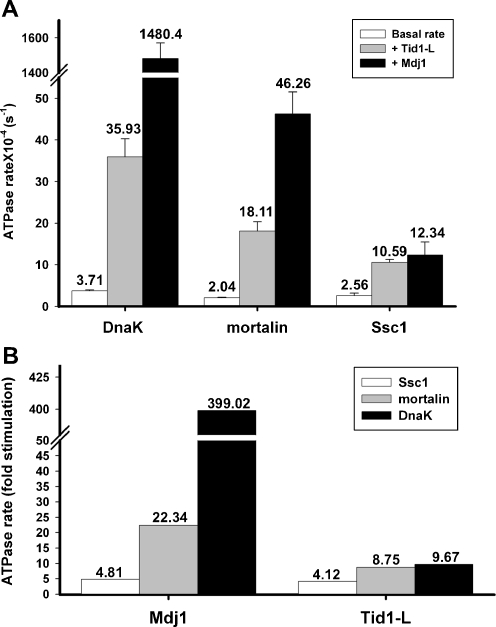
As mentioned above, yeast mitochondria harbor three Hsp70 family members while human mitochondria have only one member, mortalin. Thus, one aspect of this work was to examine the ability of yeast and human Hsp70 chaperones to functionally interact with endogenous and heterologous counterparts. Because it has been extensively characterized, the closely related bacterial Hsp70 chaperone, DnaK, was also examined. We first addressed the basal ATP hydrolysis activity of the three Hsp70 chaperones using a single-turnover assay. As shown in Fig. 1, the three related Hsp70 chaperones exhibited very low ATP hydrolysis rates, of the same order of magnitude. DnaK had the highest ATP hydrolysis rate (3.71∙10−4 s−1), while the basal ATPase activity of mortalin and Ssc1 was slightly lower (2.04∙10−4 and 2.56∙10−4 s−1, respectively).
Fig. 1.
Stimulation of Hsp70 ATPase activity by Mdj1 or Tid1-L. a The indicated Hsp70/ATP complexes were incubated in the presence or absence of either Mdj1 or Tid1-L. The indicated turnover values, expressed as the rate of ATP hydrolysis per second, were obtained from the slope of the equation describing the linear portion of the reaction. b The ATPase activity is presented as fold stimulation of Hsp70 by either Mdj1 or Tid1-L
A functional interaction between Mdj1 and the chaperones DnaK, mortalin, and Ssc1 is clearly revealed (Fig. 1) by the fact that the ATPase activity of all Hsp70s is significantly enhanced by the co-chaperone (399.0-, 22.3- and 4.8-fold, respectively). Tid1-L was also able to functionally interact with the three Hsp70 chaperones. However, the ATPase of DnaK and mortalin, was enhanced to a much less degree in the presence of Tid1-L than in the presence of Mdj1 (9.6 and 8.7, respectively). The increase in ATPase rate that was obtained with Tid1-L was specific since no enhancement was observed using the inactive Tid1-L mutant H121Q (Supplemental Fig. 5). Thus, we demonstrated that the bacterial, yeast and human Hsp70s are able to functionally interact with both Mdj1 and Tid1-L, results which reflect the evolutionary conservation of this interaction.
DnaK, Ssc1, and mortalin are able to reactivate protein aggregates when assisted by Mdj1
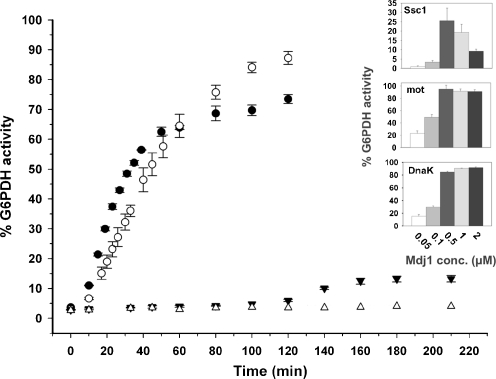
Previous studies have shown that DnaK and Ssc1 can reactivate stable protein aggregates when assisted by bacterial DnaJ and GrpE co-chaperones (De Los Rios et al. 2006; Mogk et al. 1999). However, the ability of mortalin to reactivate preformed protein aggregates has not been demonstrated so far. The time-dependent reactivation of G6PDH by DnaK, Ssc1 and mortalin was first examined in the presence of yeast Mdj1 and Mge1. As shown in Fig. 2, when examined with Mdj1, DnaK, and mortalin were both able to efficiently reactivate G6PDH aggregates, with DnaK showing slightly faster initial rates of activity (7.3·10−4 and 5.7·10−4 min−1, respectively). Interestingly, mortalin and DnaK, when assisted by yeast Mdj1, were more efficient at reactivating preformed G6PDH aggregates than Ssc1, the yeast partner. When we examined the dependence of Ssc1 reactivation activity on Mdj1 concentration, we found that it reached a maximum at 0.5 μM of the J-protein (Fig. 2, inset). Upon further increase in Mdj1 concentration, however, the reactivation activity decreased, similar to what was shown previously for DnaK and DnaJ (Laufen et al. 1999). In contrast to Ssc1 activity, DnaK and mortalin activity did not decrease in the presence of the higher Mdj1 concentrations. Still, at the optimal Mdj1 concentration examined in this experiment, the maximal yield of reactivated G6PDH in the presence of Ssc1 reached only 30%.
Fig. 2.
Time-dependent reactivation of stable G6PDH aggregates by DnaK, Ssc1, and mortalin. Stable G6PDH aggregates were reactivated at 30°C in the presence of 16 μM of either DnaK (filled circles), Ssc1 (filled inverted triangles) or mortalin (empty circles), and yeast co-chaperones Mdj1 (2 μM) and Mge1 (1 μM). The spontaneous refolding of the aggregates in the presence of the reaction buffer is also shown (empty triangles). The insets describe the G6PDH activity after reactivation in the presence of 16 μM of either DnaK, Ssc1 or mortalin, the indicated concentrations of Mdj1, and 1 μM of Mge1. G6PDH activity was measured after 230 min at 30°C. Background activity was subtracted
Tid1-L can assist DnaK and mortalin, but not Ssc1, in mediating the ATP-dependent reactivation of G6PDH aggregates
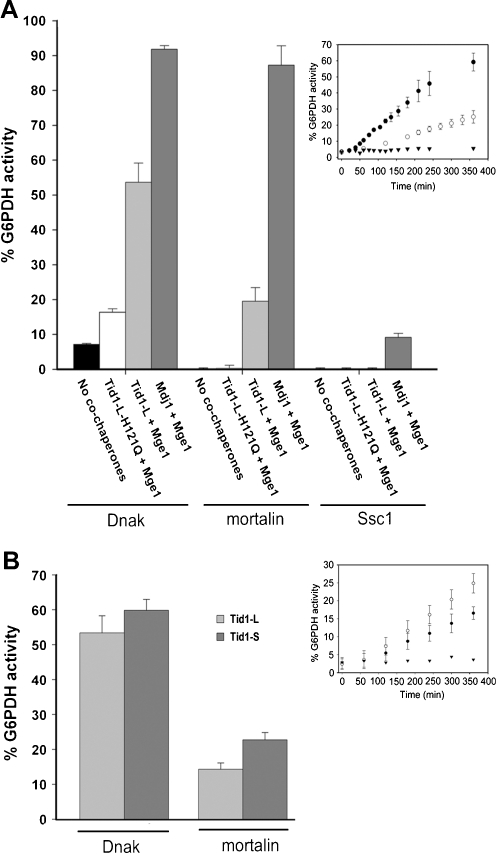
The above result demonstrated for the first time that mortalin can reactivate G6PDH aggregates, albeit only in the presence of yeast Mdj1. We next examined whether mortalin’s partner, Tid1-L, is also able to assist the three chaperones in the reactivation of preformed G6PDH aggregates. The specificity of the reactivation activity was confirmed by examining the inactive mutant of Tid1-L, H121Q. In addition, mortalin-V482F (Iosefson and Azem 2010), a mutant exhibiting a defective interaction with client proteins, failed to reactivate G6PDH preformed aggregates (Supplemental Fig. 6). As shown in Fig. 3, Tid1-L was efficiently able to assist both DnaK and mortalin in mediating the reactivation of aggregates. Interestingly, mortalin was more efficient at reactivating G6PDH when it was assisted by Mdj1 than when assisted by its own endogenous co-chaperone, Tid1-L. Noticeably, Ssc1 was much less efficient in its ability to reactivate protein aggregates even when assisted with its endogenous co-chaperone. Moreover, Ssc1 was unable to reactivate preformed G6PDH aggregates at all in combination with Tid1-L.
Fig. 3.
Yield of G6PDH refolding (after 6 h) obtained by DnaK, Ssc1, and mortalin. a Stable G6PDH aggregates were reactivated at 30°C in the presence of DnaK, Ssc1, or mortalin (16 μM), either alone or assisted by the indicated J-protein (2 μM) and Mge1 (1 μM). The presented values are averages of at least three different experiments with the background activity subtracted. The inset describes the time-dependent G6PDH activity in the presence of 16 μM DnaK (filled circles) or mortalin (empty circles) assisted by 2 μM Tid1-L and 1 μM Mge1. The spontaneous refolding of the aggregates in the presence of the reaction buffer is also shown (filled inverted triangles). b Same conditions as in (a), except that 1 μM of Tid1-L or Tid1-S was used. The inset describes the time-dependent G6PDH activity in the presence of 16 μM mortalin assisted by either 1 μM of Tid1-L (filled circles) or 1 μM of Tid1-S (empty circles) and 1 μM Mge1
In summary, we found that among the three chaperones, Ssc1 acted as a poor reactivation machinery as compared with DnaK and mortalin, whereas, Tid1-L was a less efficient co-chaperone of the reactivation reaction than yeast Mdj1.
As mentioned above, in addition to Tid1-L human cells express also another spliced form, Tid1-S. It was shown that Tid1-L is much more stable in the cytosol than Tid1-S, while both were equally stable inside the mitochondria (Lu et al. 2006). Thus, it was very interesting to examine whether Tid1-S and Tid1-L exhibit different reactivation activities. To this end, we also examined the ability of Tid1-L and Tid1-S to reactivate preformed G6PDH aggregates. Interestingly, Tid1-S was more active in the reactivation of aggregates than Tid1-L, with rates of reactivation 3.12∙10−5 and 1.56∙10−5 min−1, respectively (Table 2 and Fig. 3b).
Table 2.
Comparison of disaggregation efficiencies between Tid1 splice variants
| J-protein | DnaK | Mortalin | |
|---|---|---|---|
| ATPase (min−1) | Tid1-L | 1.33 ± 0.085 | 1.99 ± 0.16 |
| Tid1-S | 2.06 ± 0.057 | 1.93 ± 0.14 | |
| G6PDH disaggregation (min−1) | Tid1-L | 7.29·10−5 ± 9·10−6 | 1.56·10−5 |
| Tid1-S | 9.3·10−5 | 3.12·10−5 | |
| nmol ATP/nmol G6PDH refolded | Tid1-L | 18,244 | 127,564 |
| Tid1-S | 22,150 | 61,858 |
Turnover numbers of steady-state ATPase and G6PDH disaggregation were determined as described under “Materials and methods”; 16 μM of the indicated Hsp70 proteins, 1 μM of the relevant Tid1 construct, and 1 μM of Mge1 were used
The efficiency of disaggregation is determined by chaperone/co-chaperone combination
Finally, we examined the efficiency of the disaggregation activity exerted by the three chaperones when assisted by either Mdj1 or Tid1-L and analyzed the steady-state ATPase rate of Hsp70s under the same conditions as the disaggregation reaction. As presented in Table 1, DnaK and mortalin worked with a nearly similar efficiency when Mdj1 was included in the disaggregation reaction, hydrolyzing almost ∼3,150 molecules of ATP for each reactivated G6PDH molecule. Interestingly, DnaK and mortalin exhibited lower efficiency in the presence of Tid1-L, hydrolyzing 12,800 and 151,282 molecules of ATP for each refolded G6PDH molecule, respectively. Since mortalin exhibited similar steady-state ATP hydrolysis rates in the presence of both Tid1-L and Tid1-S, the immediate outcome should be that mortalin functions more efficiently in the presence of Tid1-S than Tid1-L, in terms of number of ATP molecules hydrolyzed per reactivated G6PDH molecules (Table 2).
Table 1.
Disaggregation efficiencies of the studied chaperones
| J-protein | DnaK | Mortalin | Ssc1 | |
|---|---|---|---|---|
| ATPase (min−1) | Mdj1 | 2.3 ± 0.2 | 1.82 ± 0.1 | 1.58 ± 0.08 |
| Tid1-L | 1.6 ± 0.17 | 2.36 ± 0.24 | 1.77 | |
| G6PDH disaggregation (min−1) | Mdj1 | 7.3·10−4 ± 9·10−5 | 5.7·10−4 ± 9·10−5 | 6.25·10−5 |
| Tid1-L | 1.25·10−4 ± 2.2·10−5 | 1.56·10−5 | NA | |
| nmol ATP/nmol G6PDH refolded | Mdj1 | 3,150 | 3,193 | 25,280 |
| Tid1-L | 12,800 | 151,282 | – |
Turnover numbers of steady-state ATPase and G6PDH disaggregation were determined as described under “Materials and methods”; 16 μM of the indicated Hsp70 proteins, 2 μM of the relevant J-domain protein, and 1 μM of Mge1 were used
NA not active
Concluding remarks
Molecular chaperones serve as a first line of defense against uncontrolled unfolding and aggregation of proteins (Broadley and Hartl 2009). Mitochondria serve as the major site of ATP production, a process which involves electron transfer and free radical production. Thus, its proteins face continuous stress, which if not countered, may lead to protein aggregation and cell death (de Castro et al. 2010; Pedersen et al. 2003; Schapira 1998; Xie et al. 2010; Vijayvergiya et al. 2005). Expression of aggregation-prone mutant form of ornithine transcarbamylase in the mitochondrial matrix leads to the induction of the mUPR (Zhao et al. 2002). Interestingly, the mammalian homologue of Tid1, but not mortalin, was among the proteins that were induced upon mUPR (Zhao et al. 2002).
While an interaction between Tid1 and mortalin has been demonstrated previously (Goswami et al. 2010), the ability of these proteins to cooperate in reactivating preformed proteins aggregates has not been demonstrated so far. In situations of folding malfunction, reactivation of protein aggregates is clearly a critical function for continued viability of the cell.
Using G6PDH as a model substrate, we showed for the first time that mortalin and Tid1 splice variants, can mediate disaggregation of preformed G6PDH protein aggregates and refolding of the enzyme to a functional state. Interestingly, in vitro, Tid1-L was more active with the bacterial Hsp70, DnaK, than with its endogenous chaperone mortalin, and it was unable to assist Ssc1 in reactivating protein aggregates in and ATP-dependent manner. This probably reflects the fact that DnaK is much stronger ATPase than mortalin. Nevertheless, in terms of efficiency, both DnaK and mortalin consumed similar amounts of ATP per refolded G6PDH when examined with yeast Mdj1 (Table 1).
Of the two spliced forms found in human cells Tid1-S was more efficient in mediating the reactivation of protein aggregates than Tid1-L. This is in accordance with previous study, in which Tid1-S was more efficient in its ability to prevent aggregation of denatured rhodanese than Tid1-L (Goswami et al. 2010). More work is required in order to show whether this observation is related to the fact that Tid1-L is more stable in the cytosol than Tid1-S (Lu et al. 2006).
A recent, single-molecule analysis of DnaK and Ssc1 showed that, despite their evolutionary conservation, these proteins exhibited different conformational changes upon their interaction with ADP and their co-chaperones (Mapa et al. 2010). These differences were suggested to help the chaperones to meet specific functions in different organisms and cellular compartments (Mapa et al. 2010). It is tempting to speculate that the conformational differences between the various chaperones may also be reflected by distinct abilities to reactivate misfolded protein aggregates.
Because mammalian mitochondria contain a single Hsp70 (mortalin), a single Hsp40 (Tid1) and no ClpB-like disaggregating co-chaperones, mortalin remains the only known mitochondrial chaperone that, together with Tid1, could carry the active detoxification of harmful protein aggregates by unfolding them and, thus, provide stressed human mitochondria with a precious protective mechanism against apoptosis. It is tempting to speculate that non-toxic drugs that could induce the accumulation of mortalin and Tid1 could also reduce toxic protein aggregation in mitochondria and thus block apoptotic signals in inflammatory protein misfolding diseases and aging. Conversely, drugs directly blocking mortalin and Tid1 would favor apoptosis in cancer cell.
Materials and methods
Cloning of mortalin or Ssc1 and yeast Hep1 into the pETDuet co-expression vector
The nucleotide sequence encoding residues 51–679 of mature mortalin was generated by PCR from a human cDNA genomic library and cloned between the BamHI (forward primer-I; Supplemental Table 1) and NotI (backward primer-II) restriction sites. The nucleotide sequence encoding amino acid residues 49–171 of mature S. cerevisiae Hep1 was cloned between the NdeI (forward primer-III) and XhoI (backward primer-ΙV) restriction sites. The two PCR products were digested with suitable restriction enzymes and ligated into a modified pETDuet vector (Iosefson and Azem 2010). The resulting plasmid co-expresses mortalin and yeast Hep1. Both proteins were engineered to contain a His-tag at their N terminus, however; only mortalin’s His-tag can be cleaved by the TEV protease. The pETDuet plasmid was transformed into E. coli which served as a host for co-expression of mortalin and Hep1. Similar strategy was used to create co-expression vectors for Ssc1 and Hep1. A mortalin-V482F mutant was generated using the pETDuet plasmid as a template for mutagenesis according to the i-Pfu DNA polymerase protocol (Intron).
Purification of mortalin and Ssc1
The E. coli BL-21 Tuner strain carrying the pETDuet plasmid was grown for 3–4 h at 37°C in LB media, containing 200 μg/ml ampicillin and 34 μg/ml chloramphenicol. Protein expression was induced with 1 mM of IPTG for 16 h at 16°C. Next, cells were harvested and resuspended (1:10, w/v) in buffer A (20 mM MES, pH 6.6, and 5% glycerol) containing 45 U/ml of DNase, 2 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mg/ml lysozyme, and one tablet of complete EDTA-free protease inhibitor cocktail (Roche). After incubation for 15 min at 4°C, bacteria were homogenized and disrupted using a micro-fluidizer (Microfluidic). The soluble fraction was loaded on an anion-exchange column (RESOURCE Q, ∼30 ml bead volume, Amersham) pre-equilibrated with buffer A. The column was washed with ∼2 L of buffer A until the majority of Hep1 was removed. Fractions were eluted with a linear gradient (0–50%, 90 ml) of buffer B (buffer A containing 1 M NaCl). Fractions enriched with the desired protein were pooled, diluted 1:5 with buffer C (50 mM Tris–HCl, pH 7.7, 200 mM NaCl, 5% glycerol and 20 mM imidazole) and loaded on a Ni-NTA column (∼15 ml bead volume) pre-equilibrated with buffer C. The column was washed with buffer C until a stable baseline was observed. Fractions were eluted with a linear gradient (0–50%, 90 ml) of buffer D (buffer C containing 1 M imidazole). Fractions enriched with the protein were pooled and treated with TEV protease (TEV/protein 1:25, w/w) overnight at 4°C, in a tubular dialysis membrane (12–14 kDa MWCO) against 4 L of buffer E (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 5% glycerol, and 20 mM imidazole). After TEV digestion, the protein was loaded onto a second nickel column to remove: (1) TEV protease and the residual Hep1, both containing an uncleavable His-tag; (2) the uncleaved protein of interest that still contained the His-tag; and (3) other nonspecifically bound proteins. Relevant fractions were concentrated using protein concentrators (Mw cutoff of 30,000, Millipore) and desalted using PD10 columns (Pharmacia) with buffer F (20 mM Na-HEPES, pH 7.4, 200 mM NaCl, and 5% glycerol). The protein was divided into aliquots, flash-frozen in liquid N2 and stored at −80°C until use. All purification procedures were carried out at 4°C. We confirmed the absence of residual Hep1 protein in the purified mortalin preparations using western blotting with anti-His-tag antibodies. Mortalin-V482F was purified according to the same protocol.
Cloning of Tid1-L and Tid1-S into the pET21d expression vector
The nucleotide sequence encoding mature Tid1-L (residues 66–480) or Tid1-S (residues 66–453) were generated by PCR from a human cDNA genomic library with BamHI (forward primer-VII) and XhoI (backward primers-VIII and XV, respectively) restriction sites. The PCR product was cloned into the pGEM-T Easy vector, transformed into XL1-Blue E. coli competent cells and plated onto LB-plates supplemented with 200 μg/ml ampicillin. Plasmids were isolated from the positive colonies and sequenced to rule out PCR-induced sequence errors. The construct was digested with BamHI and XhoI and ligated into a modified pET21d (+) vector. The protein was engineered to contain a His-tag at its N terminus that is cleavable by TEV protease. However, in the function experiments described in this work, the tag was not removed. The pET21d (+) plasmid was transformed into BL-21 E. coli competent cells for subsequent expression. A Tid1-L-H121Q mutant was generated using the pET21d plasmid as a template for mutagenesis according to the i-Pfu DNA polymerase protocol (Intron).
Purification of Tid1 proteins
E. coli BL-21 tuner strain, carrying the pET21d (+) plasmid with the relevant Tid1 construct, was grown for 3–4 h at 37°C in LB media, containing 200 μg/ml ampicillin and 34 μg/ml chloramphenicol. Protein expression was induced with 1 mM of IPTG. After 4–5 h, at 16°C, cells were harvested and suspended (1:10, w/v) in buffer A (20 mM Tris–HCl, pH 7.4, 0.5 M NaCl, 10% glycerol, 1% Triton X-100, 0.05% Brij-58, and 2 mM β-mercaptoethanol) containing 45 U/ml of DNase, 2 mM PMSF, 0.1 mg/ml lysozyme, and one tablet of complete EDTA-free protease inhibitor cocktail. After incubation for 15 min at 4°C, bacteria were homogenized and disrupted using a micro-fluidizer and the sample was centrifuged at 14,000 rpm for 1 h at 4°C. Tid1 was found in the pellet fraction (inclusion bodies) together with a small number of additional proteins. Pure inclusion bodies were obtained by three sequential cycles of pellet resuspension in buffer A and centrifugation at 14,000 rpm for 30 min. In a preliminary experiment, we found that 3 M urea rescues most of Tid1-L from the inclusion bodies. Therefore, in the next step the inclusion bodies were resuspended in buffer B (buffer A containing 3 M urea and 5 mM imidazole) and incubated for 30 min on an end-over-end rotator. The insoluble material was removed by centrifugation for 30 min at 14,000 rpm. The soluble fraction was incubated for 1 h at 4°C with Ni-NTA beads that had been pre-equilibrated with buffer B (∼2 ml bead volume), on an end-over-end rotator. The beads were centrifuged for 2 min at 2,000 rpm and supernatant containing unbound proteins was removed. Next, the beads were washed twice with buffer B and twice more with buffer C (buffer B lacking Triton X-100). Bound proteins were eluted by incubating the Ni-NTA beads for 30 min at 4°C with buffer D (buffer C containing 300 mM imidazole). The eluted fraction, containing Tid1, was separated from the beads by centrifugation for 2 min at 2,000 rpm. The urea was removed from the Tid1 solution by overnight dialysis in a tubular dialysis membrane against 2 L of buffer E (20 mM Tris, pH 7.4, 0.5 M NaCl, 10% glycerol, 0.05% Brij-35, 2 μM β-mercaptoethanol, and 5 mM imidazole). A significant amount of protein aggregated after the urea removal. The insoluble material was removed by centrifugation at 14,000 rpm for 1 h. The soluble fraction (∼50 ml) was purified by a second incubation with Ni-NTA beads (∼1 ml bead volume), in a batch procedure. The following few modifications were made compared with the first Ni-NTA purification: (1) the Ni-NTA beads were pre-equilibrated with buffer E; (2) after protein binding, the beads were washed four times with buffer E; (3) the proteins were eluted with 2.5 ml buffer F (buffer E containing 300 mM imidazole). The eluted fraction was desalted (PD-10 columns) in 3.5 ml of buffer G (buffer F without imidazole), divided into aliquots, and flash-frozen in liquid N2. All purification procedures were carried out at 4°C.
Cloning and purification of DnaK
DnaK was cloned into a modified pET21d that contained a TEV protease site between the protein and the His-tag. The engineered plasmid expresses the protein containing an octa-His-tag at its N terminus that can be removed after treatment with TEV protease. In general, DnaK was purified in four steps: (1) purification on Ni-NTA agarose beads, (2) cleavage of the tag with TEV protease, (3) second Ni-agarose to remove uncut protein and the His-tagged TEV protease, and (4) gel filtration using a Superdex 200 column.
Protein concentration determination
The concentrations of Ssc1, mortalin, DnaK, and Mge1 were determined using the bicinchoninic acid protein assay with BSA as a standard. The concentrations of Tid1 constructs and Mdj1 were obtained through their absorbance at 280 nm and using the predicted molar extinction coefficient of the proteins. These values were determined using the ProtParam ExPASy proteomic tool (http://www.expasy.org/tools/protparam.html). The molar extinction coefficients (M−1 cm−1) at 280 nm were: 29,380 for Mdj1 and 31,330 for Tid1.
Formation of the Hsp70·ATP complex and single-turnover analysis
Mortalin, DnaK, or Ssc1 were incubated with 10 μCi [α-32P] ATP (3,000 Ci/mmol, Izotop) in buffer A (50 mM Tris acetate, 100 mM potassium acetate, 10 mM magnesium acetate, pH 7.4, and 2 mM DTT) containing 25-μM cold ATP. Mortalin and Ssc1 were incubated at room temperature for 5 and 2.5 min, respectively, while DnaK was incubated for only 2 s at 4°C. Then, the complex was isolated by size exclusion chromatography column (PD-10, Amersham) at 4°C, pre-equilibrated with buffer A; ∼250 μl fractions were collected. The radioactivity of the fractions was monitored via a β-counter (PerkinElmer). The first peak of radioactivity that contained the Hsp70/ATP complex was collected, divided into aliquots, and flash-frozen in liquid N2.
For single-turnover experiments, the complex was quickly thawed and 10–12 μl was added to an equal amount of buffer B (20 mM Na-HEPES, pH 7.4, 100 mM KCl, 100 mM NaCl, 10 mM MgCl2, and 2 mM DTT) containing various factors and incubated at 30°C. At the indicated time points, 3 μl of the reaction mixture was withdrawn and mixed with 3 μl buffer C (250 mM EDTA, pH 8, and 25 mM ATP) to stop the reaction. The mixture was loaded on a TLC membrane and developed for 30 min by using 0.5 M sodium phosphate, pH 3.5, as the mobile phase. The membranes were dried and exposed in BAS CASSETTE 2040 (FUJIX) overnight. The ADP and ATP bands were detected using phosphor imager (FLA-5100, FUJI) and quantified by the Image Gauge (version 4) program (FUJIFILM).
In vitro refolding of heat-denatured G6PDH
The refolding of heat-denatured glucose-6-phosphate dehydrogenase (G6PDH) was carried out as described previously (Diamant et al. 2000) with minor modifications. G6PDH (0.85 μM) from Leuconostoc mesenteroides (Sigma) was incubated for ∼15 min at 52°C in buffer A (100 mM Tris, pH 7.4, 100 mM KCl, 10 mM MgCl2, and 10 mM DTT). The remaining activity after the denaturation step was <2%. The stable aggregates were incubated for 10 min at 4°C before adding the ATP regenerating mixture (0.2 M glycine betaine monohydrate, 10 μg/ml pyruvate kinase, 5 mM phosphoenol pyruvate, and 4 mM ATP). Then the chaperone system was added (16 μM of the indicated Hsp70 proteins, 2 or 1 μM of the relevant J-domain protein, and 1 μM of the nucleotide exchange factor). The final concentrations of G6PDH and ATP after these dilutions were 0.64 μM and 3 mM, respectively. The entire mixture was incubated at 30°C, and G6PDH enzymatic activity was measured at room temperature as previously described (Hansen and Gafni 1993).
Steady-state ATPase measurments
The same conditions used in G6PDH refolding assays were applied to the steady-state ATPase measurements, but in the absence pyruvate kinase; 16 μM of the various Hsp70s, 2 or 1 μM of the relevant J-domain protein, and 1 μM Mge1 were incubated in buffer A (100 mM Tris–HCl, pH 7.4, 100 mM KCl, 10 mM MgCl2, 10 mM DTT, 0.2 M glycine betaine monohydrate, 5 mM phosphoenol pyruvate, and 3 mM ATP) in the presence of 0.64 μM G6PDH and 0.01 μCi [γ-32P] ATP (6,000 Ci/mmol, Izotop) at 30°C. Samples from the required time points were taken and separated on TLC as described for single-turnover experiments.
Miscellaneous
Mdj1 and yeast Mge1 were cloned and purified as described previously (Deloche et al. 1997; Horst et al. 1997).
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Sequence alignment of human mortalin with yeast mtHsp70s (Ssc1, Ssq1, and Ecm10) and E. coli DnaK. (DOC 842 kb)
Sequence alignment of human Tid1-L and Tid1-S, yeast mtHsp40 (Mdj1). and E coli DnaJ. (DOC 530 kb)
Purification steps of mortalin. (DOC 1,255 kb)
Purification steps of Tid1-L. (DOC 1,388 kb)
The ATPase activity of mortalin is not stimulated by Tid1-L H121Q, under single-turnover conditions. (DOC 52 kb)
Time-dependent reactivation of stable G6PDH aggregates by mortalin and mortalin-V482F. (DOC 56 kb)
Sequence of primers (DOC 36 kb)
Acknowledgments
We thank Zvi Fishelson and Moran Saar for plasmids of mortalin’s domains. A.A. is supported by the German-Israeli Foundation for Scientific Research and Development (GIF-1012/08) and Israel Science Foundation (452/09). O.I. was partially supported by a grant from Philip Morris International.
Footnotes
Ohad Iosefson and Shelly Sharon contributed equally to this study.
References
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2(9):e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann F, Milisav I, Neupert W, Herrmann JM. Ecm10, a novel hsp70 homolog in the mitochondrial matrix of the yeast Saccharomyces cerevisiae. FEBS Lett. 2000;487(2):307–312. doi: 10.1016/S0014-5793(00)02364-4. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Karnezis AN, Murphy SP, Hoang T, Freeman BC, Phillips B, Morimoto RI. Cloning and subcellular localization of human mitochondrial hsp70. J Biol Chem. 1995;270(4):1705–1710. doi: 10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- Blamowska M, Sichting M, Mapa K, Mokranjac D, Neupert W, Hell K. ATPase domain and interdomain linker play a key role in aggregation of mitochondrial Hsp70 chaperone Ssc1. J Biol Chem. 2010;285(7):4423–4431. doi: 10.1074/jbc.M109.061697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583(16):2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Burbulla LF, Schelling C, Kato H, Rapaport D, Woitalla D, Schiesling C, Schulte C, Sharma M, Illig T, Bauer P, Jung S, Nordheim A, Schols L, Riess O, Kruger R (2010) Dissecting the role of the mitochondrial chaperone mortalin in Parkinson’s disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum Mol Genet. doi:10.1093/hmg/ddq370 [DOI] [PMC free article] [PubMed]
- D’Silva P, Liu Q, Walter W, Craig EA. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat Struct Mol Biol. 2004;11(11):1084–1091. doi: 10.1038/nsmb846. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581(19):3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Castro IP, Martins LM, Tufi R. Mitochondrial quality control and neurological disease: an emerging connection. Expert Rev Mol Med. 2010;12:e12. doi: 10.1017/S1462399410001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA. 2006;103(16):6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena L, Coto E, Sanchez-Ferrero E, Ribacoba R, Guisasola LM, Salvador C, Blazquez M, Alvarez V. Mutational screening of the mortalin gene (HSPA9) in Parkinson’s disease. J Neural Transm. 2009;116(10):1289–1293. doi: 10.1007/s00702-009-0273-2. [DOI] [PubMed] [Google Scholar]
- Deloche O, Liberek K, Zylicz M, Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiae Mdj1p, the mitochondrial DnaJ homologue. J Biol Chem. 1997;272(45):28539–28544. doi: 10.1074/jbc.272.45.28539. [DOI] [PubMed] [Google Scholar]
- Deocaris CC, Widodo N, Ishii T, Kaul SC, Wadhwa R. Functional significance of minor structural and expression changes in stress chaperone mortalin. Ann N Y Acad Sci. 2007;1119:165–175. doi: 10.1196/annals.1404.007. [DOI] [PubMed] [Google Scholar]
- Deocaris CC, Kaul SC, Wadhwa R. From proliferative to neurological role of an hsp70 stress chaperone, mortalin. Biogerontology. 2008;9(6):391–403. doi: 10.1007/s10522-008-9174-2. [DOI] [PubMed] [Google Scholar]
- Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275(28):21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J. Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J Biol Chem. 2003;278(32):29719–29727. doi: 10.1074/jbc.M303527200. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66(4):840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem Sci. 2007;32(8):372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96(24):13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami AV, Chittoor B, D’Silva P. Understanding the functional interplay between mammalian mitochondrial Hsp70 chaperone machine components. J Biol Chem. 2010;285(25):19472–19482. doi: 10.1074/jbc.M110.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JE, Gafni A. Thermal switching between enhanced and arrested reactivation of bacterial glucose-6-phosphate dehydrogenase assisted by GroEL in the absence of ATP. J Biol Chem. 1993;268(29):21632–21636. [PubMed] [Google Scholar]
- Hinault MP, Farina-Henriquez-Cuendet A, Mattoo RU, Mensi M, Dietler G, Lashuel HA, Goloubinoff P (2010) Stable {alpha}-synuclein oligomers strongly inhibit chaperone activity of the HSP70 system by weak interactions with J-domain co-chaperones. J Biol Chem. doi:10.1074/jbc.M110.127753 [DOI] [PMC free article] [PubMed]
- Horst M, Oppliger W, Rospert S, Schonfeld HJ, Schatz G, Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16(8):1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iosefson O, Azem A (2010) Reconstitution of the mitochondrial Hsp70 (mortalin)-p53 interaction using purified proteins—identification of additional interacting regions. FEBS Lett. doi:10.1016/j.febslet.2010.02.019 [DOI] [PubMed]
- Kaul SC, Deocaris CC, Wadhwa R. Three faces of mortalin: a housekeeper, guardian and killer. Exp Gerontol. 2007;42(4):263–274. doi: 10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Koren J, 3rd, Jinwal UK, Lee DC, Jones JR, Shults CL, Johnson AG, Anderson LJ, Dickey CA. Chaperone signalling complexes in Alzheimer’s disease. J Cell Mol Med. 2009;13(4):619–630. doi: 10.1111/j.1582-4934.2008.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96(10):5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA. Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem. 2001;276(9):6112–6118. doi: 10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- Lu B, Garrido N, Spelbrink JN, Suzuki CK. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J Biol Chem. 2006;281(19):13150–13158. doi: 10.1074/jbc.M509179200. [DOI] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CA, Neupert W, Lamb DC, Mokranjac D. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell. 2010;38(1):89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Mitra A, Shevde LA, Samant RS. Multi-faceted role of HSP40 in cancer. Clin Exp Metastasis. 2009;26(6):559–567. doi: 10.1007/s10585-009-9255-x. [DOI] [PubMed] [Google Scholar]
- Mizzen LA, Chang C, Garrels JI, Welch WJ. Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp 58, a homolog of the bacterial groEL protein. J Biol Chem. 1989;264(34):20664–20675. [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18(24):6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Bross P, Winter VS, Corydon TJ, Bolund L, Bartlett K, Vockley J, Gregersen N. Misfolding, degradation, and aggregation of variant proteins. The molecular pathogenesis of short chain acyl-CoA dehydrogenase (SCAD) deficiency. J Biol Chem. 2003;278(48):47449–47458. doi: 10.1074/jbc.M309514200. [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77(2):249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial dysfunction in neurodegenerative disorders. Biochim Biophys Acta. 1998;1366(1–2):225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: an unfolding story. Curr Protein Pept Sci. 2009;10(5):432–446. doi: 10.2174/138920309789351930. [DOI] [PubMed] [Google Scholar]
- Sharma SK, De Los Rios P, Christen P, Lustig A, Goloubinoff P (2010) The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. doi:10.1038/nchembio.455 [DOI] [PubMed]
- Shi M, Jin J, Wang Y, Beyer RP, Kitsou E, Albin RL, Gearing M, Pan C, Zhang J. Mortalin: a protein associated with progression of Parkinson disease? J Neuropathol Exp Neurol. 2008;67(2):117–124. doi: 10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- Sichting M, Mokranjac D, Azem A, Neupert W, Hell K. Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 2005;24(5):1046–1056. doi: 10.1038/sj.emboj.7600580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26(1):27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syken J, De-Medina T, Munger K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci USA. 1999;96(15):8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvergiya C, Beal MF, Buck J, Manfredi G. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J Neurosci. 2005;25(10):2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowsky B, Major T, Knapp K, Voos W. The disaggregation activity of the mitochondrial ClpB homolog Hsp78 maintains Hsp70 function during heat stress. J Mol Biol. 2006;357(3):793–807. doi: 10.1016/j.jmb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Ikawa Y, Sugimoto Y. Identification of a novel member of mouse hsp70 family. Its association with cellular mortal phenotype. J Biol Chem. 1993;268(9):6615–6621. [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol Chem. 2005;386(8):739–744. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- Xie W, Wan OW, Chung KK. New insights into the role of mitochondrial dysfunction and protein aggregation in Parkinson’s disease. Biochim Biophys Acta. 2010;1802(11):935–941. doi: 10.1016/j.bbadis.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Zhai P, Stanworth C, Liu S, Silberg JJ. The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J Biol Chem. 2008;283(38):26098–26106. doi: 10.1074/jbc.M803475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21(17):4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Sequence alignment of human mortalin with yeast mtHsp70s (Ssc1, Ssq1, and Ecm10) and E. coli DnaK. (DOC 842 kb)
Sequence alignment of human Tid1-L and Tid1-S, yeast mtHsp40 (Mdj1). and E coli DnaJ. (DOC 530 kb)
Purification steps of mortalin. (DOC 1,255 kb)
Purification steps of Tid1-L. (DOC 1,388 kb)
The ATPase activity of mortalin is not stimulated by Tid1-L H121Q, under single-turnover conditions. (DOC 52 kb)
Time-dependent reactivation of stable G6PDH aggregates by mortalin and mortalin-V482F. (DOC 56 kb)
Sequence of primers (DOC 36 kb)