The article reviews the issues that must be considered when combining targeted therapies in early clinical drug development and proposes various novel trial designs that are logical for determining the efficacy of a drug or drug combination for personalized treatment.
Keywords: Phase I trials, Clinical trial design, Drug combinations
Abstract
Numerous practical issues must be considered when combining targeted therapies in early clinical drug development. These include tumor resistance mechanisms, the existence of multiple, redundant signaling pathways, and the failure of single-agent therapies to achieve cures. The strategies adopted to examine combinatorial therapy include the goal of hitting more than one target by specifically inhibiting signal transduction cascades and suppressing specific mechanisms of action with the use of multitargeted kinase inhibitors made possible by high-throughput screening techniques, combinatorial chemistry, and chemoinformatics. Two complex considerations are: which agents to combine given the heterogeneity of tumors and their various underlying perturbations, including secondary mutations and feedback loops, and how to translate findings from the bench to the bedside or directly from the bedside. Another consideration is: When is there enough information to provide a rationale for instituting a phase I trial? Various strategies have been used in combining molecules, including targeting diverse pathways, inhibiting upstream and downstream signals, and adopting a synthetic lethality paradigm. Other issues are: determining appropriate target populations for treatment, how to combine therapeutics with diagnostics, and the frequency of targets in patients referred to clinical trials. Here, we review these issues and we propose various novel trial designs that are logical for determining the efficacy of a drug or drug combination for personalized treatment. A difficult issue that must be answered is how many and which drugs to combine. Recent technologies, such as multiplexed assay platforms and bioinformatics, will shape the future of clinical trials and help answer these questions surrounding combinatorial treatment.
Introduction
The recent approval of targeted therapies, such as imatinib for the treatment of chronic myeloid leukemia and trastuzumab for human epidermal growth factor receptor 2 (HER-2)+ breast cancer, heralded the era of personalized cancer treatment [1, 2].
The first wave of these targeted therapies was directed at growth receptors or their downstream signals [3, 4]. It soon became apparent that single-agent application of these therapies, although sometimes showing remarkable antitumor effects, was still often not enough to eradicate disease. In addition, some of these treatments have had limited efficacy, and only a few of them have attained regulatory approval. What success there has been has, by and large, been observed primarily in subpopulations of patients with a given disease [5–7], such as erlotinib for mutated non-small cell lung cancer [8].
Resistance mechanisms are being explored, focused on the recognition that secondary mutations or other aberrations may abrogate the salutary effects of targeted agents. For example, KIT mutations that are associated with decreased drug binding may attenuate the beneficial effects of imatinib in gastrointestinal stromal tumors [9]; other aberrations may activate downstream proteins, such as phosphatidyl-inositol-3-kinase (PI3K), which then circumvent the effects of the targeted agent [10].
It now appears that, for many cancers, multiple, redundant aberrant signaling pathways are at play as a result of genetic perturbations at different levels [11]. These realities mandate that a combinatorial treatment approach is needed to cure malignancies. The complexity of safely combining numerous agents and matching diverse aberrations in tumors with the right combination of drugs is considerable. Although the preclinical data driving the use of specific drug combinations have been the subject of numerous papers, the practical issues that may ultimately lead to their success have received less attention and are the focus of this review.
First Question: Do I Really Need to Combine Agents?
Combining therapies has been successful in many areas of medicine, among them, hypertension, hypercholesterolemia, tuberculosis, AIDS, and cancer.
Hodgkin's disease exemplifies the successful use of combination therapy to achieve a cancer cure. As early as the 1970s, the combination of mustargen, vincristine, procarbazine, and prednisone was proven to be curative, whereas response rates to the individual agents were unimpressive. Superior response rates have also been achieved from combining 5-fluorouracil with radiation therapy for treating rectal cancer and combining different hormonal agents such as analogs of luteinizing hormone–releasing hormone and antiandrogens for prostate cancer or combined with antiestrogens for breast cancer in premenopausal women [12–14]. Combinations of molecularly targeted agents together with cytotoxic agents have also been shown to produce higher response rates than single agents. 5-Fluorouracil, leucovorin, and oxaliplatin plus bevacizumab has been effective in colorectal cancer [15], as has radiation therapy combined with cetuximab for head and neck cancer [16].
It seems rational, then, to design drug combinations for cancer treatment that will target various loci in underlying aberrant signal transduction pathways in order to enhance the antiproliferative effect of treatment. When two or more drugs are combined, and one drug does not influence the other, an additive effect may be produced; in contrast, if the drugs influence each other, a synergistic or an antagonistic effect, depending upon whether the overall outcome of the combination is more or less potent than the sum of the effect of either agent alone, may be observed. Table 1 classifies different possible outcomes of combining molecular therapies.
Table 1.
Possible outcomes of combining therapeutic modalities
Abbreviations: 5-FU, 5-fluorouracil; EGFR, epidermal growth factor receptor; HER-2, human epidermal growth factor receptor 2; PARP, poly ADP-ribose polymerase.
Adapted from Bentzen SM, Harari PM, Bernier J. Exploitable mechanisms for combining drugs with radiation: Concepts, achievements and future directions. Nat Clin Pract Oncol 2007;4:172–180.
“Dirty” Drugs or a Cocktail of “Clean” Drugs?
Several strategies evolved from both empirical and rational drug development with the goal of hitting more than one target.
One approach has been using highly selective drugs (“clean” drugs) designed to specifically inhibit signal transduction cascades. This group includes monoclonal antibodies, antisense oligonucleotides, any gene therapy strategy, and a minority of tyrosine kinase inhibitors, such as erlotinib and lapatinib (which are mainly selective for the tyrosine kinase domains of epidermal growth factor receptor [EGFR]-1 and HER-2).
At the other extreme are drugs that were rationally developed to inhibit a specific mechanism of action, irrespective of the selective consequences of this inhibition, rather than having a unique molecular effect. These include histone deacetylase inhibitors, proteasome inhibitors, heat shock protein inhibitors, demethylating agents, and others. Because these agents target multiple client proteins, their intercellular effects are highly unpredictable because they lack the ability to comprehensibly identify their cellular targets [17–19]. Current technology cannot address the entire proteome vis-à-vis these agents, yet because these drugs affect many cancer gene products, they have been validated as anticancer treatments.
Somewhere in the midst of these two extremes are two more strategies. One, the use of multitargeted kinase inhibitors (tyrosine and serine/threonine kinase inhibitors), is a result of either empirical, high-throughput screening of large compound libraries against one target or selective profiling against several or a full panel of protein kinases. Each compound may have a unique inhibition profile against many kinases and, in fact, these drugs have been called “promiscuous.” Drugs such as sorafenib were discovered using this approach.
On the other hand, combinatorial chemistry and chemoinformatics (computational chemistry) have allowed drugs to be engineered with rationally designed selectivity for specific kinases by composing the drug with different pharmacophores. An example is the efforts of Apsel et al. [20], aimed at inhibiting PI3K and the mammalian target of rapamycin (mTOR) by designing a drug that fills the “chemical space” between those two kinases. In an analogous approach, a drug with two specific domains was developed by producing bispecific antibodies [21, 22].
Although using multitargeted agents may seem a more pragmatic approach, a scientifically appealing approach is to use cocktails of “clean” drugs. This strategy has several theoretical advantages over single-agent therapy:
It can maximize cell kill while minimizing host toxicities by using agents with nonoverlapping dose-limiting toxicities.
It can increase the range of activity against tumor cells with endogenous resistance to specific types of therapy.
Finally, it may prevent or slow the development of newly resistant tumor cells [23].
In summary, the approach of combining highly specific agents can be more precisely targeted because one knows, at least to a degree, what is being inhibited, thus abrogating off-target effects such as toxicities. However, clinical experience so far suggests that the effects of multitargeted “dirty” drugs are as good or better than those of highly specific agents.
Okay, Let's Combine … What Do We Combine?
Although combinations of targeted therapies have a commonality with combinations of chemotherapeutic agents, targeted therapies often do not have a direct dose–response–toxicity correlation.
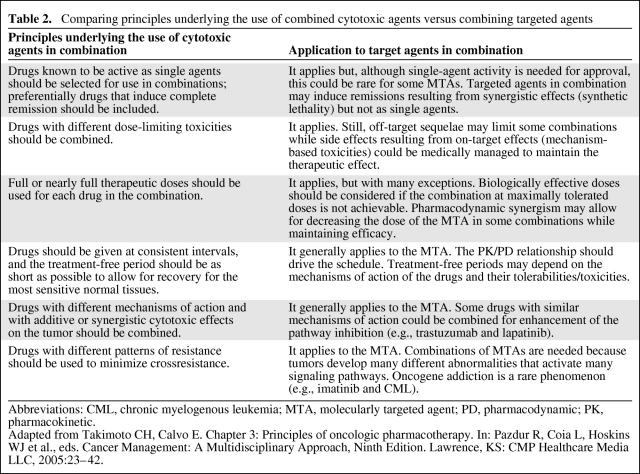
Thus, the recommended dose of a targeted therapy may be below the maximally tolerated dose and may be guided by target inhibition, that is, a biologically effective dose [24]. Table 2 provides a comparison of the principles underlying the use of combined cytotoxic agents versus combining targeted agents.
Table 2.
Comparing principles underlying the use of combined cytotoxic agents versus combining targeted agents
Abbreviations: CML, chronic myelogenous leukemia; MTA, molecularly targeted agent; PD, pharmacodynamic; PK, pharmacokinetic.
Adapted from Takimoto CH, Calvo E. Chapter 3: Principles of oncologic pharmacotherapy. In: Pazdur R, Coia L, Hoskins WJ et al., eds. Cancer Management: A Multidisciplinary Approach, Ninth Edition. Lawrence, KS: CMP Healthcare Media LLC, 2005:23–42.
Again: What to Combine?
The impetus underlying the combined inhibition of different targets generally springs from diverse early observations. A simple and pragmatic example is the observation that tumors are heterogeneous and have different mechanisms by which they can achieve a malignant status. As a consequence, two drugs inhibiting two different underlying pathways should theoretically be more effective than one, such as combining cetuximab with bevacizumab [25], or trastuzumab with bevacizumab [26].
Similarly, one observes that survival (antiapoptotic) mechanisms are frequently activated in cancer. Thus, inhibiting these pathways, for example, by activating apoptosis, inhibiting Akt, or modulating p53 expression or DNA repair, may sensitize cells to a cytotoxic or cytostatic agent. Given this hypothesis, it is rational to combine a poly ADP-ribose polymerase inhibitor with platinum drugs [27] or a mitogen-activated protein kinase (MAPK)/extracellular signal–regulated kinase (MEK) inhibitor with paclitaxel [28].
There is also a growing understanding that feedback loops that mediate escape or resistance mechanisms are activated once a signaling molecule is inhibited so that a reasonable approach is one that modulates this response by using a second agent, for example, insulin-like growth factor receptor (IGFR)-1 inhibition after mTOR inhibition [29, 30], a MEK inhibitor plus a PI3K inhibitor [31], an EGFR inhibitor with a hepatocyte growth factor receptor (MET) inhibitor [32], and so on.
Stemming from the observation that secondary mutations are at play as a mechanism of resistance, additional combination treatments can be generated by using two drugs aimed at those mutations, following the paradigm used in treating tuberculosis, and thus avoiding the development of resistance [33].
Another approach harnesses the concept of oncogene addiction and the empirical quest for inducing synthetic lethality via small interfering RNA (siRNA) libraries [34, 35]. When translating these propositions from theory to the clinic, the underlying rationale for combining any two or more drugs should be kept in mind so that mechanisms for testing such underlying hypotheses can be built into clinical trials.
This caveat can be effected through the following strategies:
Targeting diverse pathways to produce an additive effect through synergism, which may be achieved by inhibition at the receptor level (EGFR and vascular endothelial growth factor receptor [VEGFR], estrogen receptor and HER-2) or, alternatively, by effecting dual inhibition of parallel signaling pathways (i.e., inhibition of MAPK and mTOR) [36–39].
Superinhibition: Inhibiting a single receptor using two different strategies, such as combining a monoclonal antibody against the extracellular domain with small molecule inhibition of the tyrosine kinase domain (cetuximab plus erlotinib, trastuzumab plus lapatinib) [40]. Although this strategy evolved from the observation of incomplete signal transduction interruption with a single agent, the use of an antibody and a small molecule may have unexpected extra benefits [41], especially because recent data suggest that some receptor kinases, such as EGFR, also signal through nonkinase pathways [42].
Inhibiting both upstream and downstream signals. The goal of this strategy is to interrupt signaling at its origin (the receptor) as well as to disrupt the intermediate relays that amplify it (e.g., combining EGFR and mTOR inhibition) [43–45]. This approach has the benefit of overcoming known mechanisms of resistance—KRAS mutations and sensitivity to EGFR inhibition [5] and phosphatase and tensin homolog loss and PI3K mutations and resistance to HER-2–targeted therapies [46]. Additionally, two discrete cell populations may be targeted this way, for example, HER-2+ cells versus p95HER-2 cells and EGFR+ cells versus EGFRvIII cells [47–49].
Empiric combinations or “let the data talk” approach. This strategy is based on the fact that a clear understanding of how various molecular pathways interact is frequently lacking. An example is the use of a synthetic lethality paradigm, wherein cells are treated with an initial drug and a siRNA library is used to define potential targets for synergistic combinations. Alternatively, one can limit the plethora of possible empirical combinations by mathematical modeling, such as through directed discovery algorithms [50, 51] in which second generation combinations are built upon the results of a first set of testing, and so on.
Now, How Do I Proceed in the Clinic?
Before clinical testing, a drug must be evaluated preclinically, and information about its pharmacology (pharmacokinetics and metabolism, pharmacodynamics, and toxicology in at least two different species) and efficacy must be appraised by the U.S. Food and Drug Administration (FDA), a procedure that is, interestingly, not necessary for many drug combinations.
In fact, some phase II trials have been initiated with full doses of each of the compounds, without a run-in to assess the safety of the combination. This approach seems most feasible when the drugs have nonoverlapping toxicities or little toxicity, and is often safe and successful. However, at times, unexpected serious toxicities have been encountered, for example, several cases of microangiopathic hemolytic anemia in patients with solid tumors receiving bevacizumab and sunitinib were recently reported and the FDA shut down the trials with this combination [52]. Such toxicities could likely be avoided by starting with lower doses. Indeed, in our own trial of bevacizumab and sunitinib, using low doses, we saw no significant side effects (Kurzrock, unpublished data).
The ultimate arbiter regarding the need for preclinical studies for combination treatment seems to be the reigning culture among different academic centers and institutional review boards that govern decisions about whether there is sufficient rationale and presumptive safety to warrant testing a combination.
It appears clear that, for many drugs, the clinical experience with each compound provides sufficient data to safely combine the drugs in the clinic and to be a powerful predictor of efficacious rational drug combinations. Further, combining two or more drugs, each of which has activity in a specific type of cancer, often significantly improves response rates. Whether it does so because cumulative subsets of disease are impacted or because individuals with cancer have multiple aberrations, or both, remains unclear, and may vary from tumor to tumor.
When Do We Have Enough Information to Provide A Rationale for Instituting A Phase I Trial?
Various lines of evidence can address this question.
Literature Based or Based on Basic Science Theory
Combinations are based on the different pathways that are relevant to cancer and the way they interact, as viewed by basic science, for example, HER-2 activation upregulates VEGF so the combined inhibition of HER-2 and VEGF is likely to be effective [53]. The main problem with this approach is that responses to genetic alterations and pharmacological inhibition of the pertinent protein product can diverge considerably [35].
Empirical Approach
A plus B act synergistically in a panel of diverse cell lines (chemosensitivity assays) or as seen after screening a library of many combinations of compounds. The main difficulty with this approach is that the mechanism of action of targeted therapies may require testing in an animal model, rather than in isolated cell cultures, if the microenvironment or immunity is affected.
Observation-Based Approach
This approach is based on preclinical in vitro and in vivo tests of the candidate drugs in different tumor models (cell lines, xenografts, orthotopic tumors, primary cultures, and animal models). The sequence of thought is: when we treat with A preclinically, we observe upregulation of X. B is an inhibitor of X. Preclinically A plus B works synergistically. A clinical trial of A plus B is therefore justified (e.g., EGFR and IGFR inhibitors [54]). This is the most extensive approach, but currently, because most cell lines are well profiled, there is a great risk of biasing the results by selecting “adequate” models that “prove” the hypothesis, that is, asking the answer.
Highly Sophisticated Approach
The rationale for this strategy is based on transgenic mice harboring specific mutations or engineered models with conditional expression or suppression of target genes. The main problem for drug development is that this kind of model, which is ideal for describing mechanisms of action, is vastly removed from “real” tumors. There is a disjuncture between the bench and the bedside.
Pragmatic Approach
This approach is based on clinical experience more than on the results from preclinical assays. The progression of thought underlying this strategy is: A is active in a specific disease (at least in some cases). B is also active in the same disease (at least in some cases). A and B are well tolerated and do not appear to have overlapping toxicities, so A plus B should be more effective (or at least effective in more cases) and a trial is warranted (e.g., sorafenib plus temsirolimus in renal cell carcinoma [55]). The main difficulty with this approach is that there is the risk of missing potentially beneficial activity in diseases for which A or B is not active as a single agent. However, a significant advantage to this approach is that it is possible to impact the clinical setting in a short period of time.
Beginning the Clinical Trial: Feasibility and Clinical Trial Design
Once there is a strong rationale that supports clinical testing, two key parameters have to be considered for success.
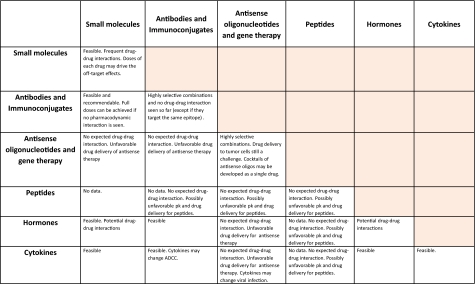
First is to select the most appropriate drugs for clinical testing, considering the pharmacology of each drug, routes of delivery, and potential drug–drug interactions (Fig. 1).
Figure 1.
Potential drug interactions.
Abbreviations: ADCC, antibody-dependent cell-mediated cytotoxicity; PK, pharmacokinetic.
The interaction between two given drugs could be a result of pharmacodynamic interaction or pharmacokinetics (via inhibition or induction of metabolic enzymes or transporters). The latter occur when one drug influences the absorption, distribution, metabolism, and/or excretion of another. Today, it is common practice and FDA mandatory [56] to test preclinically for potential drug interactions of novel combinations. Several in vitro methods assess drug interaction issues pertaining to transportation and enzyme inhibition and/or induction. The major paradigm shift in the field is associated with the use of modern human tissue preparations such as human liver slices, freshly isolated human liver cells, human hepatocyte primary cultures, subcellular fractions such as microsomes, cytosol, and S9 fractions, recombinant human enzymes (cytochrome P-450 and UDP-glucuronosyltransferase), transgenic cell lines, and cell-based reporter assays (reviewed in [57–59]).
Still, the prediction of in vivo drug interactions from in vitro metabolic data remains highly controversial because the in vitro data do not necessarily translate directly into relative extents of inhibition in vivo. The mentioned assays, therefore, are more likely to be useful to halt the further development of potentially problematic drug combinations if alternative ones are available.
Information regarding the interaction of drugs with monoclonal antibodies is scarce, and a formal assessment of this type of relationship is inherently complicated. Still, one may expect these interactions to be not clinically significant (reviewed in [60]).
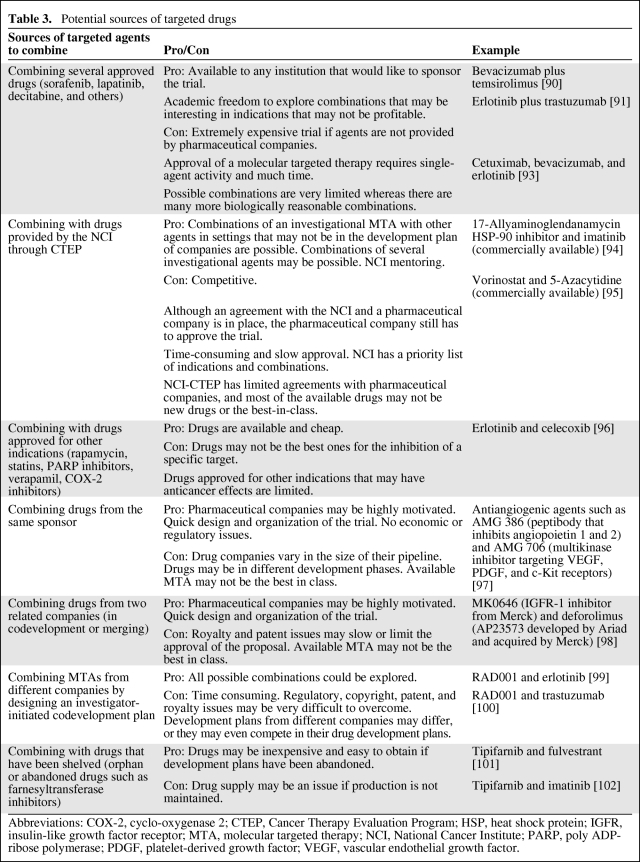
Obtaining the desired drug(s) for the trial can be problematic (Table 3) because there are limited options surrounding approved targeted therapies. For one, in order for a drug to be approved in the first place, significant activity has to be demonstrated, requiring time, and also it may be difficult to prove the activity of single agents. For combining investigational agents, the Cancer Therapy Evaluation Program of the National Cancer Institute has access to some of these experimental drugs and it has championed clinical trials combining targeted therapies [61].
Table 3.
Potential sources of targeted drugs
Abbreviations: COX-2, cyclo-oxygenase 2; CTEP, Cancer Therapy Evaluation Program; HSP, heat shock protein; IGFR, insulin-like growth factor receptor; MTA, molecular targeted therapy; NCI, National Cancer Institute; PARP, poly ADP-ribose polymerase; PDGF, platelet-derived growth factor; VEGF, vascular endothelial growth factor.
Trial design is another important consideration. The design of the clinical trial may be key for proof of concept (“hitting the target”) or may eliminate, early in development, combinations that will eventually fail.
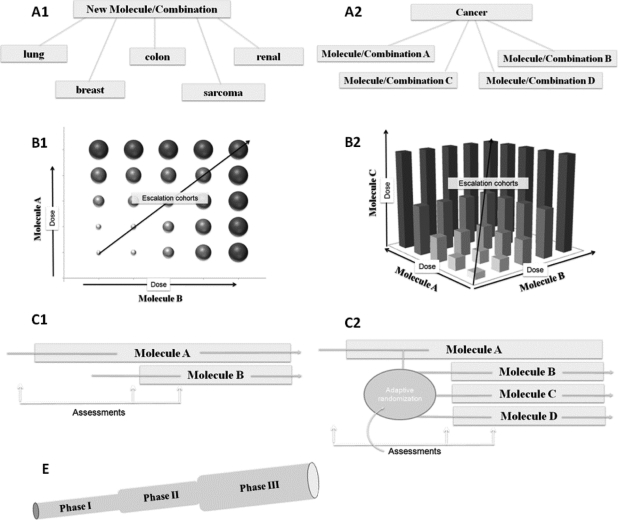
Since the realization that molecularly targeted therapies likely behave differently from classical cytotoxic chemotherapeutic agents, many proposals have advocated changing how early clinical trials are conducted when targeted therapies are being tested [62]. Some of these proposals have been incorporated into the design of phase I and phase II trials and have been proven to be successful. Still, the way that we perform combination trials has rarely changed, and few guidelines or proposals of novel designs for testing combinations have been discussed. In our organization, some of these proposals, with variations, are being considered for clinical trials (Fig. 2).
Figure 2.
Proposal of novel trial design. Some of these designs are nicknamed “octopus trials” (A1, A2, C2) because they can assess several options in a single study.
Broad Trial Indication Designated Finder with Multiple Cancers in One Study
This is a proposal for a phase I/II clinical trial in which drug activity in many different tumors is quickly assessed. Once the recommended doses of drugs are determined, multiple expansion cohorts, one for each indication, are opened, each of them analogous to the first part of a Simon two-stage design (Fig. 2A1). This can be performed with either single agents or combination therapy.
Multiple Agents Compared in a Single Study
This is also designated “complete phase I” and uses the same approach as above. Here, an array of different combinations is assessed. In this trial design, one drug could remain the same in each arm with the second drug being variable (Fig. 2A2).
Multiple Permissible Maximum-Tolerated Doses
This clinical trial design explores various possible combinations of dose levels. The model of Plimack and Berry (Fig. 2B1) is a two-dimensional escalation model that allows several cohorts to be opened at the same time if only one drug is escalated (high doses of drug A and low doses of drug B or low doses of drug A and high doses of drug B or intermediate doses of both drugs). Rodon's model (Fig. 2B2) is a three-dimensional extension of Plimack's schema, combining three drugs (e.g., bevacizumab, sorafenib, and temsirolimus) with rules based on mechanism-related toxicities required to stop escalation in one of the directions.
Synergy Finder
Here, the treatment leads in with drug A, and after assessment drug B is added (Fig. 2C1). Assessments at different time points (biomarker, efficacy, toxicity) are used to determine molecular synergy. If several drug candidates are to be evaluated, an arm for each combination is developed and patients are allocated by adaptive randomization based on such assessments (Fig. 2C2). For example, an EGFR inhibitor plus dasatinib versus an EGFR inhibitor plus a MET inhibitor, with the endpoint being overcoming resistance to EGFR inhibitors. A similar objective could be achieved with a phase 0 trial combining targeted therapies for biomarker testing (for this, the doses of both drugs of the combination must be well known, there must be no expectation of interaction between the drugs, and the biomarker must already have been validated) [63, 64].
Telescoped Trial
This is when a phase I trial leads to an embedded phase II trial that ultimately adds a control arm and becomes a phase III trial (Fig. 2E). Intermediate assessments of feasibility are incorporated at each stage (safety or biomarker based in the phase I stage and Simon's stopping rules in the phase II stage) [65].
Histology-Independent Clinical Trial
Patients are selected on the basis of the presence of a molecular marker, regardless of the anatomic origin of the tumor. This approach is based on the concept that tumors with a similar genetic background (and oncogene addiction) may respond similarly, and specific targeted therapies may be active for specific genetic backgrounds, for example, mutation or amplifications of PI3K-α (mutations are frequent in breast and bowel cancers, whereas amplifications are present in >50% of ovarian, cervical, and lung cancers) [66–68]. This approach is applicable to the study of single targeted agents as well as combination regimens.
Early clinical trials are becoming an arena for hypothesis testing, and ideas such as mechanism(s) of action and proof of concept, optimal biologic dose, and the incorporation of pharmacodynamic endpoints will need to be incorporated into their design. Whether these concepts change how we test drug combinations has yet to be determined.
Clinical Trials and Personalized Medicine
The ultimate design for demonstrating the efficacy of a drug or drug combination is the randomized controlled trial, which has important limitations. One can answer only a very limited number of questions per trial, such as the appropriate use of drugs or comparing various (but not many) drug regimens. Currently, in clinical trials, the regimens to be compared are preset and patients are assigned to different arms randomly, regardless of their genetic drivers.
“Personalized medicine,” on the other hand, refers to tailoring medical treatment to the individual characteristics of each patient or tumor [69]. It is based on the ability to classify individuals into subpopulations that differ in their responses to a specific treatment. Inherent in the concept of personalized medicine is the ability to distinguish in advance those patients who will benefit from a given treatment or drug combination. It is thus necessary to develop and validate markers for identifying patients who will benefit from particular interventions or to tailor therapeutic regimens to individual variations [70, 71]. Tailoring drug combinations to specific patient subpopulations requires designing clinical trials in which patients' tumors are biopsied and aberrant pathways, defined either by functional assays (gene expression profiling, proteomics, or metabolomics) or by determining particular genetic aberrations, are identified. Successful whole-genome sequencing is a near reality [72], thus defining the context of vulnerability [73] and the consequent ability to select a drug or drug combination with a high chance of efficacy given that context.
“Theragnostics” combines therapeutics with diagnostics [74], so that a diagnostic test that identifies patients most likely to be helped or harmed by a new medication is linked with a targeted drug therapy based on the test results. In oncology, there are few, but illustrative, examples of how to apply this therapeutic approach.
TargetNow is a pilot trial designed to identify the frequency of targets occurring in patients referred for phase I studies whose tumors have progressed on all standard therapies [75]. Patients' tissues are submitted for molecular profiling both by immunohistochemistry and oligonucleotide microarray modalities, identifying several potential targets for which a putative therapeutic agent is available. The Molecular Profiling Protocol trial (SCRI-CA-001, also known as Bisgrove Trial) [76] is a prospective clinical trial whose purpose is to validate this approach by comparing time on therapy (TOT) using a treatment regimen selected by molecular profiling with TOT for the most recent regimen on which the patient has just progressed.
The Biomarker integrated Approaches of Targeted Therapy of Lung cancer Elimination program is an “umbrella trial” plus four phase II clinical trials for patients with stage IIIB, stage IV, or advanced, incurable non-small cell lung cancer. Patients have biopsy samples taken for biomarker profile assessment before randomization. The expression levels of four types of biomarkers are assessed. Patients are then classified into one of the five marker groups (EGFR mutation/amplification, KRAS and/or BRAF mutation, VEGF and/or VEGFR expression, RXR and/or cyclin D1 expression, and none of the above). Patients are allocated to one of four regimens (with erlotinib, sorafenib, vandetanib, and the combination of erlotinib and bexarotene) following outcome-based adaptive randomization [77].
Whether or not these innovative proposals satisfy their primary endpoint(s), their novel designs evade the stagnation in clinical research produced by time-worn, large, and expensive randomized trials whose endpoints are miniscule gains in survival in large patient populations [78]. Their designs reflect a response to the increasingly obvious need for flexible trial designs whose objectives are to match patients with appropriate drug combinations. This is a far cry from the “one size fits all” randomized clinical trials, and they well might be early progenitors of a personalized medicine approach for cancer treatment.
The Third Wave
In 2006, Jose Baselga, in a perspective paper in Science, described how the experience with “first-generation” tyrosine kinase inhibitors (imatinib, trastuzumab, cetuximab, erlotinib) had guided the development of drugs such as Src, MEK, or PI3K inhibitors, the second generation of kinase inhibitors [79]. Given the complexity and heterogeneity of tumors and the crosstalk among multitudinous signaling pathways, it is reasonable to anticipate that combinations of drugs will be more potent than single-agent therapies.
One question is: How many drugs do we need to combine? To dedifferentiate adult cells into stem cells (thought to be analogous to the generation of cancer stem cells), two different labs modified four genes [80, 81]. Minn et al. [82] showed that four genes were necessary for invasion and metastasis in a breast cancer model and that those processes could be inhibited by combining three drugs (an anti-EGFR antibody, a matrix metalloprotease inhibitor, and a cyclo-oxygenase 2 inhibitor). Stommel et al. [11] showed that multiple kinases were coactivated in glioblastoma models and that combining three drugs (an EGFR inhibitor, a MET inhibitor, and the platelet-derived growth factor receptor/c-Kit/Abl kinase inhibitor imatinib) was highly efficacious.
The human kinome includes >90 proteins with tyrosine kinase domains [83], and many of these are frequently aberrantly activated in tumors by alterations at the genetic (DNA, RNA), proteomic, or epigenetic level, which is also true for alterations in other types of protein and lipid kinases. Despite these advances in delineating specific underlying mechanisms and perturbations of cancer, how best to inhibit multiple targets remains largely empirical, even with efforts aimed at identifying tumor subtypes and subpopulations of patients who would most benefit from these drugs and their incorporation into novel clinical trial designs.
Recent and continuing developments in high-throughput and multiplexed assay platforms as well as in disciplines such as bioinformatics and biostatistics will surely shape the future of clinical trials. Application of novel techniques in a comprehensive approach, revealing the interrelations among targets and the mechanisms of action underlying cancer (systems biology) [51, 84], may lead to comprehensive diagnostic tools (systems pathology) [85] and specific combinations of drugs (cocktails of monoclonal antibodies, RNA therapeutics, or others) in what has been called the actualization of personalized medicine. We know that momentum in the era of targeted therapy will continue to accelerate, bringing new hope to our patients with cancer and their families.
Acknowledgments
We thank Elisabeth Plimack and Don Berry for sharing with us their experience and new ideas on novel trial designs. We thank the NCI-CTEP team for their continuous support in the development of clinical trials combining targeted therapies. We also thank Judit Anido for reviewing this manuscript.
Author Contributions
Conception/Design: Jordi Rodon, Razelle Kurzrock, Jose Perez
Financial support: Razelle Kurzrock
Administrative support: Razelle Kurzrock
Collection and/or assembly of data: Jordi Rodon, Razelle Kurzrock
Data analysis and interpretation: Jordi Rodon
Manuscript writing: Jordi Rodon, Jose Perez
Final approval of manuscript: Jordi Rodon, Razelle Kurzrock
Joann Aaron, Investigational Cancer Therapeutics Department, M.D. Anderson Cancer Center, provided writing and editorial assistance.
References
- 1.O'Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 4.Tibes R, Trent J, Kurzrock R. Tyrosine kinase inhibitors and the dawn of molecular cancer therapeutics. Annu Rev Pharmacol Toxicol. 2005;45:357–384. doi: 10.1146/annurev.pharmtox.45.120403.100124. [DOI] [PubMed] [Google Scholar]
- 5.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 6.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 7.Seidman AD, Berry D, Cirrincione C, et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: Final results of Cancer and Leukemia Group B protocol 9840. J Clin Oncol. 2008;26:1642–1649. doi: 10.1200/JCO.2007.11.6699. [DOI] [PubMed] [Google Scholar]
- 8.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup phase III trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichhorn PJ, Gili M, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 12.Prostate Cancer Trialists' Collaborative Group. Maximum androgen blockade in advanced prostate cancer: An overview of the randomised trials. Lancet. 2000;355:1491–1498. [PubMed] [Google Scholar]
- 13.Bosset JF, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 14.Klijn JG, Beex LV, Mauriac L, et al. Combined treatment with buserelin and tamoxifen in premenopausal metastatic breast cancer: A randomized study. J Natl Cancer Inst. 2000;92:903–911. doi: 10.1093/jnci/92.11.903. [DOI] [PubMed] [Google Scholar]
- 15.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 16.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 17.Adams J. The proteasome: A suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 18.Goetz MP, Toft DO, Ames MM, et al. The Hsp90 chaperone complex as a novel target for cancer therapy. Ann Oncol. 2003;14:1169–1176. doi: 10.1093/annonc/mdg316. [DOI] [PubMed] [Google Scholar]
- 19.Mork CN, Faller DV, Spanjaard RA. A mechanistic approach to anticancer therapy: Targeting the cell cycle with histone deacetylase inhibitors. Curr Pharm Des. 2005;11:1091–1104. doi: 10.2174/1381612053507567. [DOI] [PubMed] [Google Scholar]
- 20.Apsel B, Blair JA, Gonzalez B, et al. Targeted polypharmacology: Discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiewe P, Hasmuller S, Kahlert S, et al. Phase I trial of the trifunctional anti-HER2 x anti-CD3 antibody ertumaxomab in metastatic breast cancer. Clin Cancer Res. 2006;12:3085–3091. doi: 10.1158/1078-0432.CCR-05-2436. [DOI] [PubMed] [Google Scholar]
- 22.Lu D, Zhang H, Koo H, et al. A fully human recombinant IgG-like bispecific antibody to both the epidermal growth factor receptor and the insulin-like growth factor receptor for enhanced antitumor activity. J Biol Chem. 2005;280:19665–19672. doi: 10.1074/jbc.M500815200. [DOI] [PubMed] [Google Scholar]
- 23.Bauer S, Yu LK, Demetri GD, et al. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer Res. 2006;66:9153–9161. doi: 10.1158/0008-5472.CAN-06-0165. [DOI] [PubMed] [Google Scholar]
- 24.Parulekar WR, Eisenhauer EA. Phase I trial design for solid tumor studies of targeted, non-cytotoxic agents: Theory and practice. J Natl Cancer Inst. 2004;96:990–997. doi: 10.1093/jnci/djh182. [DOI] [PubMed] [Google Scholar]
- 25.Saltz LB, Lenz HJ, Kindler HL, et al. Randomized phase II trial of cetuximab, bevacizumab, and irinotecan compared with cetuximab and bevacizumab alone in irinotecan-refractory colorectal cancer: The BOND-2 study. J Clin Oncol. 2007;25:4557–4561. doi: 10.1200/JCO.2007.12.0949. [DOI] [PubMed] [Google Scholar]
- 26.Pegram M, Chan D, Dichmann R, et al. Phase II combined biological therapy targeting the HER2 proto-oncogene and the vascular endothelial growth factor using trastuzumab and bevacizumab as first line treatment of HER2-amplified breast cancer. Presented at the 2006 San Antonio Breast Cancer Symposium; December 14–17, 2006; San Antonio, Texas. [Google Scholar]
- 27.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKeigan JP, Collins TS, Ting JP. MEK inhibition enhances paclitaxel-induced tumor apoptosis. J Biol Chem. 2000;275:38953–38956. doi: 10.1074/jbc.C000684200. [DOI] [PubMed] [Google Scholar]
- 29.O'Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, Harkavy B, Shen N, et al. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26:1932–1940. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 31.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah NP, Skaggs BJ, Branford S, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner NC, Lord CJ, Iorns E, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bukowski RM, Kabbinavar FF, Figlin RA, et al. Randomized phase II study of erlotinib combined with bevacizumab compared with bevacizumab alone in metastatic renal cell cancer. J Clin Oncol. 2007;25:4536–4541. doi: 10.1200/JCO.2007.11.5154. [DOI] [PubMed] [Google Scholar]
- 37.Carracedo A, Ma L, Teruya-Feldstein J, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemens M, Kaufman B, Mackey J, et al. Trastuzumab plus anastrozole may prolong overall survival in postmenopausal women with HER2-positive, hormone-dependent metastatic breast cancer: Results of a post-hoc analysis from the TAnDEM study. San Antonio Breast Cancer Symposium. 2007;46 [Google Scholar]
- 39.Johnston S, Pegram M, Press M, et al. First-Line Therapy With Lapatinib Combined with Letrozole vs Letrozole Alone for Postmenopausal Hormone Receptor Positive MBC: First Results from the Phase III Double-Bind EGF 30008 Trial. Presented at the 2008 San Antonio Breast Cancer Symposium; December 10–14, 2008; San Antonio, Texas. [Google Scholar]
- 40.Huang S, Armstrong EA, Benavente S, et al. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): Combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–5362. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- 41.Scaltriti M, Verma C, Guzman M, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 42.Weihua Z, Tsan R, Huang WC, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianco R, Garofalo S, Rosa R, et al. Inhibition of mTOR pathway by everolimus cooperates with EGFR inhibitors in human tumours sensitive and resistant to anti-EGFR drugs. Br J Cancer. 2008;98:923–930. doi: 10.1038/sj.bjc.6604269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck E, Eyzaguirre A, Brown E, et al. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol Cancer Ther. 2006;5:2676–2684. doi: 10.1158/1535-7163.MCT-06-0166. [DOI] [PubMed] [Google Scholar]
- 45.Ramalingam S, Forster J, Naret C, et al. Dual inhibition of the epidermal growth factor receptor with cetuximab, an IgG1 monoclonal antibody, and gefitinib, a tyrosine kinase inhibitor, in patients with refractory non-small cell lung cancer (NSCLC): A phase I study. J Thorac Oncol. 2008;3:258–264. doi: 10.1097/JTO.0b013e3181653d1b. [DOI] [PubMed] [Google Scholar]
- 46.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 47.Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 49.Scaltriti M, Rojo F, Ocaña A, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 50.Miller J, Zinner R, Barrett B. Santa Fe, New Mexico: Santa Fe Institute; 2005. Directed Discovery of Novel Drug Cocktails. Santa Fe Institute Working Paper No. 05–07-031. [Google Scholar]
- 51.Nelander S, Wang W, Nilsson B, et al. Models from experiments: Combinatorial drug perturbations of cancer cells. Mol Syst Biol. 2008;4:216. doi: 10.1038/msb.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.U.S. Food and Drug Administration. Microangiopathic Hemolytic Anemia (MAHA) in Patients Treated with Avastin® (Bevacizumab) and Sunitinib Malate. [accessed January 7, 2010]. Available at http://www.fda.gov/downloads/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm126434.pdf.
- 53.Wen XF, Yang G, Mao W, et al. HER2 signaling modulates the equilibrium between pro- and antiangiogenic factors via distinct pathways: Implications for HER2-targeted antibody therapy. Oncogene. 2006;25:6986–6996. doi: 10.1038/sj.onc.1209685. [DOI] [PubMed] [Google Scholar]
- 54.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patnaik A, Ricart A, Cooper J, et al. A phase I, pharmacokinetic and pharmacodynamic study of sorafenib (S), a multi-targeted kinase inhibitor in combination with temsirolimus (T), an mTOR inhibitor in patients with advanced solid malignancies. J Clin Oncol. 2007;25(18 suppl):3512. [Google Scholar]
- 56.Zhang L, Zhang YD, Zhao P, et al. Predicting drug-drug interactions: An FDA perspective. AAPS J. 2009;11:300–306. doi: 10.1208/s12248-009-9106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hariparsad N, Sane RS, Strom SC, et al. In vitro methods in human drug biotransformation research: Implications for cancer chemotherapy. Toxicol In Vitro. 2006;20:135–153. doi: 10.1016/j.tiv.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 58.Aszalos A. Drug-drug interactions affected by the transporter protein, P-glycoprotein (ABCB1, MDR1) I. Preclinical aspects. Drug Discov Today. 2007;12:833–837. doi: 10.1016/j.drudis.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 59.Gómez-Lechón MJ, Donato MT, Castell JV, et al. Human hepatocytes in primary culture: The choice to investigate drug metabolism in man. Curr Drug Metab. 2004;5:443–462. doi: 10.2174/1389200043335414. [DOI] [PubMed] [Google Scholar]
- 60.Seitz K, Zhou H. Pharmacokinetic drug-drug interaction potentials for therapeutic monoclonal antibodies: Reality check. J Clin Pharmacol. 2007;47:1104–1118. doi: 10.1177/0091270007306958. [DOI] [PubMed] [Google Scholar]
- 61.Goldman B. For investigational targeted drugs, combination trials pose challenges. J Natl Cancer Inst. 2003;95:1744–1746. doi: 10.1093/jnci/95.23.1744. [DOI] [PubMed] [Google Scholar]
- 62.El-Maraghi RH, Eisenhauer EA. Review of phase II trial designs used in studies of molecular targeted agents: Outcomes and predictors of success in phase III. J Clin Oncol. 2008;26:1346–1354. doi: 10.1200/JCO.2007.13.5913. [DOI] [PubMed] [Google Scholar]
- 63.Doroshow JH, Parchment RE. Oncologic phase 0 trials incorporating clinical pharmacodynamics: From concept to patient. Clin Cancer Res. 2008;14:3658–3663. doi: 10.1158/1078-0432.CCR-07-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kummar S, Kinders R, Rubinstein L, et al. Compressing drug development timelines in oncology using phase '0′ trials. Nat Rev Cancer. 2007;7:131–139. doi: 10.1038/nrc2066. [DOI] [PubMed] [Google Scholar]
- 65.Rogatko A, Schoeneck D, Jonas W, et al. Translation of innovative designs into phase I trials. J Clin Oncol. 2007;25:4982–4986. doi: 10.1200/JCO.2007.12.1012. [DOI] [PubMed] [Google Scholar]
- 66.Bertelsen BI, Steine SJ, Sandvei R, et al. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: Frequent PIK3CA amplification and AKT phosphorylation. Int J Cancer. 2006;118:1877–1883. doi: 10.1002/ijc.21461. [DOI] [PubMed] [Google Scholar]
- 67.Levine DA, Bogomolniy F, Yee CJ, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 68.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 69.Woodcock J. The prospects for “personalized medicine” in drug development and drug therapy. Clin Pharmacol Ther. 2007;81:164–169. doi: 10.1038/sj.clpt.6100063. [DOI] [PubMed] [Google Scholar]
- 70.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 71.Peck RW. Driving earlier clinical attrition: If you want to find the needle, burn down the haystack. Considerations for biomarker development. Drug Discov Today. 2007;12:289–294. doi: 10.1016/j.drudis.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Sundquist A, Ronaghi M, Tang H, et al. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS One. 2007;2:e484. doi: 10.1371/journal.pone.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Von Hoff DD, Gray PJ, Dragovich T. Pursuing therapeutic targets that are and are not there: A tumor's context of vulnerability. Semin Oncol. 2006;33:367–368. [Google Scholar]
- 74.Pene F, Courtine E, Cariou A, et al. Toward theragnostics. Crit Care Med. 2009;37(1 suppl):S50–S58. doi: 10.1097/CCM.0b013e3181921349. [DOI] [PubMed] [Google Scholar]
- 75.Von Hoff DD, Penny R, Shack S, et al. Frequency of potential therapeutic targets identified by immunohistochemistry (IHC) and DNA microarray (DMA) in tumors from patients who have progressed on multiple therapeutic agents. J Clin Oncol. 2006;24(18 suppl):3071. [Google Scholar]
- 76.U.S. National Institutes of Health. ClinicalTrials.gov. Molecular Profiling Protocol (SCRI-CA-001) [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00530192.
- 77.U.S. National Institutes of Health. ClinicalTrials.gov. BATTLE Program: Umbrella Protocol for Patients With NSCLC. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00409968.
- 78.Stewart DJ, Kurzrock R. Cancer: The road to Amiens. J Clin Oncol. 2009;27:328–333. doi: 10.1200/JCO.2008.18.9621. [DOI] [PubMed] [Google Scholar]
- 79.Baselga J. Targeting tyrosine kinases in cancer: The second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 80.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 81.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 82.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 84.van der Greef J, Hankemeier T, McBurney RN. Metabolomics-based systems biology and personalized medicine: Moving towards n = 1 clinical trials? Pharmacogenomics. 2006;7:1087–1094. doi: 10.2217/14622416.7.7.1087. [DOI] [PubMed] [Google Scholar]
- 85.Saidi O, Cordon-Cardo C, Costa J. Technology insight: Will systems pathology replace the pathologist? Nat Clin Pract Urol. 2007;4:39–45. doi: 10.1038/ncpuro0669. [DOI] [PubMed] [Google Scholar]
- 86.Bentzen SM, Harari PM, Bernier J. Exploitable mechanisms for combining drugs with radiation: Concepts, achievements and future directions. Nat Clin Pract Oncol. 2007;4:172–180. doi: 10.1038/ncponc0744. [DOI] [PubMed] [Google Scholar]
- 87.U.S. National Institutes of Health. ClinicalTrials.gov. AZD2281 and Carboplatin in Treating Patients With BRCA1/BRCA2-Associated, Hereditary, or Triple Negative Metastatic or Unresectable Breast Cancer or Ovarian Epithelial Cancer. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00647062.
- 88.Bourhis J, Rosine D. Radioprotective effect of amifostine in patients with head and neck squamous cell carcinoma. Semin Oncol. 2002;29(suppl 19):61–62. doi: 10.1053/sonc.2002.37349. [DOI] [PubMed] [Google Scholar]
- 89.Harari PM, Huang SM. Head and neck cancer as a clinical model for molecular targeting of therapy: Combining EGFR blockade with radiation. Int J Radiat Oncol Biol Phys. 2001;49:427–433. doi: 10.1016/s0360-3016(00)01488-7. [DOI] [PubMed] [Google Scholar]
- 90.U.S. National Institutes of Health. ClinicalTrials.gov. Study Comparing Bevacizumab + Temsirolimus Vs Bevacizumab + Interferon-Alfa In Advanced Renal Cell Carcinoma Subjects (INTORACT) [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00631371.
- 91.U.S. National Institutes of Health. ClinicalTrials.gov. Trastuzumab and Erlotinib as First-Line Therapy in Treating Women With Metastatic Breast Cancer Associated With HER2/Neu Overexpression. [accessed January 7, 2010]; Available at http://clinicaltrials.gov/ct2/show/NCT00033514. [Google Scholar]
- 92.Takimoto CH, Calvo E. Chapter 3: Principles of oncologic pharmacotherapy. In: Pazdur R, Coia L, Hoskins WJ, et al., editors. Cancer Management: A Multidisciplinary Approach. Ninth Edition. Lawrence, KS: CMP Healthcare Media LLC; 2005. pp. 23–42. [Google Scholar]
- 93.Lin CC, Calvo E, Papadopoulos KP, et al. Phase I study of cetuximab, erlotinib, and bevacizumab in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2009;63:1065–1071. doi: 10.1007/s00280-008-0811-x. [DOI] [PubMed] [Google Scholar]
- 94.U.S. National Institutes of Health. ClinicalTrials.gov. Imatinib Mesylate and 17-N-Allylamino-17-Demethoxygeldanamycin in Treating Patients With Chronic Myelogenous Leukemia. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00066326.
- 95.U.S. National Institutes of Health. ClinicalTrials.gov. Vorinostat and Azacitidine in Treating Patients With Locally Recurrent or Metastatic Nasopharyngeal Cancer or Nasal Natural Killer T-Cell Lymphoma. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00336063.
- 96.Reckamp KL, Krysan K, Morrow JD, et al. A phase I trial to determine the optimal biological dose of celecoxib when combined with erlotinib in advanced non-small cell lung cancer. Clin Cancer Res. 2006;12:3381–3388. doi: 10.1158/1078-0432.CCR-06-0112. [DOI] [PubMed] [Google Scholar]
- 97.The University of Texas MD Anderson Cancer Center. Study Summary: No. 2005–0794. [accessed January 7, 2010]. Available at http://utm-ext01a.mdacc.tmc.edu/dept/prot/clinicaltrialswp.nsf/Index/2005–0794.
- 98.U.S. National Institutes of Health. ClinicalTrials.gov. A Combination Study With Ridaforolimus (MK8669) and Dalotuzumab (MK0646) in Patients With Advanced Cancer. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00730379.
- 99.Johnson BE, Jackman D, Jänne PA. Rationale for a phase I trial of erlotinib and the mammalian target of rapamycin inhibitor everolimus (RAD001) for patients with relapsed non small cell lung cancer. Clin Cancer Res. 2007;13:s4628–s4631. doi: 10.1158/1078-0432.CCR-07-0717. [DOI] [PubMed] [Google Scholar]
- 100.U.S. National Institutes of Health. ClinicalTrials.gov. Trastuzumab and RAD001 in Patients With Human Epidermal Growth Receptor 2 (HER-2) Overexpressing Breast Cancer. [accessed January 7, 2010]. Available at http://clinicaltrials.gov/ct2/show/NCT00317720.
- 101.Li T, Christos PJ, Sparano JA, et al. Phase II trial of the farnesyltransferase inhibitor tipifarnib plus fulvestrant in hormone receptor-positive metastatic breast cancer: New York Cancer Consortium Trial P6205. Ann Oncol. 2009;20:642–647. doi: 10.1093/annonc/mdn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cortes J, Quintás-Cardama A, Garcia-Manero G, et al. Phase 1 study of tipifarnib in combination with imatinib for patients with chronic myelogenous leukemia in chronic phase after imatinib failure. Cancer. 2007;110:2000–2006. doi: 10.1002/cncr.23006. [DOI] [PubMed] [Google Scholar]