Abstract
The manuscript examines the role of isocitrate dehydrogenase 1 and the IDH1 gene in the prognosis and therapy of glial tumors.
Keywords: Glioma, Isocitrate dehydrogenase, Apoptosis resistance, Anticancer treatments
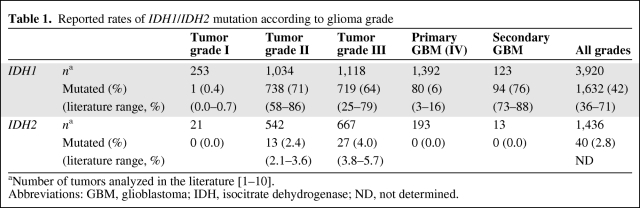
Despite intensive therapies, including surgery, radiotherapy, and chemotherapy, the outcome of glioma patients remains dismal, with a median overall survival time for patients with the most aggressive subtype (glioblastoma [GBM]) reaching only 15 months. Current research efforts focused on identifying genetic alterations implicated in GBM might help to define subgroups of patients with different prognoses and different responses to specific treatments. In this context, a recent genomewide mutational analysis conducted on 22 GBM samples identified a recurrent point mutation affecting codon 132 of the isocitrate dehydrogenase 1 gene (IDH1), located on chromosome locus 2q33. This mutation was present in 12% of GBMs and resulted in an Arg to His substitution in >90% of the cases [1]. IDH1 encodes cytosolic isocitrate dehydrogenase 1, which is involved in the control of oxidative cellular damage [1]. Subsequent sequencing studies on larger glioma patient cohorts have been conducted by our lab and others [2–7]. These studies showed that the IDH1 mutation is inversely associated with grade in diffuse glial tumors, affecting 71% of grade II, 64% of grade III, and 6% of primary glioblastomas (Table 1). It is of interest to note that IDH1 is markedly more mutated in secondary glioblastomas (76%), probably because these tumors are derived from lower grade gliomas [8, 9]. Moreover, the IDH1 mutation was tightly associated with a 1p19q codeleted genotype [5, 8]. To date, five different IDH1 mutations have been described, with the R132H mutation being the most prominent one (>90%).
Table 1.
Reported rates of IDH1/IDH2 mutation according to glioma grade
Moreover, mutation of IDH1 appears to be a very strong prognostic factor in diffuse gliomas, whatever the grade. Indeed, patients whose tumor harbored an IDH1 mutation had a significantly longer survival time than patients with a tumor of the same grade but wild type (wt) for IDH1. For example, in grade III tumors, the median survival time of patients with mutated IDH1 is about fourfold higher than that for patients with wt IDH1 (81.1 months versus 19.4 months, respectively). Multivariate analysis confirms that mutations in the IDH1 gene are independent factors of better outcome in patients with gliomas [5]. A few studies have also reported the frequency of IDH1 mutations in other malignancies: only rare mutations were detected in prostate carcinomas and B-acute lymphoblastic leukemia (<3%) [10]. In acute myeloid leukemia, Mardis et al. [11] reported that IDH1 mutations were found in 8% of samples and were associated with a normal cytogenetic status. No IDH1 mutation was reported in a broad range of other tumor types, including gastrointestinal stromal tumors, melanoma, and bladder, breast, colorectal, lung, ovarian, pancreas, and thyroid carcinomas [1, 3], and in brain metastases of colorectal cancers [12]. Taken together, these results suggest that mutations in IDH1 play a unique role in the pathogenesis of gliomas. Although rare, mutations of the related IDH2 gene were also detected in astrocytic and oligodendroglial gliomas lacking IDH1 mutations [7] (Table 1).
Mutations in IDH1 appear as a new molecular feature in gliomas, encouraging investigations of the role of cellular metabolic pathways in brain tumors. Indeed, very few proteins involved in these pathways have been implicated in the mechanisms of oncogenesis and tumor development. Only mutations in succinate dehydrogenase, fumarate hydratase, and N-acetylgalactosaminyltransferase 12 have been reported to play crucial roles in paraganglioma, leiomyosarcoma, and colon cancer, respectively. These studies underscore the idea that alterations in cellular metabolism could contribute to tumorigenesis [13–15]. These results raise a wealth of questions. What are the cellular and molecular consequences of IDH1 mutations in the context of brain tumors? What is the link between IDH1 mutation and better outcome? What role does IDH1 play in response to chemotherapy and/or radiotherapy? Should IDH1 status be used as a stratification factor in clinical trials?
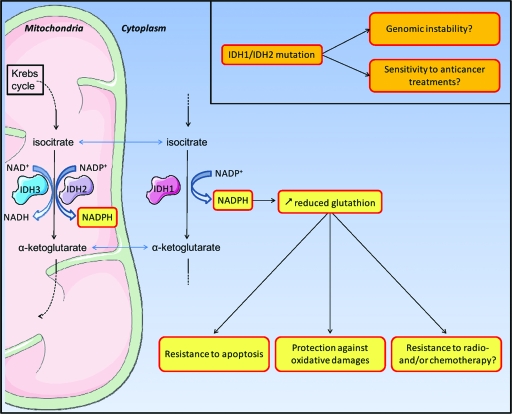
Whereas the functions of IDH1 have been extensively studied in yeast [16, 17], very few studies have aimed at characterizing human IDH1. The human genome possess five IDH genes, coding for three distinct IDH enzymes whose activities are either nicotinamide adenine dinucleotide phosphate (NADP+)-dependent (IDH1 and IDH2) or NAD+-dependent (IDH3). Whereas IDH2 an IDH3 are located in the mitochondria, IDH1 is located in the cytoplasm and in peroxisomes. IDH1 catalyzes the oxidative carboxylation of isocitrate to α-ketoglutarate, which allows the reduction of NADPH. The production of NADPH is essential for the regeneration of reduced glutathione, which functions as the main antioxidant in mammalian cells and promotes resistance to apoptosis [15, 18] (Fig. 1). The R132 residue, located in the active site of the enzyme, is conserved in all NADP+-dependent IDHs. This residue is involved in substrate binding, forming three hydrogen bonds with isocitrate, whereas other residues involved in isocitrate binding form no more than two hydrogen bonds [19]. Thus, mutation of the R132 residue may impair the interaction of the enzyme with isocitrate. This hypothesis was confirmed by the work of Yan and colleagues. Indeed, those authors demonstrated that IDH1 mutation results in the inactivation of the enzyme with a dominant negative effect on the IDH1 heterodimer [7, 8, 19]. This translates into a decrease in α-ketoglutarate, with, among other consequences, the inhibition of hypoxia-inducible factor 1 degradation [19]. Even more recently, Dang et al. [20] demonstrated that IDH1 mutation results not only in a loss of oxidative decarboxylation of isocitrate to α-ketoglutarate but also in a gain of enzyme function for the NADPH-dependent reduction of α-ketoglutarate to 2-hydroxyglutarate, with production of NADP+ (Fig. 2). This new enzymatic activity results in an accumulation of 2-hydroxyglutarate, which has been linked to a higher risk for developing brain tumors [21]. The accumulation of this metabolite also induces an increase in oxidative stress in the brain [22]. Under these conditions, both greater reduced glutathione, resulting from the decrease in NADPH, and greater 2-hydroxyglutarate may raise the oxidative stress level in mutant IDH1 tumor cells.
Figure 1.
Roles of IDH1/IDH2 in cellular resistance to apoptosis, oxidative stress and anticancer treatments.
Abbreviations: IDH, isocitrate dehydrogenase; NADP, nicotinamide adenine dinucleotide phosphate.
Figure 2.
IDH1 mutations result in both loss and gain of enzyme function.
A robust antioxidant system is particularly critical for tumor cell survival. Tumor cells evolve in an oxidizing microenvironment as a result of reactive oxygen species (ROS) generated by impaired vascularization and the resulting hypoxia [23]. Tumor development also creates local inflammatory conditions that participate in the production of ROS [24]. IDH1 plays a prominent protective role against oxidative damage induced by ROS via the regeneration of reduced glutathione. In this context, mutation of IDH1 appears paradoxical: on one hand, mutant IDH1 cells may be more sensitive to genetic instability caused by an oxidative environment, and thus IDH1 mutations may contribute to tumor development, and on the other hand, mutant IDH1 cells are less protected against oxidative cellular damage. Anticancer therapies are also a source of ROS, which contribute to the cytotoxic activities of these treatments. These effects have been extensively demonstrated for ionizing radiation and to a lesser extent for chemotherapy [25]. This may partly explain the long survival duration observed in patients with mutated IDH1. Given the broad range of therapeutic strategies used to treat these patients, IDH1 mutation may modulate the efficacy of antitumor treatments. To date, the impact of IDH1 mutations on both radio- and chemotherapy responses has not been reported. Preclinical studies are needed to determine whether IDH1 status could influence the antitumor activity of drugs and/or radiation. Greater therapeutic sensitivity is expected for IDH1 mutant cells than for wt IDH1 tumor cells. If this hypothesis is true, the antitumor efficacy of both conventional and targeted chemotherapy should be considered in regard to IDH1 status. For example, a drug evaluated in a nonselected population of patients with high-grade gliomas could be considered globally inefficient because it did not work on wt IDH1 tumors, but this agent may benefit the mutant IDH1 subgroup. Currently, the available data suggest that knowledge of the IDH1 status should be considered in the design of preclinical and clinical studies.
Finally, the close association between mutated IDH1 and a higher survival rate also suggests that IDH1 could be a relevant target in glioma treatment, especially in primary GBM, a tumor type in which IDH1 is rarely mutated. The challenge will be to design a therapeutic strategy able to inhibit the activity of IDH1. Such an approach could contribute to sensitize wt IDH1 tumor cells to the cytotoxic effects of both radio- and chemotherapy and to improve patient outcome.
Though many questions remain unanswered, IDH1 mutations appear to be very promising factors in both the diagnosis and treatment of glioma patients.
Acknowledgments
We are grateful to the Association pour la Recherche sur les Tumeurs Cérébrales (ARTC) and to the Ligue Nationale contre le Cancer for financial support.
Author Contributions
Conception/Design: Marianne Labussiere, Jean-Yves Delattre, Ahmed Idbaih, Marc Sanson
Data analysis and interpretation: Marianne Labussiere, Jean-Yves Delattre, Ahmed Idbaih, Marc Sanson
Manuscript writing: Marianne Labussiere, Jean-Yves Delattre, Ahmed Idbaih, Marc Sanson
Final approval of manuscript: Marianne Labussiere, Jean-Yves Delattre, Ahmed Idbaih, Marc Sanson
References
- 1.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balss J, Meyer J, Mueller W, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 3.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 5.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nobusawa S, Watanabe T, Kleihues P, et al. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 10.Kang MR, Kim MS, Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 11.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdhoff M, Parsons DW, Diaz LA., Jr Mutations of IDH1 and IDH2 are not detected in brain metastases of colorectal cancer. J Neurooncol. 2009;94:297. doi: 10.1007/s11060-009-9855-y. [DOI] [PubMed] [Google Scholar]
- 13.Guda K, Moinova H, He J, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci U S A. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 15.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin AP, McAlister-Henn L. Isocitrate binding at two functionally distinct sites in yeast NAD+-specific isocitrate dehydrogenase. J Biol Chem. 2002;277:22475–22483. doi: 10.1074/jbc.M202534200. [DOI] [PubMed] [Google Scholar]
- 17.Taylor AB, Hu G, Hart PJ, et al. Allosteric motions in structures of yeast NAD+-specific isocitrate dehydrogenase. J Biol Chem. 2008;283:10872–10880. doi: 10.1074/jbc.M708719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SM, Koh HJ, Park DC, et al. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: A case report and literature review. J Neurooncol. 2009;91:233–236. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- 22.Latini A, Scussiato K, Rosa RB, et al. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur J Neurosci. 2003;17:2017–2022. doi: 10.1046/j.1460-9568.2003.02639.x. [DOI] [PubMed] [Google Scholar]
- 23.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266:37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Yi J. Cancer cell killing via ROS: To increase or decrease, that is the question. Cancer Biol Ther. 2008;7:1875–1884. doi: 10.4161/cbt.7.12.7067. [DOI] [PubMed] [Google Scholar]