An update on the most recent data on promising biological prognostic and/or predictive markers in patients with colorectal cancer is provided.
Keywords: Colorectal cancer, Biomarkers, Prognostic, Overall survival, Disease-free survival
Abstract
Rapidly growing insights into the molecular biology of colorectal cancer (CRC) and recent developments in gene sequencing and molecular diagnostics have led to high expectations for the identification of molecular markers to be used in optimized and tailored treatment regimens. However, many of the published data on molecular biomarkers are contradictory in their findings and the current reality is that no molecular marker, other than the KRAS gene in the case of epidermal growth factor receptor (EGFR)- targeted therapy for metastatic disease, has made it into clinical practice. Many markers investigated suffer from technical shortcomings, resulting from lack of quantitative techniques to capture the impact of the molecular alteration. This understanding has recently led to the more comprehensive approaches of global gene expression profiling or genome-wide analysis to determine prognostic and predictive signatures in tumors. In this review, an update of the most recent data on promising biological prognostic and/or predictive markers, including microsatellite instability, epidermal growth factor receptor, KRAS, BRAF, CpG island methylator phenotype, cytotoxic T lymphocytes, forkhead box P3–positive T cells, receptor for hyaluronic acid–mediated motility, phosphatase and tensin homolog, and T-cell originated protein kinase, in patients with CRC is provided.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed type of cancer in men and women worldwide. An estimated 148,810 new cases of colon/rectum cancer were forecast to be diagnosed in the U.S. in 2008 [1], while 376,400 cases were forecast to be diagnosed in Europe in 2004 [2]. In addition, CRC continues to be one of the most common fatal types of cancer.
CRC develops slowly over several years and progresses through cytologically distinct benign and malignant stages of growth ranging from single crypt lesions through adenoma to malignant carcinoma with the potential for invasion and metastasis [3, 4]. Colorectal carcinogenesis is characterized by the successive accumulation of mutations in genes controlling epithelial cell growth and differentiation leading to genomic instability whereby widespread loss of DNA integrity is perpetuated [5]. The development of genomic instability is an important event in the multistep progression of colorectal carcinogenesis. Two apparently independent pathways of genomic instability have been identified. The suppressor pathway is found in the majority of cancers (85%) and these tumors have a molecular profile characterized by specific chromosomal amplifications and transformations, aneuploidy, and loss of heterozygosity (LOH). Cancers originating from the mutator pathway have a defective DNA mismatch repair mechanism (MMR), which allows mutations to be accumulated at many times the normal rate. This inability to repair DNA mismatches can easily be demonstrated because it results in cell-to-cell variability in the length of DNA microsatellites, called microsatellite instability (MSI) [6]. Colorectal malignancies demonstrating MSI have a very heterogeneous histological appearance, better prognosis, and altered response to chemotherapy and radiotherapy; see further below [7, 8]. In addition, in recent years, it has become apparent that promoter methylation is as important in shutting down tumor suppressor genes, as are the various mechanisms of somatic mutation. More than half of the tumor suppressor genes that are involved in familial cancer syndromes, because of germline mutation, have been found to be silenced in sporadic colorectal cancer by promoter hypermethylation. Careful characterization of the epigenetic factors, particularly promoter sequence methylation, has led to the definition of CpG island methylator phenotype (CIMP) cancer, which is proposed as a novel, third pathway [9].
Treatment of CRC consists of complete surgical removal of the primary tumor and the regional lymph nodes [10]. At the time of resection, approximately 30%–40% of patients with CRC are diagnosed with stage II disease [11]. Despite improvements in surgical techniques and the dosing and scheduling of adjuvant and neoadjuvant therapy, the 5-year survival rate for patients with early-stage CRC, that is, without invasion or lymph node metastasis, is about 90%, but this falls of to 65% for tumors with regional spread and to 10% for late-stage disease in which the cancer has metastasized to distant sites [12]. Prognostication relies on the stage or anatomic extent of disease based on the International Union Against Cancer tumor–node–metastasis (TNM) and American Joint Committee on Cancer staging classifications. The function of TNM staging has expanded from predicting prognosis to aiding in the choice of treatment [10, 12]. Accordingly, all node-positive cases (T3/4NposM0, stage III) receive adjuvant therapy while, despite several large randomized trials (Intergroup trials, Multicenter International Study of Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of Colorectal Cancer; Immediate Preoperative Arimidex®, Tamoxifen, or Combined with Tamoxifen; Surveillance, Epidemiology, and End Results; and National Surgical Adjuvant Breast and Bowel Project trials) conducted over the past decades, the value of adjuvant therapy for node-negative cases (T3/4N0M0, stage II) is controversial [10, 13]. There may well be a subgroup of patients with stage II CRC who would benefit from adjuvant chemotherapy [14]. Patients with stage IIB (T4N0M0) CRC have a worse prognosis than patients with stage IIIA (T1–2N1M0) CRC, although the latter are usually treated with adjuvant therapy. The predictive value of TNM staging is limited because even the outcome within each stage group is not homogeneous.
Indeed, CRC should be regarded as a heterogeneous, multipathway disease, an observation sustained by the fact that histologically identical tumors may have neither a similar prognosis nor a similar response to therapy [12, 15]. Therefore, particularly in stage II CRC, there is a need for markers capable of selecting those patients with aggressive disease that might benefit from adjuvant chemotherapy [16].
With recent developments in gene sequencing, molecular diagnostics, and targeted therapies, cancer treatment is beginning to move from the traditional “trial and error” approach to a position involving a personalized approach. There is clearly a need for strong and independent prognostic markers that can reliably differentiate patients into subgroups for which different treatment options, including the possibility of no adjuvant treatment, are appropriate. In addition, markers to prospectively predict response or resistance to specific therapies and markers to identify patients who are likely to develop severe toxic side effects from these specific treatments are warranted [16]. However, in practice, the distinction between prognostic and predictive factors is not straightforward, and many factors are a mixture of the two [17].
Consequently, in recent years a huge amount of research has been devoted to the study of new biological prognostic/predictive markers (Table 1). Several criteria must be met to ensure that a biomarker is clinically useful. In addition, evidence needs to be derived from multiple independent studies, which ideally should include a prospective trial. Most importantly, the marker concentration or status must be able to affect patient management [16]. Although hundreds of these markers have been proposed in the last two to three decades, the current reality is that no molecular marker, other than the KRAS gene in the case of epidermal growth factor receptor (EGFR)-targeted therapy for metastatic disease, has made it into clinical practice [16, 17].
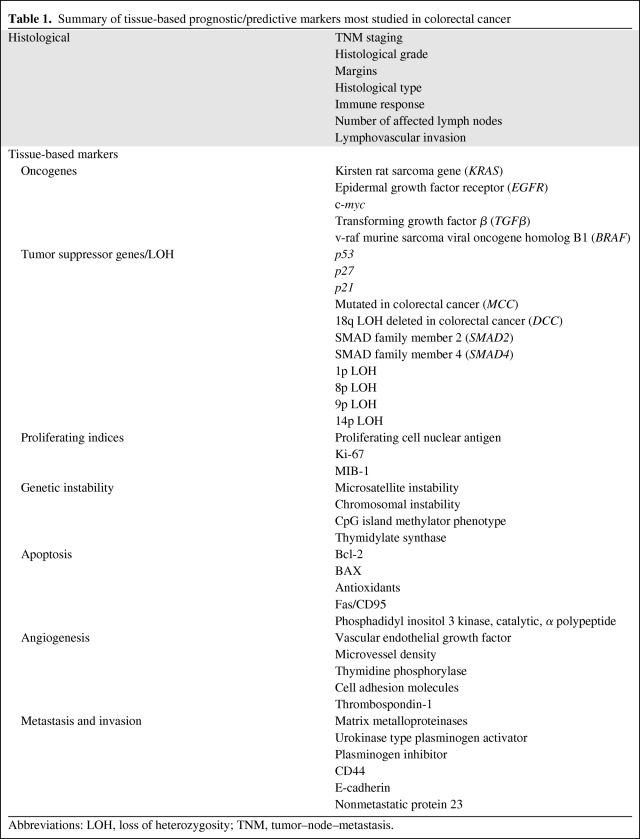
Table 1.
Summary of tissue-based prognostic/predictive markers most studied in colorectal cancer
Abbreviations: LOH, loss of heterozygosity; TNM, tumor–node–metastasis.
The aim of this review is to provide an update of the most recent data on biological prognostic and or predictive markers in patients with CRC that show promise in the clinic.
Tissue-Based Biomarkers
MSI
As mentioned above, MSI reflects the presence of a defective mismatch repair (MMR) mechanism and is characterized by somatic alterations in the size of simple repeat microsatellite nucleotide sequences common throughout the genome [18, 19]. As a consequence, genes containing simple repeat sequences, such as TGFβ RII, EGFR, or BAX, are often mutated in these tumors [20]. While 10%–15% of sporadic CRC cases display MSI, predominantly caused by epigenetic hypermethylation of the MLH1 mismatch repair gene, the majority of hereditary nonpolyposis colorectal cancer (HNPCC) tumors are characterized by this MSI phenotype. Individuals with a high-frequency MSI (MSI-H) tumor have phenotypically distinct features that are substantially different from those following the chromosomal instability pathway, though not all characteristics have consistently been demonstrated in all studies [21, 22]. MSI-H tumors tend to be more proximal, poorly differentiated, and mucinous, and show marked lymphocytic infiltration [23]. In addition, these tumors tend to retain the native diploid state [24]. These distinct features might be useful as diagnostic, predictive, or prognostic markers. In addition, MSI-H CRCs have different behavior patterns and responses to chemotherapy, and possibly different outcomes [25]. Therefore, MSI is considered one of the most promising markers studied to date.
MSI as a Prognostic Factor
Although several studies indicated that MSI is an independent prognostic factor in CRC, uncertainty remains. Controversial data have been reported (Table 2), leaving an area of uncertainty on the usefulness of this molecular marker to indicate a requirement for adjuvant therapy in clinical practice (as reviewed in [18, 20, 24, 26–34]). Graziano and Cascinu [18] focused on the role of MSI in early-stage sporadic CRC. Among the 26 selected studies, 17 retrospective investigations showed a significant association between MSI-H or abrogated hMLH1 and/or hMSH2 expression and a superior prognosis. However, available data were insufficient to support the prognostic role of MSI in Dukes' stage B CRC patients [18]. A recently published systematic review of MSI and CRC prognosis, pooling data from 32 studies and a total of 7,642 unselected patients, demonstrated a significant survival advantage for patients with MSI tumors, compared with patients with microsatellite stable (MSS) CRC [24]. The findings of that systematic review indicate that MSI status has the potential to identify patients who might be treated with surgery alone, particularly in those patients who already show indications of a favorable prognosis.
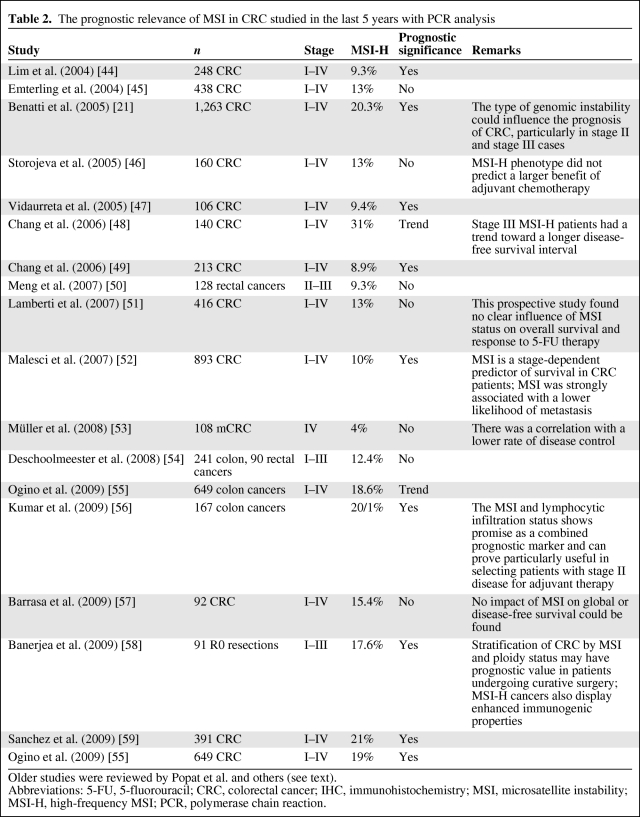
Table 2.
The prognostic relevance of MSI in CRC studied in the last 5 years with PCR analysis
Older studies were reviewed by Popat et al. and others (see text).
Abbreviations: 5-FU, 5-fluorouracil; CRC, colorectal cancer; IHC, immunohistochemistry; MSI, microsatellite instability; MSI-H, high-frequency MSI; PCR, polymerase chain reaction.
However, before MSI status can be routinely used to influence patient management, validation in the context of a prospective clinical trial is required. The relatively low frequency of MSI in sporadic CRC cases is a major limitation for planning such large prospective trials with MSI-based identification of high-risk patients [18]. Nevertheless, currently the impact of MSI and 18q LOH is being tested prospectively in stage II CRC patients (Intergroup E5202 trial, see below) [23]. In contrast, in the metastatic setting, MSI-H is not a common feature and does not seem to play a role in stratifying good versus poor prognosis in these patients [35].
MSI and Adjuvant Therapy: Predictive Potential
MSI-H has been associated with a favorable prognosis in most studies; however, it is not clear whether this is because MSI-H tumors are inherently less aggressive or because they are more sensitive to chemotherapy [36].
Since the early 1990s, 5-fluorouracil (5-FU) has been the mainstay of chemotherapeutic treatment for patients with CRC in the adjuvant setting. Nowadays, almost all adjuvant chemotherapy regimens involve the use of 5-FU, typically in combination with leucovorin and more recently with oxaliplatin [21]. Warusavitarne and Schnitzler [25] recently reviewed the role of chemotherapy in MSI-H CRC. In several studies, the impact of 5-FU chemotherapy in CRC patients led to a survival advantage for MSI+ disease [37–39]. However, contrary to this, most studies found that adjuvant chemotherapy with 5-FU itself did not benefit patients with tumors exhibiting MSI (as reviewed in [5, 31, 34, 36]) [40–42]. In addition, an overall lesser benefit from adjuvant therapy in MSI+ CRC patients could not be demonstrated in recent systematic reviews and a meta-analysis [24, 32, 36, 43]. There is evidence to suggest that different chemosensitivity exists in MSI-H tumors and MSS CRCs. The most likely mechanisms by which 5-FU resistance is conferred are possibly reduced thymidilate synthase activity and inability of the MMR genes to bind 5-FU–modified DNA [25]. Although the clinical evidence is conflicting, the in vitro data suggest a strong association between MSI-H and resistance to 5-FU. However, closer analysis of the clinical data appears to suggest no benefit from 5-FU treatment in patients with MSI-H CRC, which is supported by in vitro studies. The studies that showed a benefit from 5-FU chemotherapy in MSI-H CRCs relied on retrospective reviews of consecutive stage III CRC cases, for which no randomization to chemotherapy occurred. Because patients with MSI-H tumors have a better prognosis than those with MSS CRCs, analysis of nonselected patients results in a selection bias in the absence of appropriate controls [25].
Because the role of chemotherapy in patients with MSI-H tumors has not been well determined in the literature, more randomized controlled trials are required to evaluate its predictive potential in the adjuvant setting [25].
EGFR
EGFR Pathway
EGFR is a member of the transmembrane glycoprotein receptor tyrosine kinase family known as the ErbB or HER receptor family [60]. It is composed of an extracellular ligand-binding domain, a hydrophobic transmembrane region, and an intracellular tyrosine kinase domain. When activated, EGFR phosphorylates and activates other intracellular proteins that affect cell signaling pathways, cellular proliferation, and control of apoptosis and angiogenesis [60, 61]. A major downstream signaling route of the ErbB family is via the RAS–RAF–mitogen-activated protein kinase (MAPK) pathway. Another important target in EGFR signaling is phosphatidyl inositol 3-kinase (PI3K) and the downstream protein serine/threonine kinase Akt [62].
EGFR has been implicated in colorectal tumorigenesis, tumor progression, and metastasis, as reviewed in Lockhart and Berlin [63, 64]. Overexpression of EGFR has been described in up to 65%–70% of human colon tumors and has been associated with advanced stage disease [64]. In fact, the highest EGFR reactivity by immunohistochemistry (IHC) is seen in the deepest, most invasive regions of the tumor [65]. Differences in EGFR expression are also seen between stage II and stage IV tumors, implying that the pathway is potentially implicated in the progression of CRC to a more advanced stage [64]. Therefore, EGFR not only represents a possible prognostic marker in the adjuvant setting of primary tumors but primarily a rational molecular target for a new class of anticancer agents, especially in the setting of metastatic CRC (mCRC) [66–68].
EGRF as a Prognostic Factor
Based on the importance of EGFR in many aspects of CRC pathogenesis, a multitude of studies has evaluated the prognostic relevance of EGFR in primary tumors [60, 61, 68–74], but the impact of its expression on survival remains controversial, and a convincing link to poorer survival has not been demonstrated to date [75]. As reviewed by Spano et al. [62], some studies showed evidence of correlation between EGFR expression and advanced stage, worse histological grade, and lymphovascular invasion [76–78]. In contrast, other evaluations, including a recent large-scale study examining 249 CRC cases, have indicated no relation between EGFR status and histological type, tumor grade, stage, or survival [65, 68, 73, 74, 79]. In addition, Deng et al. [80] and Goldstein et al. [81] showed a trend toward shorter survival for patients with EGFR expression in metastatic lymph nodes in stage III and stage IV patients, suggesting that EGFR expression in metastatic lymph nodes may be more accurate in predicting survival than its expression in primary tissues or metastases at distant sites. The heterogeneity of the results may partly be attributed to noncomparable study populations, variability in laboratory protocols, fixation of tissue samples, choice of antibodies, and the lack of a uniform scoring system [82]. Additionally, the lack of a clear relationship between EGFR expression and prognosis is to be expected given that its activity, and therefore its influence on cancer cell survival, can be amplified by a number of mechanisms other than greater receptor expression [62].
Despite the lack of a definitive association between EGFR dysregulation and clinical outcome in the adjuvant setting, pharmacological inhibitors of EGFR have resulted in significant benefit in certain CRC patient populations [64].
EGFR-Targeting Strategies
In preclinical models, it was found that the inhibition of EGFRs had antitumor activity, and available data suggest synergy with both chemotherapy and radiotherapy [69]. EGFR inhibition can therefore be considered an attractive approach for cancer treatment. EGFR signaling can be targeted by either monoclonal antibodies (mAbs) (such as cetuximab and panitumumab), which interfere directly with receptor signaling, or tyrosine kinase inhibitors (TKIs) (such as gefitinib), which interfere with the catalytic activity of the cytoplasmatic domain and alter downstream signal propagation [62, 68, 69]. Nowadays, EGFR-targeted therapy is undergoing extensive clinical evaluation [83] and while TKIs overall generated disappointing results in CRC, mAbs have reached the clinic for mCRC. Cetuximab, a mouse chimeric IgG1 mAb, and panitumumab, a fully human IgG2 mAb, are clearly active agents and have been investigated in several clinical trials as single agents and in combination with chemotherapy for the treatment of recurrent or first-line mCRC as well as in the adjuvant setting [62, 83–85]. Results of these studies have demonstrated a manageable and acceptable toxicity profile and a promising level of activity [67, 83]. It is believed that the cellular mechanism of action of cetuximab is related to inhibition of cell cycle progression, increased apoptosis, inhibition of angiogenesis, and possibly amplification of antineoplastic cytotoxic effects of chemotherapy [86]. At present, the antibodies cetuximab and panitumumab are the only anti-EGFR therapies the U.S. Food and Drug Administration has approved for the treatment of patients with EGFR-expressing mCRC in combination with chemotherapy or as a single agent in patients who have failed oxaliplatin- and irinotecan-based therapy and who are intolerant to irinotecan [62, 83]. The European Medicine Agency (EMEA) has restricted the use of cetuximab and panitumumab to patients with EGFR-expressing, wild-type KRAS mCRC (see below).
KRAS
Biology of RAS
The RAS gene family encodes low molecular weight (21 kDa) membrane-associated, guanine nucleotide-binding proteins (p21) that function as binary molecular switches that control intracellular signaling networks and are involved in the control of cellular proliferation and differentiation by transduction of extracellular mitogenic signals [18]. Similar to other guanine-binding proteins, the RAS proteins switch between an active guanosine triphosphate–bound form and an inactive, guanosine diphosphate–bound form. The activation of RAS proteins is dependent on post-translational modification via prenylation, a critical step mediated by farnesyl transferase, which allows for RAS protein docking in the plasma membrane [87]. The action of the RAS proto-oncogene lies in the RAS–RAF–MAPK pathway downstream of EGFR and is a major pathway for tumor cell proliferation in CRC [88]. Oncogenic mutations of the RAS gene are prevalent in a large array of human cancers and are correlated with tumor type, although absolute specificity is not seen. The most commonly seen KRAS mutations occur at hot spots that are critical for KRAS regulation (codons 12, 13, and 62). Mutations in the KRAS oncogene result in constitutive activation, even in the absence of growth factor receptor–ligand binding, which is associated with unregulated proliferation and impaired differentiation [89, 90]. Overexpression of KRAS also plays a role in the ability of cells to metastasize, in part by increased production of proteases that degrade the extracellular matrix and promote angiogenesis [88]. KRAS mutations are believed to be early events in colorectal tumorigenesis and are noted in 40%–50% of CRC cases [30]. The functions of KRAS support its putative prognostic role in the adjuvant CRC setting, and several studies have been performed in recent years in this setting [18].
KRAS as a Prognostic Marker
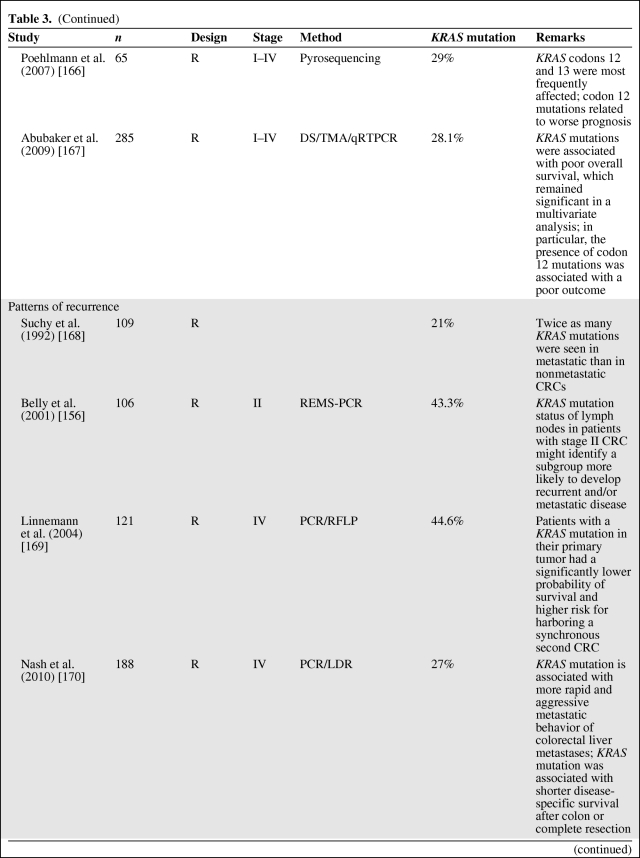
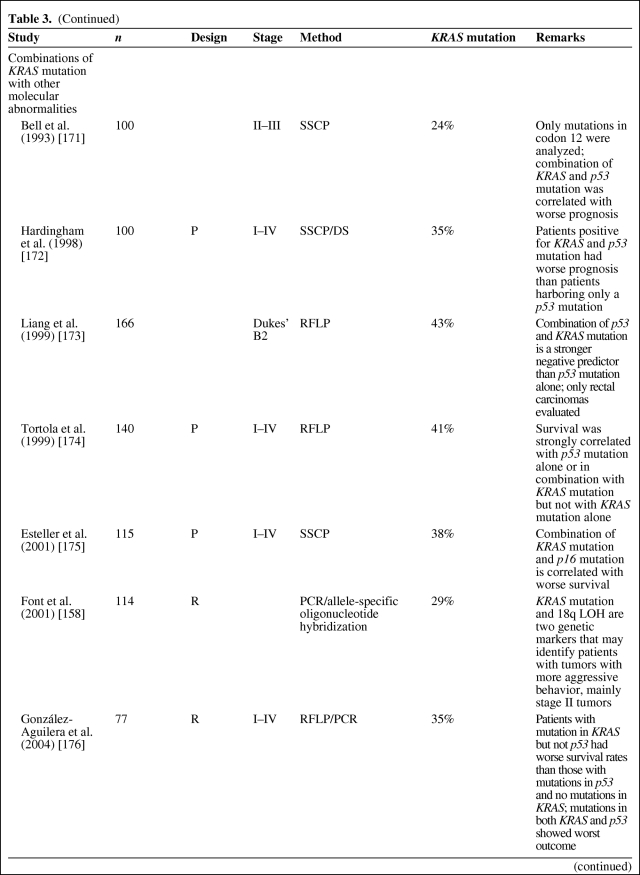
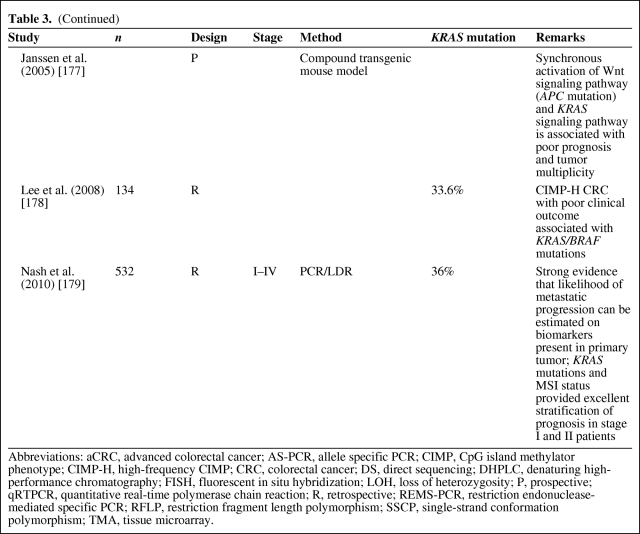
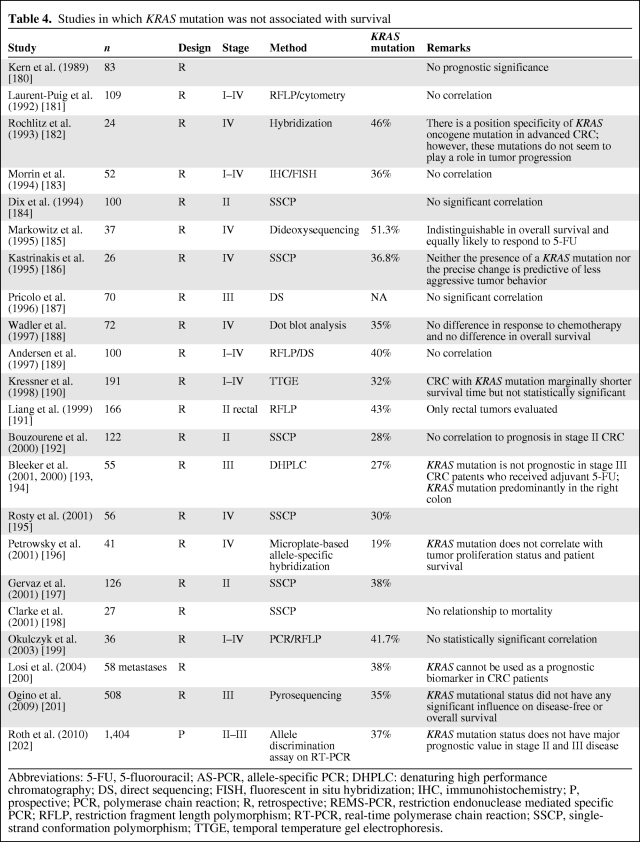
The prognostic significance of both KRAS mutations (by polymerase chain reaction [PCR]) and p21 staining (by IHC) has been assessed in a multitude of studies, with conflicting results (as reviewed in [19, 27–29, 91–93]). The majority of reported studies show KRAS mutation as an adverse prognostic indicator, indicating the need for adjuvant therapy, but these studies have wide variability in their specific results [19, 27, 91, 94]. As reviewed in [19, 27, 92, 95], some investigations have shown that KRAS mutation is prognostic only in some stages of the disease or only when associated with specific mutation types (transition or transversion, specific codons) [26], or when related to specific types of recurrence or in combination with other molecular abnormalities (p53 mutation) (Table 3). In contrast, the study of Conlin et al. [96] indicated that the presence of KRAS mutations predicts poor patient prognosis in CRC, independently of tumor stage. Elnatan et al. [97], Samowitz et al. [98], and Pajkos et al. [99] described tumor site–dependent (proximal versus distal) prognostic relevance of KRAS mutation. In contrast, a substantial number of reported studies has found no association between KRAS gene mutation and survival, either in isolation or in combination with other tumor suppressor genes [19, 27, 28, 94] (Table 4).
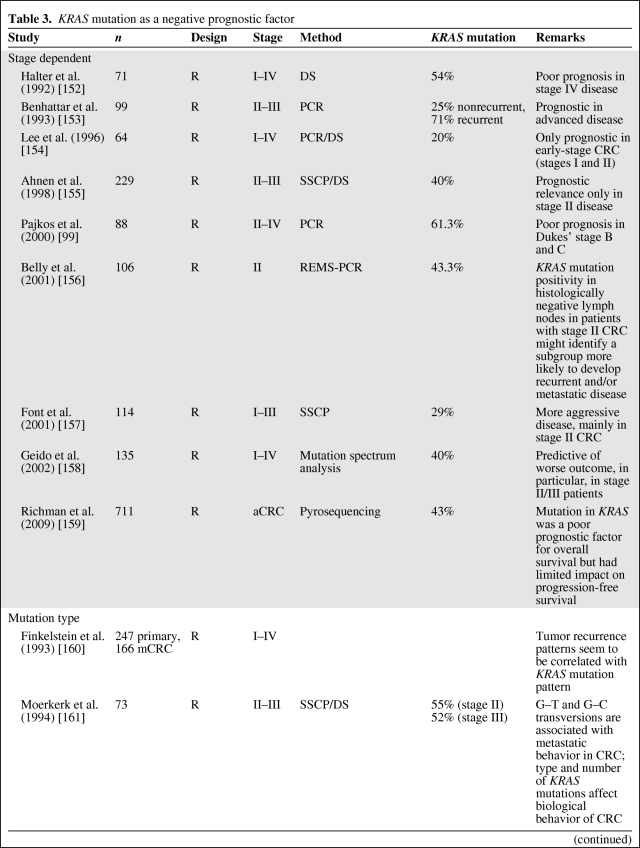
Table 3.
KRAS mutation as a negative prognostic factor
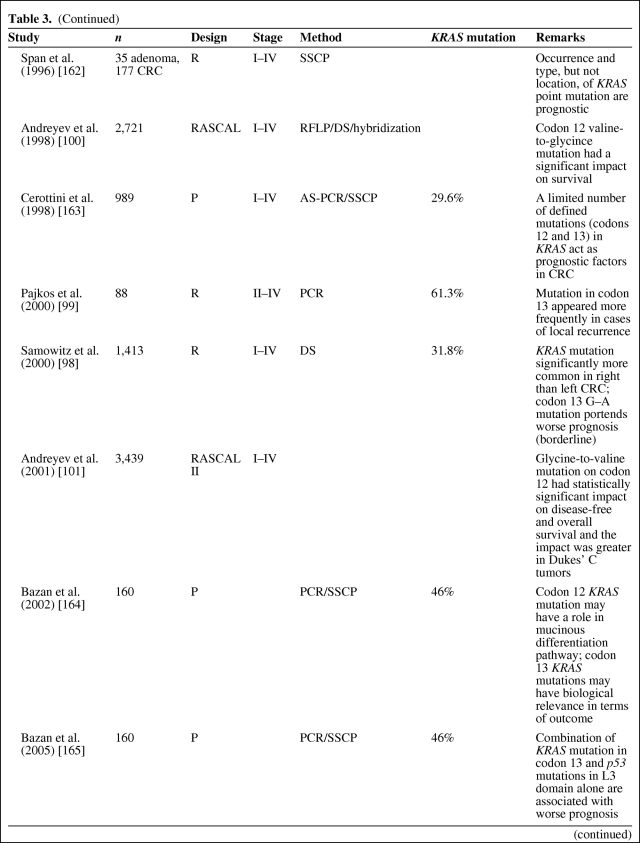
Table 3.
(Continued)
Table 3.
(Continued)
Table 3.
(Continued)
Table 3.
(Continued)
Abbreviations: aCRC, advanced colorectal cancer; AS-PCR, allele specific PCR; CIMP, CpG island methylator phenotype; CIMP-H, high-frequency CIMP; CRC, colorectal cancer; DS, direct sequencing; DHPLC, denaturing high-performance chromatography; FISH, fluorescent in situ hybridization; LOH, loss of heterozygosity; P, prospective; qRTPCR, quantitative real-time polymerase chain reaction; R, retrospective; REMS-PCR, restriction endonuclease-mediated specific PCR; RFLP, restriction fragment length polymorphism; SSCP, single-strand conformation polymorphism; TMA, tissue microarray.
Table 4.
Studies in which KRAS mutation was not associated with survival
Abbreviations: 5-FU, 5-fluorouracil; AS-PCR, allele-specific PCR; DHPLC: denaturing high performance chromatography; DS, direct sequencing; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; P, prospective; PCR, polymerase chain reaction; R, retrospective; REMS-PCR, restriction endonuclease mediated specific PCR; RFLP, restriction fragment length polymorphism; RT-PCR, real-time polymerase chain reaction; SSCP, single-strand conformation polymorphism; TTGE, temporal temperature gel electrophoresis.
However, as described by Klump et al. [28], if one considers only the five largest studies, including the RASCAL studies and the study by Samowitz et al. [98], prognostic relevance is reported by four of them. In the collaborative RASCAL studies [100, 101], researchers worldwide were invited by the Kirsten Ras in Colorectal Cancer Collaborative Group to share data on the clinical, histological, and outcome parameters of their patients with CRC in whom the KRAS status was known. This resulted in a collaborative database called “RASCAL.” In the first RASCAL study, the mutational status of the KRAS gene was analyzed in 2,721 patients collected from 22 centers from 13 different nations. In that study, the authors showed that mutations in the KRAS gene are important for the progression and outcome of established CRC, although some specific KRAS mutations (glycine–valine at codon 12) seem to have a more important prognostic role than others. The results of this collaborative study showed conclusively, for the first time, that different gene mutations have different impacts on outcome, even when the mutation occurs at the same site in the genome [100].
To explore the effect of KRAS mutations at different CRC stages, more patients were recruited to the database. The RASCAL II study analyzed data regarding 3,439 cases of CRC collected from 35 centers from 19 different nations, with a mean follow-up of 55 months. Thirty-five percent of the cases analyzed showed mutations in the KRAS gene, 26% of which were in codon 12 and 9% of which were in codon 13. About 9% of the overall mutations brought about the replacement of the amino acid glycine with valine in codon 12. No association was found between KRAS mutations and other clinicopathological variables. Multivariate analysis showed that advanced Dukes' stage, age, and the codon 12 glycine-to-valine mutation were significantly associated with a poorer prognosis. A separate analysis of the effects of codon 12 glycine–valine mutation in patients with Dukes' stage B or stage C tumors showed that the mutation brought about a significantly shorter disease-free interval and lower survival rate only in patients with Dukes' stage C tumors [100–102]. This collaborative study suggested that the presence of a codon 12 glycine-to-valine mutation is not only important for cancer progression but it may also predispose to more aggressive biological behavior in patients with advanced CRC.
Samowitz et al. [98] performed the first population-based study and largest nonmeta-analysis (1,413 individuals) of KRAS mutation in colon cancer patients. They showed that KRAS mutation, in general, was not associated with a higher cancer-related mortality rate. However, mutations in codon 13 were associated with a 40% greater likelihood of dying, although this difference was of borderline statistical significance. In addition, a small but statistically significant relationship between codon 12 KRAS mutation and tumor stage, proximal location, and male gender was demonstrated.
Interpretation of the various studies published on the prognostic role of KRAS might be difficult because of the different mutations of the KRAS gene that are being investigated, differences in data collection among the different studies, staging techniques, and different methodologies used to detect KRAS mutations [91]. Therefore, future studies on the prognostic role of KRAS mutations in CRC should be conducted prospectively with standardized assays to define those patients in whom routine mutation analysis could be of benefit for determining adjuvant treatment options [103]. Only a few studies have already been performed prospectively (Table 3).
Recently, the focus on KRAS has shifted from a prognostic to a predictive marker. Although KRAS mutations presently cannot be used to select patients who need adjuvant chemotherapy [104], they are strong predictive markers in mCRC patients treated with anti-EGFR mAbs [105].
Predictive Markers in Anti-EGFR Therapy: EGFR Expression, EGFR Amplification, KRAS Mutations, and Skin Rash
As mentioned above, cetuximab and panitumumab have been shown to be effective in mCRC treatment, either as monotherapy or in combination with cytotoxic agents [106]. Data (as reviewed in [62]) have shown no consistent association between tumor expression of the EGFR protein and response or survival following anti-EGFR–targeted therapy [107, 108]). The EGFR signaling pathway is complex, and it is possible that the level of expression of the receptor ligands, the level of tyrosine phosphorylation of the receptor, and the expression of other downstream molecules are critically involved in the action of cetuximab and therefore are more predictive of treatment response than the total level of the receptor per se. A higher EGFR gene copy number, as determined by fluorescence in situ hybridization, has been described in mCRC as being associated with a better response to anti-EGFR antibody treatment [109–113]. However, reproducibility, methodological concerns, and uncertainty on the clinical cutoff values published thus far currently prevent its routine use in clinical practice [106, 108–110, 112–117].
Whereas tumor EGFR expression or gene copy number may not be a reliable predictor of response, the appearance of treatment-related rash and, particularly, the presence of KRAS mutation might be of significance [62, 84, 105, 106, 118, 119].
Several retrospective analyses (Table 5) of tumor samples from mCRC patients receiving anti-EGFR antibody treatment have shown that patients with mutated KRAS did not benefit from this therapy (as reviewed in [83, 108, 120–131]), independently of EGFR expression status [62, 132, 133]. The first large study to confirm the negative predictive value of KRAS mutation was the pivotal randomized phase III study of panitumumab monotherapy in the relapsed or refractory setting [134]. In addition, KRAS data from large randomized phase II–III cetuximab studies—Cetuximab Combined With Irinotecan in First-Line Therapy for Metastatic Colorectal Cancer [107], Oxaliplatin and Cetuximab in First-Line Treatment of mCRC [135], National Cancer Institute of Canada Clinical Trials Group [136], and Evaluation of Velcade® Employed as Retreatment for Efficacy, Safety, and Tolerability [137]—have recently been published. The data from these studies strongly support that mutant KRAS is to be considered a predictive marker for anti-EGFR therapy resistance in mCRC. Lièvre et al. [106, 138] were the first to suggest that activating KRAS mutations could be responsible for acquired activation of the RAS–RAF–MAPK pathway independently of the ligand-induced activation of EGFR, leading to resistance to cetuximab treatment. This has been confirmed by others [134, 139], and the mutational status of KRAS is now routinely requested at the instigation of the EMEA to select mCRC patients who might benefit from the use of cetuximab and panitumumab [120]. This is also endorsed by the recent provisional opinion of the American Society of Clinical Oncology (ASCO), stating that if a KRAS mutation in codon 12 or codon 13 is detected then the patient should not receive anti-EGFR therapy as part of their treatment [130].
Table 5.
The role of KRAS mutation as a predictive marker in anti-EGFR therapy with monoclonal antibodies
Table 5.
(Continued)
Table 5.
(Continued)
Abbreviations: 5-FU, 5-fluorouracil; AREG, amphiregulin; COX-2, cyclo-oxygenase 2; CRC, colorectal cancer; DS, direct sequencing; ELISA, enzyme-linked immunosorbent assay; EGFR, epidermal growth factor receptor; EREG, epiregulin; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; MA, microassay; mAb, monoclonal antibody; MAPK, mitogen-activated protein kinase; mCRC, metastatic colorectal cancer; MSI, microsatellite instability; OS, overall survival; P, prospective; PCR-LCR, polymerase chain reaction-ligase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fraction length polymorphism; PFS, progression-free survival; PI3KCA, phosphatidylinositol 3-kinase α catalytic subunit; PTEN, phosphatase and tensin homolog; qPCR, quantitative polymerase chain reaction; R, retrospective; RTPCR, reverse transcriptase polymerase chain reaction.
However, only half of wild-type KRAS patients benefit from treatment, suggesting the need to identify additional biomarkers for anti-EGFR mAb-based treatment efficacy. Alterations in other EGFR effectors, including members of the RAS–MAPK or PI3K pathways together with alternative KRAS mutations (in codons 61 and 146), could drive resistance to anti-EGFR therapy and are currently being investigated [140–148]. Furthermore, Sartore-Bianchi et al. [149] described that when expression of phosphatase and tensin homologue (PTEN) and mutation of KRAS, BRAF, and PI3KCA are concomitantly ascertained, up to 70% of mCRC patients who are unlikely to respond to anti-EGFR therapies can be identified.
Additionally, acneiform rash is a class effect of EGFR-targeted agents and occurs frequently and in a dose-dependent manner in treated patients. Skin toxicity has previously been shown to correlate with clinical benefit (response rate, progression, and overall survival) in patients with advanced CRC receiving anti-EGFR antibodies [23, 62, 117, 131]. Of note, patients not developing a skin reaction may still achieve a tumor response. The mechanism underlying the correlation between skin toxicity and tumor response is currently unclear. However, some research groups have hypothesized that the rash is a surrogate indicator of an adequate degree of receptor saturation by cetuximab. Another hypothesis to explain the predictive value of skin toxicity is germinal genetic polymorphisms among individuals [62].
KRAS-Targeted Therapy
Because of its pivotal role in oncogenesis, various strategies have been developed to target KRAS for the treatment of human cancers. These strategies have ranged from inhibiting protein expression via antisense oligonucleotides to blocking post-translational modifications with farnesyltransferase inhibitors (FTIs) to inhibiting downstream effectors [150, 151].
A number of reviews have described (pre)clinical data obtained with FTIs, and although an extensive body of literature supports preclinical efficacy, they still do not represent a meaningful strategy to target KRAS for cancer therapy as yet [151].
Alternatively, immunological approaches are being investigated. Mutated KRAS proteins can be considered tumor specific and could therefore represent a highly specific technique to target cytotoxic T cells to tumors. Vaccination with mutant RAS peptides can stimulate an immune response that involves both CD4+ and CD8+ T lymphocytes, in both cancer patients and normal individuals. Full activation of T cells requires multiple signals, and it seems that mutant KRAS is not highly immunogenic in the native tumor setting based on the low frequency of immunoreactions in nonvaccinated patients. Therefore, these peptides are often given in conjunction with an immunostimulatory adjuvant. In another technique, isolated antigen-presenting cells are loaded with RAS peptides ex vivo before reintroduction into the patient. An alternative immunological approach to targeting mutant KRAS uses whole yeast expressing mutant proteins. Because whole yeast cells are typically recognized as pathogens, they can elicit a cell-mediated immune response directed against the recombinant protein. Yeast-based immunotherapy may represent an alternative to peptide-based vaccines directed at mutant KRAS, but the technique has not yet been tested in humans [151].
BRAF
BRAF is a member of the RAF kinase family that encodes kinases that are regulated by RAS and mediate cellular responses to growth factor signals [178]. Activating mutations in BRAF have been reported in 5%–15% of CRC cases, and >80% of all known mutations involve a thymine-to-adenine transversion in nucleotide 1799, which leads to a substitution of valine by glutamic acid at amino acid residue 600 (V600E), and this results in the upregulation of the RAS–RAF–MAPK pathway independently of KRAS mutation [178, 212, 213].
BRAF mutations are frequently found in MSI-H tumors, in which the MMR system is epigenetically inactivated [178]. Some groups speculate that the activation of BRAF is related to the inactivation of the MMR system [214]. However, alterations in the BRAF gene are not involved in the tumorigenesis of MSI-H tumors with germline mutations in hMLH1 and hMSH2 [215], and in addition they also occur in MSS tumors [216].
BRAF as a Prognostic and Predictive Factor
The presence of BRAF mutations in tumors has been characteristically associated with a worse clinical outcome, suggesting a need for adjuvant therapy in these circumstances [55, 142, 159, 217]. Whereas BRAF mutation is associated with worse survival in MSS tumors, the presence of high-grade CIMP (CIMP-H) appears to eliminate, at least in part, the adverse effect of BRAF mutation [55]. Ogino et al. [55] also found that the good prognosis associated with MSI-H tumors was abrogated in the presence of a BRAF mutation. In contrast, Samowitz et al. [218] and Roth et al. [202] found that BRAF mutations were associated with significantly shorter survival in patients with MSS tumors, but had no effect on the excellent prognosis of patients with MSI tumors. Therefore, it has been postulated that it is not the BRAF mutation itself that confers a poor prognosis but rather that the mutation has different effects depending on the type of genetic pathway in which it is produced [212]. In addition, patients whose tumors bear the BRAF V600E allele are not likely to experience significant benefit from either cetuximab or panitumumab treatment. Therefore, like KRAS mutation, BRAF mutation analysis presently cannot be used to select patients to receive adjuvant therapies but might be used as an additional predictive factor in the metastatic setting for the selection of patients who might benefit from EGFR-targeted mAb therapies [122, 143, 149, 219].
Although KRAS- and BRAF-encoded proteins are mutated in many of the same types of cancer, concomitant mutations are extremely rare. Intuitively, this is because both genes undergo gain-of-function mutations and thus represent different mechanisms of activating the same pathway. More recently, however, the most active (V600E) BRAF mutation was described together with a KRAS mutation in advanced CRCs and their lymph node metastases [220]. It seems that concomitant KRAS and BRAF mutations also increase the progression of MSS tumors, suggesting that the activation of both genes is likely to harbor a synergistic effect [221]. All these findings need to be further elucidated by additional independent prospective studies that should consider a joint examination of CIMP, MSI, KRAS mutation, and BRAF mutation to decipher the role of these molecular features in the biological and clinical behavior of tumors [55].
Epigenetics: CIMP
Epigenetics describes changes in phenotype or gene expression that do not involve DNA sequence changes. Among these, alteration in DNA methylation patterns is known to be a key component for the altered gene expression associated with human cancers [222]. Promoter CpG island hypermethylation acts as an important mechanism for inactivation of tumor suppressor genes and tumor-related genes [178]. Promoter CpG island hypermethylation and its associated histone modifications render the chromatin structure of a gene promoter into a closed compact structure inaccessible to transcription factors, which results in the inactivation of gene transcription [222].
Recent studies have demonstrated that promoter CpG island hypermethylation is more frequent than genetic changes in human CRCs, which suggests that promoter CpG island hypermethylation is a potential mechanism of colorectal carcinogenesis. In addition to two known molecular pathways in colorectal carcinogenesis, which involve chromosomal instability and MSI, a third epigenetic instability pathway, the promoter CIMP was proposed by the group of Dr. Issa [223]. CIMP+ CRCs are characterized by widespread hypermethylation of promoter CpG island loci, which results in the inactivation of the involved genes. A growing number of studies has consistently demonstrated a close association between CIMP-H CRC and proximal colon location, MSI, and a high frequency of BRAF mutation, regardless of the methodology and CIMP marker panels used [178, 224]. In contrast, a phenotype with less widespread promoter methylation, CIMP-low (CIMP-L), has not been well characterized. Promoter hypermethylation and silencing of the DNA repair gene O6-methylguanine-DNA-methyltransferase (MGMT) has been associated with G>A mutations and is a common event in colorectal tumorigenesis [225]. MGMT hypermethylation is associated with MSI-L tumors and KRAS mutations, and seems to be limited to CIMP-L tumors, supporting the suggestion that CIMP-H and CIMP-L might be different molecular phenotypes in CRC [226, 227].
CIMP as a Prognostic and/or Predictive Factor
Consequently, marked controversy exists over the prognostic implications of CIMP. Some studies suggest an adverse effect of CIMP on survival in CRC patients [55, 228–230], whereas other studies suggest little prognostic value for CIMP [231] or even a low cancer-specific mortality rate [55]. This issue is considerably complicated by the described associations between methylation and factors known to affect prognosis in CRC [59, 229].
Despite the close pathological similarity between MSI tumors and CIMP-H/MSS lesions, they have significantly different prognoses [59, 228]. Patients with CIMP-H/MSS tumors have a significantly worse outcome than patients with CIMP-0/MSS and MSI tumors [178, 227–229]. In addition, accumulating evidence suggests that the development of MSI may act as an antidote to the adverse prognostic effects of widespread methylation [55, 59, 178, 218, 228, 230, 232]. It is also controversial whether CIMP confers a survival benefit from chemotherapy in CRC patients. Van Rijnsoever et al. [233] provided evidence for the chemosensitivity of stage III tumors with aberrant DNA methylation. In contrast, Ogino et al. [227] and Shen et al. [229] suggested that CIMP may predict worse outcome among advanced CRC patients who receive 5-FU and irinotecan-based chemotherapy. Randomized trials are necessary to definitively assess treatment efficacy [34].
Lee et al. [178] and others [222] pointed out that the poor clinical outcome of patients with CIMP+/MSS tumors is closely associated with the presence of KRAS/BRAF mutation. Therefore, it is possible that the relationship between prognosis and CIMP-H tumors is actually a result of the relationship between prognosis and KRAS/BRAF mutation [34]. In addition, there was little prognostic value of CIMP in studies in which multivariate analyses were performed [55, 178, 218].
18q LOH/DCC
Biology of 18q
Another promising prognostic marker is allelic loss of chromosome 18q, which is highly prevalent in CRC [92, 234, 235]. The long arm of chromosome 18 contains several genes of potential importance in CRC pathogenesis and progression. Among the genes located on 18q are the DCC tumor suppressor gene, which codes for a neutrin-1 receptor important in cell adhesion and apoptosis; the SMAD4 gene, which codes for a downstream signal transducer in transforming growth factor (TGF)-β1 signaling involved in tumor suppression; and the SMAD22 gene, involved in endodermal differentiation [19, 92]. Most clinical studies on the DCC gene have examined 18q LOH by PCR amplification of polymorphic microsatellite markers at or near 18q21. Chromosomal loss in this region is thought to result in haploinsufficiency in DCC, and therefore reduced protein expression. However, the results of these studies are somewhat controversial [5].
18q LOH as a Prognostic Factor
In 1992, O'Connell et al. [236] suggested a possible relationship between 18q LOH in Duke's stage B and stage C tumors and poor survival. In 1994, Jen et al. [237] also showed that 18q LOH was associated with an adverse prognosis, suggesting a need for adjuvant therapy. Since then, several large series have accomplished the assessment of DCC/allelic variance of 18q and prognosis in resected CRC cases [238]. Although most subsequent studies have consistently agreed that 18q LOH and decreased DCC mRNA are associated with poor survival, estimates of the prognostic value have varied considerably among studies, as have the methods used (as reviewed in [18, 27–30, 92, 239, 240]).
An important meta-analysis of data concerning the prognostic significance of 18q LOH was performed by Popat et al. [239]. Those authors examined 27 studies (29 data sets) assessing survival by chromosome 18q allelic imbalance (AI) and DCC expression eligible for systematic review and meta-analysis. They found that, although different methods were used to assess chromosome 18q status, estimates of the frequency of AI were similar across all methods, including the rate of DCC loss of expression (LOE). Of the 27 studies, 17 provided suitable data to be included in the meta-analysis, that is, data from 2,189 patients for overall survival and from 683 patients for disease-free survival. Using these pooled estimates of outcome in CRC patients supported the notion that chromosome 18q AI and DCC LOE is a negative predictive factor for survival. Whereas CRC with chromosomal 18q AI/DCC LOE seem to be associated with a poorer prognosis, one caveat to this conclusion is that there was significant evidence for heterogeneity among studies because the optimal method and thresholds for assessing this phenotype are unclear at present. In addition, publication bias is a major concern in all forms of meta-analyses. Although there was some evidence of bias, 18q AI/DCC LOE still maintained its prognostic utility after correction. Popat et al. [239] concluded that their findings indicate that chromosome 18q AI/DCC status has the potential to define a group of patients who may benefit from adjuvant chemotherapy following potential curative surgery.
In addition, Locker et al. [19] performed a literature review on 18q LOH for the ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. Sixteen studies were considered to be of sufficient quality to evaluate. Although there was suggestive evidence of an association of 18q loss and survival, the small number of patients and the retrospective nature of the studies made it premature to use this marker as a prognosticator [19]. Moreover, the large prospective study of 555 patients with non-MSI-H CRC conducted by Ogino et al. [241] found no association between 18q LOH or AI and patient survival. Validation in the context of prospective clinical trials using consistent methodology is required before introducing assessment of this phenotype routinely into CRC patient management strategies.
Prospective Clinical Adjuvant Trial
At this time, retrospective analysis of tumor biologic laboratory observations has been the dominant research methodology [39, 242]. In stage III colon cancer, a retrospective analysis by Watanabe t al. [39] and O'Connell et al. [242], both using formalin-fixed, paraffin-embedded material, suggested a statistically significant difference in survival favoring patients who retained the 18q allele and those who demonstrated MSI and a TGF- βRII mutation. These results and those from other investigators support the design of the current Gastrointestinal Intergroup stage II study (E5202 trial) coordinated by the Eastern Cooperative Oncology Group. The current E5202 stage II adjuvant colon cancer trial is a randomized study in which a total of 3,610 patients will be stratified according to stage and tumor biological characteristics (MSI and 18q LOH) to determine treatment strategy [243–245]. This is the first trial in colon cancer to use prognostic and predictive markers prospectively [19, 23, 235, 243, 245]. Stage II patients enrolled in the trial will have tumor block assessment for MSI and LOH on 18q to determine their risk for tumor recurrence and their likelihood of benefiting from adjuvant 5-FU. Patients at low risk—those with retention of 18q alleles or high levels of MSI—will be observed; on the other hand, patients at high risk—those with MSS or low levels of MSI with 18q LOH—will be assigned randomly to receive 5-FU, leucovorin, and oxaliplatin with or without bevacizumab.
Although this study is powered to compare the 3-year disease-free survival rates of stage II patients randomized into two chemotherapy arms, one of the secondary objectives is to prospectively determine the impact of tumor biological characteristics on the survival of patients with stage II colon cancer (http://clinicaltrials.gov/ct2/show/NCT00217737) [235]. Although of interest, there are limitations to the study, that is, the addition of a control arm in which patients with an MSS/18q-stable tumor are stratified to receive postoperative 5-FU–based chemotherapy would be necessary to establish the prognostic value of MSI and 18q LOH [92]. First results are expected in April of 2011.
Host Lymphoid Response to Tumor
Biology of Cytotoxic T lymphocytes in CRC
Colorectal tumors become clinically malignant after cancer cells invade through the muscularis mucosa into the submucosa. With the invasion, various immune/inflammatory responses by competent immune effector cells, critically involved in the protection of the host organism against cancer cells, take place. These responses are more concentrated along the invasive margin [246, 247]. Tumor-associated antigens, expressed as a consequence of genetic alterations, are exposed on the tumor cells in association with human leukocyte antigen (HLA) class I proteins and can be recognized by cytotoxic T lymphocytes (CTLs) via T-cell receptors, finally resulting in a cellular immune response effective in limiting tumor growth and spread [86, 247]. Immune cells are present in the tumor stroma at the periphery of the tumor and occasionally invade cancer cell nests. CD8+ CTLs and the CD4+ T helper lymphocytes represent the adaptive, or specific, component of the immune response. CD4+ T cells mainly produce cytokines, like interleukin (IL)-2 and interferon-α, which influence the type of immune response, whereas CD8+ T cells produce perforin and granzyme B, which are cytotoxic to their target cells [248]. Consequently, CTLs are able to perform tumor-specific recognition and can mediate specific destruction of tumor cells [249, 250]. Koch et al. [251] were the first to show functional reactivity of tumor-infiltrating T cells against antigens in CRC patients. In addition, they demonstrated, for the first time, tumor-selective activation and cytotoxic activity in situ of tumor-infiltrating CD8+ T cells and tumor-selective migration of CD4+ T helper cells in CRC.
However, the induction of CTL responses takes time, leaving time for tumor cells to escape the immune system. Therefore, natural killer (NK) cells from the innate immune system, which are not HLA restricted, may also play an important role because these cells can lyse NK-sensitive tumor targets prior to antigen sensitization or clonal expansion. In addition, NK cells express several ligands of the tumor necrosis factor family and can induce apoptosis of malignant cell targets that are phagocytosed by dendritic cells and macrophages and processed for subsequent presentation to T cells. Furthermore, NK cells constitutively express the IL-2 receptor and are able to respond to IL-2 stimulation that results in augmented cytotoxic activity [252]. Nevertheless, tumors are still capable of escaping immune recognition/destruction by several mechanisms [253–255]. The potential influence of these immune-cell infiltrates in CRC on the prognosis of patients has been investigated in several studies, but remains controversial. A greater understanding of the role of the host immune response in influencing the natural history of CRC might have important implications for risk stratification and the development of adjuvant immune-based therapies [248, 256].
CTLs and Prognosis
There is accumulating evidence showing a positive correlation between the number of tumor-infiltrating lymphocytes (TILs) and longer patient survival in CRC [257]. Jass et al. [258] pointed out that conspicuous infiltration along the invasive margin of rectal cancer is an independent prognostic factor for longer survival in a multivariate analysis. Ropponen et al. [259] confirmed the concept of a prognostic impact of TILs in CRC and showed an inverse correlation between the presence of TILs and tumor stage. Several subsequent studies also demonstrated that the infiltration of CD8+ T cells around and within tumor stroma contribute to a better prognosis [260–268]. Galon et al. [269] showed that the type, density, and location of immune cells in CRC had a prognostic value that was superior to TNM classification. The assumption that not only the number of CD8+ TILs but also their cytolytic capacity determines the effectiveness of immune system–mediated tumor control was further supported by Atreya et al. [270]. They showed that the expression of the T-box transcription factor eomesodermin, which is critically involved in controlling the cytolytic activity of CD8+ CTLs, inversely correlates with the occurrence of lymph node metastasis in CRC patients [270].
Pronounced lymphocyte infiltration in CRC is more marked in MSI-H CRC and might explain the better clinical outcome for patients with these tumors [56]. It has been postulated that MSI-H CRCs are more immunogenic than MSS CRCs [271] because of the generation of a large number of abnormal peptides by frameshift mutations [272–276]. However, crosspriming of antigen-presenting cells by released intracellular antigens or the use of HLA class II machinery and T helper cell activity may provide an alternative pathway for immune stimulation [271, 272].
For a better prognostic assessment, Dolcetti et al. [277] suggested the combination of evaluation of both local lymphocytosis and MSI status. MSI in combination with a high content of intraepithelial lymphocytes was found to be related to longer overall survival in a group of patients with exclusively right-sided CRC. Consequently, it has been suggested that these lymphocytes may actually represent an immune response that contributes to longer survival in MSI-H CRC patients, and subsequent work confirmed a possible link [261, 271, 278–280]. Dolcetti et al. [277] and Michael-Robinson et al. [279] demonstrated that the greater frequency of TILs associated with MSI-H cancers are only weakly or moderately correlated with tumor apoptosis. However, although TILs might be expected to explain the higher apoptotic rate and better prognosis with MSI-H cancers, it is likely that MSI-H cancers are intrinsically more prone to apoptosis, independently of T cell attack [279]. In addition, Buckowitz et al. [281] suggested a protective role of functionally active lymphocytes directed against MSI-H CRCs that may prevent tumor cell dissemination and metastasis formation in distant organs. In contrast, Baker et al. [282] found TIL infiltration only to be of prognostic value in MMR-proficient CRCs. They also pointed out that perturbations in the TGF-β signaling pathway play an important role in the recruitment and retention of TILs within CRC epithelium [282]. Although Prall et al. [283] also found a prognostic impact of high CD8+ density in MSI-H CRCs, they showed that it was not solely restricted to this group. Therefore, they hypothesized that tumor infiltration by CD8+ lymphocytes could reflect a general principle of antitumor immunity, irrespective of MSI status [283].
With regard to the explicit prognostic relevance of these CTLs and other components of the immune system in the setting of CRC, therapeutic tools that are able to influence these key immunological mediators present promising candidates for more successful clinical control of progression, metastasis, and recurrence of CRC. These new therapeutic approaches have already been successfully tested in various animal models of CRC or even in first clinical trials, demonstrating an encouraging tumor-suppressive capacity. Therefore, better integration of immunotherapy into clinically approved concepts of standardized treatment of CRC can be expected in the near future [247].
New Promising Markers
Forkhead Box P3–Positive Regulatory T Cells
Regulatory T cells (Tregs) were initially characterized by the CD4+CD25+ phenotype and are thought to modulate the antitumor immune response. Adaptive Tregs contribute to an immunosuppressive microenvironment in CRC through a cyclooxygenase-2–prostaglandin-E2–dependent mechanism, direct cell–cell contact, or by the release of cytokines, such as TGF-β, thereby facilitating tumor growth [284, 285]. The most specific Treg marker identified to date is the nuclear transcription factor known as forkhead box P3 (FOXP3). Loddenkemper et al. [286] reported that Treg density was lower in node-positive disease but was not associated with survival. In contrast, Ling et al. [287] found no significant difference in Treg density between advanced and early-stage disease, but did not evaluate the association with patient survival. Salama et al. [288] showed that a high density of FOXP3+ Tregs in CRC was associated with longer survival and had a stronger prognostic significance than CD8+ and CD45RO+ memory T lymphocytes in CRC. Furthermore, a high density of FOXP3+ Tregs in normal colon tissue from CRC patients was associated with a worse prognosis. The worse outcome observed for these patients might be explained by the proposed role for these cells in suppressing antitumor immunity. However, the observation of better survival for patients with a high density of FOXP3+ Tregs in their tumor tissue is counterintuitive and contrasts with what has been reported for other solid tumor types, for which Tregs are generally considered to be immunosuppressive. In contrast, Suzuki et al. [289] were not able to show a significant correlation between FOXP3+ Tregs and survival, but they did find evidence that a balance of intratumoral Tregs and CD8+ T cells is a more sensitive predictor of recurrence and survival than intratumoral Tregs or CD8+ T cells alone. Functional studies of FOXP3+ Tregs in cancer and normal tissue may shed more light on their role in the antitumor response and help to explain the observed associations with prognosis [288].
Although further studies are required before changes in clinical practice can be recommended, the results of Salama et al. [288] suggest that the assessment of FOXP3+ Treg density in tumor and normal colorectal tissue in combination with vascular and perineural invasion could improve prognostication in early-stage CRC patients. Few studies analyzed the immune gene expression profile of FOXP3+ Tregs according to MMR status. Le Gouvello et al. [290] observed higher expression levels of FOXP3 mRNA in MSS CRCs. In addition, Frey et al. [291] demonstrated that a high frequency of tumor-infiltrating FOXP3+ Tregs is associated with early T stage and independently predicts longer disease-specific survival in MMR-proficient CRC patients but not in MMR-deficient patients. In contrast, Michel et al. [292] were the first to show that the density of FOXP3+ Tregs infiltrating CRC was significantly higher in MSI-H tumors using IHC, paralleling the enhanced number of CD8+ cells in these tumors. This discrepancy might be attributed to the different methodologies applied in these studies or the role of Tregs may differ according to the clinical stage and genetic background of tumors [291]. Further research is recommended to elucidate the role of FOXP3+ Tregs and MMR status in CRC.
Members of the Extracellular Signal–Related Kinase–MAPK Signaling Pathway
Receptor for Hyaluronic Acid–Mediated Motility
The receptor for hyaluronic acid–mediated motility (RHAMM) (intracellular hyaluronic acid binding protein, CD168) is a multifaceted protein with both intracellular and extracellular functions [293]. RHAMM binds hyaluronan [294], interacts with both microtubules and microfilaments [293, 295], localizes to the centrosome, and functions in maintaining spindle integrity [296], and is suggested to represent a member of the MAP family [297, 298]. In addition, RHAMM participates in cell motility, signaling, and oncogenic events [296]. The RAF–MAPK/ERK kinase–extracellular signal–related kinase (ERK) pathway belongs to the MAPK pathways and represents one of the best characterized RAS signaling pathways. The molecule ERK is activated by a cascade of phosphorylations downstream from the RAS proto-oncogene and plays a role in differentiation, secretion, and proliferation. RHAMM binds ERK and controls expression levels of ERK [298].
In addition to its role in cell migration, RHAMM expression, and overexpression, has been linked to RAS transformation, tumor progression, and metastasis [299] in different tumor types [298]. However, the prognostic significance of RHAMM in CRC is poorly understood.
RHAMM as a Prognostic Factor.
Only a few studies, primarily from the group of Dr. Lugli, have investigated the prognostic significance RHAMM in CRC. The expression profile was assessed by means of tissue microarray IHC in CRC. Interactions between RHAMM and other potential prognostic and clinicopathological factors were also established.
In normal colonic mucosa, RHAMM was diffusely but weakly expressed in the cytoplasm of columnar cells in the crypts, but apparently not in the goblet cells. RHAMM was less strongly expressed and quantitatively less extensive in these normal tissues than in the cancer cell population [298].
Lugli et al. [298] demonstrated that higher RHAMM expression is needed to induce tumor progression and that expression of RHAMM is an independent adverse prognostic factor in MMR-proficient CRC and presumed HNPCC (MSH2− and/or MSH6− at any age or MLH1− and <55 years of age).
Zlobec et al. [300] evaluated the independent prognostic effect of a panel of IHC protein markers, including RHAMM, in unselected CRC patients. They suggested that RHAMM should be considered a more important prognosticator than tumor grade, MMR status, tumor budding, and vascular invasion in CRC. In addition, diffuse RHAMM expression was second in terms of independent prognostic value behind N stage on multivariate analysis.
Moreover, the involvement of RHAMM within the RAS–MAPK pathway and its role as a receptor suggest that it might be a potential candidate for therapeutic intervention.
Multimarker Analysis.
In a study of Lugli et al. [298], nuclear phosphorylated ERK (pERK) expression was correlated with RHAMM expression in MMR-proficient CRC and in presumed HNPCC (MSH2− and/or MSH6− at any age or MLH1− and <55 years of age), whereas an association was not found in MLH1− tumors. This leads to the hypothesis that pERK is involved in the mechanism of tumor progression in MMR-proficient CRC and HNPCC by interacting with the Wnt signaling pathway and RHAMM.
In a recent analysis including only MSI-H tumors, Zlobec et al. [300] found that the combination of p21 and RHAMM led to a highly adverse prognosis. This analysis suggested that node-negative patients overexpressing RHAMM but with loss of p21 may experience a potential benefit from postoperative treatment, whereas adjuvant chemotherapy should be reconsidered for node-positive RHAMM− tumors. Expression of p21 appears to act as a modifying factor in the survival time of patients with RHAMM+ tumors.
These findings also outline the importance of evaluating multimarker phenotype combinations of IHC protein markers [300] and provide evidence for the inclusion of RHAMM as a new and promising prognostic factor in CRC. However, prospective studies are needed to further validate the prognostic role of RHAMM in CRC and its potential role in selecting patients for adjuvant therapies [301].
PI3KCA
Class 1 PI3Ks are heterodimeric lipid kinases composed of a p85 regulatory subunit and one of several p110 catalytic subunits. Among several isoforms of the catalytic subunits, only the α-type, PI3KCA, has been shown to harbor oncogenic mutations or amplifications in its gene in human malignancies [302, 303]. The regulatory subunit of PI3K can specifically bind protein factors including KRAS, integrate various signals from membrane receptors, and activate PI3KCA. Activated PI3KCA will phosphorylate phosphatidyl-inositol-4,5-biphosphate to produce phosphatidyl-inositol-3,4,5-triphosphate (PIP3), which localizes the serine threonine kinase Akt to the cell membrane where it becomes activated. Activated Akt phosphorylates downstream protein effectors and amplifies the signaling cascade, enhancing cell proliferation and survival [302]. Components of this pathway, including PI3KCA, are often altered in human malignancies.
PI3KCA as Prognostic and/or Predictive Factor.
The prognostic role of PI3KCA is still under investigation. Jehan et al. [302] reported PI3KCA amplification as an independent prognostic factor for longer survival and suggested that this might be one of the promising markers to define subsets of CRC patients who may maximally benefit from adjuvant therapy. Recent reports have highlighted the prognostic role of PI3KCA mutations as a marker of poor outcome in surgically resectable tumors [303, 304]; however, the study of Souglakos et al. [142] did not indicate PI3KCA as a useful predictor of current adjuvant treatment strategies. Although there have been reports suggesting that PI3KCA mutation (in exons 9 and 20) might be correlated with response to anti-EGFR inhibitors [140], large patient population studies are necessary to determine whether this factor will play an important role in determining response to anti-EGFR mAbs [113].
PTEN
PTEN has been identified as a critical negative regulator of the cell-survival signaling pathway initiated by PI3K. The PTEN protein acts as a phospholipid phosphatase with PIP3 as a substrate. One of the most important downstream targets of PIP3, Akt, is activated by phosphorylation and inhibited by PTEN. PTEN protein is a negative regulator of the Akt signaling pathway, and thus inactivation of PTEN, which is a common event in human malignancies, facilitates cell proliferation and apoptosis [305, 306]. In CRC, it has been demonstrated that PTEN mutations correlate with advanced and metastatic tumors [305, 307]. Moreover, epigenetic silencing of PTEN by promoter hypermethylation has been observed in sporadic CRC cases, especially in MSI-H tumors [305, 308].
PTEN as a Prognostic and/or Predictive Factor.
The prognostic role of PTEN in CRC is still under investigation, and inconclusive results have been reported. Although some groups report an association between loss of PTEN and a shorter progression-free survival interval [306, 146], others report an association with poor prognosis in stage II patients only [305] or, in contrast, in CRC patients with liver metastasis [307]. As mentioned above, PTEN shows promise as a predictive marker for wild-type KRAS patients treated with an anti-EGFR–based regimen [143, 146].
T-Cell Originated Protein Kinase
In 2000, a new member of the ERK–MAPK pathway, T-cell originated protein kinase (TOPK), also know as PZD-binding kinase, was identified and described as a MAPKK (mitogen activated protein kinase kinase). TOPK is a serine/threonine kinase and has been shown to be involved in p38 and c-Jun N-terminal kinase signaling, and more recently in the ERK–MAPK pathway [309, 310]. TOPK seems to be overexpressed in a variety of tumors in vitro, whereas inhibition of TOPK has been shown to lead to apoptosis in breast and melanoma cell lines. Zhu et al. [310] systematically assessed this novel molecule in CRC and confirmed its oncogenic potential in vitro and in vivo. More importantly, they found that TOPK could promote malignant transformation by exerting a positive feedback loop on ERK-2 activity [310].
TOPK as a Prognostic and/or Predictive Factor.
Zlobec et al. [309] were the first to assess the prognostic and predictive value of TOPK in CRC. Given its central involvement in ERK–MAPK signaling, TOPK overexpression was significantly related to KRAS and BRAF mutations. Their results showed that TOPK may be a valuable prognostic marker in patients with sporadic CRC with KRAS and BRAF gene mutations and in patients with metastatic disease with a proficient molecular profile for positive response to anti-EGFR therapies. If confirmed prospectively, the inhibition of TOPK may represent a novel avenue of investigation of targeted treatment in patients with CRC [309].
Conclusion and Future Perspectives
For CRC patients who undergo successful surgery, additional treatment is recommended, especially for those patients with a high risk for relapse. Although CRC prognosis is stage and grade dependent, many tumors with similar histopathologic features show significantly different clinical outcomes [92]. The identification of molecular factors that have prognostic and/or predictive significance in CRC is essential to improve treatment and outcome [30]. Several biomarkers have been studied over the past decades; however, results of published studies have often been conflicting and several drawbacks affect the reliability of conclusions [238]. Most published studies have used retrospective analyses of a single marker in a small series of patients. These study designs are unlikely to precisely predict progression of disease with sufficient resolution and reproducibility [92]. In spite of these problems, some biomarkers have shown promising results. Therefore, with the multitude of putative factors available for study, a combinatorial approach to molecular prognostics, similar to the prognostic profiles established for breast cancer patients, may have significance and be used in future patient management [30]. This understanding recently led to more comprehensive approaches of global gene expression profiling and genomewide analysis to determine prognostic and predictive signatures in tumors. Many advances have been made in technologies for profiling and in decreasing the requirements of the input material, although data analysis and interpretation still remain challenging. Hence, although there are immense potential implications, clinicians are currently unable to use these data in clinical practice for decision making because of a lack of definition, adequate validation, and easy implementation [17]. Therefore, the current reality is that no molecular marker, other than the KRAS gene in the case of EGFR-targeted therapy for metastatic disease, has made it into clinical practice. Large prospective randomized trails have the potential to determine the role of various putative molecular markers. Unfortunately, biomarker-embedded clinical trials do not receive the same commercial attention as those for new chemotherapeutic compounds. Furthermore, standards need to be agreed upon for what determines the validity of a biomarker before any marker can be used in these clinical trials. The introduction of new therapeutic agents and the discovery and validation of prognostic and/or predictive markers along with new screening tools will enable oncologists to tailor patient-specific chemotherapy by maximizing drug efficacy and minimizing adverse and possibly severe side effects [92].
Author Contributions
Conception/Design: Vanessa Deschoolmeester
Manuscript writing: Vanessa Deschoolmeester
Final approval of manuscript: Marc Baay, Pol Specenier, Filip Lardon, Jan B. Vermorken
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16:481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 3.Michor F, Iwasa Y, Lengauer C, et al. Dynamics of colorectal cancer. Semin Cancer Biol. 2005;15:484–493. doi: 10.1016/j.semcancer.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Jass JR. Colorectal cancer: A multipathway disease. Crit Rev Oncog. 2006;12:273–287. doi: 10.1615/critrevoncog.v12.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 5.Goel A, Arnold CN, Niedzwiecki D, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 6.Atkin NB. Microsatellite instability. Cytogenet Cell Genet. 2001;92:177–181. doi: 10.1159/000056898. [DOI] [PubMed] [Google Scholar]
- 7.Grady W, Rajput A, Lutterbaugh JD, et al. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900–902. [PubMed] [Google Scholar]
- 8.Edmonston TB, Cuesta KH, Burkholder S, et al. Colorectal carcinomas with high microsatellite instability: Defining a distinct immunologic and molecular entity with respect to prognostic markers. Hum Pathol. 2000;31:1506–1514. doi: 10.1053/hupa.2000.20383. [DOI] [PubMed] [Google Scholar]
- 9.Worthley DL, Whitehall VL, Spring KJ, et al. Colorectal carcinogenesis: Road maps to cancer. World J Gastroenterol. 2007;13:3784–3791. doi: 10.3748/wjg.v13.i28.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornmann M, Formentini A, Ette C, et al. Prognostic factors influencing the survival of patients with colon cancer receiving adjuvant 5-FU treatment. Eur J Surg Oncol. 2008;34:1316–1321. doi: 10.1016/j.ejso.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Zlobec I, Steele R, Terracciano L, et al. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol. 2007;60:1112–1116. doi: 10.1136/jcp.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zlobec I, Lugli A. Prognostic and predictive factors in colorectal cancer. J Clin Pathol. 2008;61:561–569. doi: 10.1136/jcp.2007.054858. [DOI] [PubMed] [Google Scholar]
- 13.Benson AB., 3rd New approaches to the adjuvant therapy of colon cancer. The Oncologist. 2006;11:973–980. doi: 10.1634/theoncologist.11-9-973. [DOI] [PubMed] [Google Scholar]
- 14.Merkel S, Wein A, Günther K, et al. High-risk groups of patients with stage II colon carcinoma. Cancer. 2001;92:1435–1443. doi: 10.1002/1097-0142(20010915)92:6<1435::aid-cncr1467>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Weiser MR, Landmann RG, Kattan MW, et al. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26:380–385. doi: 10.1200/JCO.2007.14.1291. [DOI] [PubMed] [Google Scholar]
- 16.Duffy MJ, Crown J. A personalized approach to cancer treatment: How biomarkers can help. Clin Chem. 2008;54:1770–1779. doi: 10.1373/clinchem.2008.110056. [DOI] [PubMed] [Google Scholar]
- 17.De Roock W, Biesmans B, De Schutter J, et al. Clinical biomarkers in oncology: Focus on colorectal cancer. Mol Diagn Ther. 2009;13:103–114. doi: 10.1007/BF03256319. [DOI] [PubMed] [Google Scholar]
- 18.Graziano F, Cascinu S. Prognostic molecular markers for planning adjuvant chemotherapy trials in Dukes' B colorectal cancer patients: How much evidence is enough? Ann Oncol. 2003;14:1026–1038. doi: 10.1093/annonc/mdg284. [DOI] [PubMed] [Google Scholar]
- 19.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 20.Liefers GJ, Tollenaar RA. Cancer genetics and their application to individualised medicine. Eur J Cancer. 2002;38:872–879. doi: 10.1016/s0959-8049(02)00055-2. [DOI] [PubMed] [Google Scholar]
- 21.Benatti P, Gafà R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 22.Raut C, Pawlik T, Rodriguez-Bigas MA. Clinicopathologic features in colorectal cancer patients with microsatellite instability. Mutat Res. 2004;568:275–282. doi: 10.1016/j.mrfmmm.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 23.Tejpar S. The multidisciplinary management of gastrointestinal cancer. The use of molecular markers in the diagnosis and treatment of colorectal cancer. Best Pract Res Clin Gastroenterol. 2007;21:1071–1087. doi: 10.1016/j.bpg.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 25.Warusavitarne J, Schnitzler M. The role of chemotherapy in microsatellite unstable (MSI-H) colorectal cancer. Int J Colorectal Dis. 2007;22:739–748. doi: 10.1007/s00384-006-0228-0. [DOI] [PubMed] [Google Scholar]
- 26.Houlston RS. What we could do now: Molecular pathology of colorectal cancer. Mol Pathol. 2001;54:206–214. doi: 10.1136/mp.54.4.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anwar S, Frayling IM, Scott NA, et al. Systematic review of genetic influences on the prognosis of colorectal cancer. Br J Surg. 2004;91:1275–1291. doi: 10.1002/bjs.4737. [DOI] [PubMed] [Google Scholar]
- 28.Klump B, Nehls O, Okech T, et al. Molecular lesions in colorectal cancer: Impact on prognosis? Original data and review of the literature. Int J Colorectal Dis. 2004;19:23–42. doi: 10.1007/s00384-003-0499-7. [DOI] [PubMed] [Google Scholar]
- 29.Pasche B, Mulcahy M, Benson AB., 3rd Molecular markers in prognosis of colorectal cancer and prediction of response to treatment. Best Pract Res Clin Gastroenterol. 2002;16:331–345. doi: 10.1053/bega.2002.0289. [DOI] [PubMed] [Google Scholar]
- 30.Kahlenberg MS, Sullivan JM, Witmer DD, et al. Molecular prognostics in colorectal cancer. Surg Oncol. 2003;12:173–186. doi: 10.1016/s0960-7404(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 31.Clark AJ, Barnetson R, Farrington SM, et al. Prognosis in DNA mismatch repair deficient colorectal cancer: Are all MSI tumours equivalent? Fam Cancer. 2004;3:85–91. doi: 10.1023/B:FAME.0000039915.94550.cc. [DOI] [PubMed] [Google Scholar]
- 32.Söreide K, Janssen EA, Söiland H, et al. Microsatellite instability in colorectal cancer. Br J Surg. 2006;93:395–406. doi: 10.1002/bjs.5328. [DOI] [PubMed] [Google Scholar]
- 33.Habermann JK, Roblick UJ, Upender M, et al. From genome to proteome in tumor profiling: Molecular events in colorectal cancer genesis. Adv Exp Med Biol. 2006;587:161–177. doi: 10.1007/978-1-4020-5133-3_15. [DOI] [PubMed] [Google Scholar]
- 34.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haddad R, Ogilvie RT, Croitoru M, et al. Microsatellite instability as a prognostic factor in resected colorectal cancer liver metastases. Ann Surg Oncol. 2004;11:977–982. doi: 10.1245/ASO.2004.03.585. [DOI] [PubMed] [Google Scholar]
- 36.Niv Y. Biologic behavior of microsatellite-unstable colorectal cancer and treatment with 5-fluorouracil. Isr Med Assoc J. 2005;7:520–524. [PubMed] [Google Scholar]
- 37.Elsaleh H, Joseph D, Grieu F, et al. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 38.Hemminki A, Mecklin JP, Järvinen H, et al. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 41.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carethers JM. Chemotherapy and survival in colorectal cancer patients with and without microsatellite instability: Can MSI be a prognostic marker? [reply] Gastroenterology. 2004;127:689–690. doi: 10.1053/j.gastro.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 43.Des Guetz G, Uzzan B, Nicolas P, et al. Microsatellite instability: A predictive marker in metastatic colorectal cancer? Target Oncol. 2009;4:57–62. doi: 10.1007/s11523-008-0103-8. [DOI] [PubMed] [Google Scholar]
- 44.Lim SB, Jeong SY, Lee MR, et al. Prognostic significance of microsatellite instability in sporadic colorectal cancer. Int J Colorectal Dis. 2004;19:533–537. doi: 10.1007/s00384-004-0596-2. [DOI] [PubMed] [Google Scholar]
- 45.Emterling A, Wallin A, Arbman G, et al. Clinicopathological significance of microsatellite instability and mutated RIZ in colorectal cancer. Ann Oncol. 2004;15:242–246. doi: 10.1093/annonc/mdh045. [DOI] [PubMed] [Google Scholar]
- 46.Storojeva I, Boulay J, Heinimann K, et al. Prognostic and predictive relevance of microsatellite instability in colorectal cancer. Oncol Rep. 2005;14:241–249. [PubMed] [Google Scholar]
- 47.Vidaurreta M, Sanz-Casla MT, Maestro ML, et al. Microsatellite instability predicts better outcome in colorectal cancer patients. Med Clin (Barc) 2005;124:121–125. doi: 10.1157/13071004. [DOI] [PubMed] [Google Scholar]
- 48.Chang EY, Dorsey PB, Johnson N, et al. A prospective analysis of microsatellite instability as a molecular marker in colorectal cancer. Am J Surg. 2006;191:646–651. doi: 10.1016/j.amjsurg.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 49.Chang SC, Lin JK, Yang SH, et al. Relationship between genetic alterations and prognosis in sporadic colorectal cancer. Int J Cancer. 2006;118:1721–1727. doi: 10.1002/ijc.21563. [DOI] [PubMed] [Google Scholar]
- 50.Meng WJ, Sun XF, Tian C, et al. Microsatellite instability did not predict individual survival in sporadic stage II and III rectal cancer patients. Oncology. 2007;72:82–88. doi: 10.1159/000111107. [DOI] [PubMed] [Google Scholar]
- 51.Lamberti C, Lundin S, Bogdanow M, et al. Microsatellite instability did not predict individual survival of unselected patients with colorectal cancer. Int J Colorectal Dis. 2007;22:145–152. doi: 10.1007/s00384-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 52.Malesci A, Laghi L, Bianchi P, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–3839. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 53.Müller CI, Schulmann K, Reinacher-Schick A, et al. Predictive and prognostic value of microsatellite instability in patients with advanced colorectal cancer treated with a fluoropyrimidine and oxaliplatin containing first-line chemotherapy. A report of the AIO Colorectal Study Group. Int J Colorectal Dis. 2008;23:1033–1039. doi: 10.1007/s00384-008-0504-2. [DOI] [PubMed] [Google Scholar]
- 54.Deschoolmeester V, Van Damme N, Baay M, et al. Microsatellite instability in sporadic colon carcinomas has no independent prognostic value in a Belgian study population. Eur J Cancer. 2008;44:2288–2295. doi: 10.1016/j.ejca.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 55.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Chang EY, Frankhouse J, et al. Combination of microsatellite instability and lymphocytic infiltrate as a prognostic indicator for adjuvant therapy in colon cancer. Arch Surg. 2009;144:835–840. doi: 10.1001/archsurg.2009.162. [DOI] [PubMed] [Google Scholar]
- 57.Barrasa Shaw A, López-Guerrero JA, Calatrava Fons A, et al. Value of the identification of microsatellite instability in colorectal cancer. Clin Transl Oncol. 2009;11:465–469. doi: 10.1007/s12094-009-0386-y. [DOI] [PubMed] [Google Scholar]
- 58.Banerjea A, Hands RE, Powar MP, et al. Microsatellite and chromosomal stable colorectal cancers demonstrate poor immunogenicity and early disease recurrence. Colorectal Dis. 2009;11:601–608. doi: 10.1111/j.1463-1318.2008.01639.x. [DOI] [PubMed] [Google Scholar]
- 59.Sanchez JA, Krumroy L, Plummer S, et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg. 2009;96:1196–1204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- 60.Tedesco KL, Lockhart AC, Berlin JD. The epidermal growth factor receptor as a target for gastrointestinal cancer therapy. Curr Treat Options Oncol. 2004;5:393–403. doi: 10.1007/s11864-004-0029-z. [DOI] [PubMed] [Google Scholar]
- 61.Harding J, Burtness B. Cetuximab: An epidermal growth factor receptor chimeric human-murine monoclonal antibody. Drugs Today (Barc) 2005;41:107–127. doi: 10.1358/dot.2005.41.2.882662. [DOI] [PubMed] [Google Scholar]
- 62.Spano JP, Milano G, Vignot S, et al. Potential predictive markers of response to EGFR-targeted therapies in colorectal cancer. Crit Rev Oncol Hematol. 2008;66:21–30. doi: 10.1016/j.critrevonc.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 63.Lockhart AC, Berlin JD. The epidermal growth factor receptor as a target for colorectal cancer therapy. Semin Oncol. 2005;32:52–60. doi: 10.1053/j.seminoncol.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 64.Ng K, Zhu AX. Targeting the epidermal growth factor receptor in metastatic colorectal cancer. Crit Rev Oncol Hematol. 2008;65:8–20. doi: 10.1016/j.critrevonc.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 65.Goldstein NS. Recent pathology related advances in colorectal adenocarcinomas. Eur J Surg Oncol. 2001;27:446–450. doi: 10.1053/ejso.2000.1107. [DOI] [PubMed] [Google Scholar]
- 66.Scartozzi M, Pierantoni C, Berardi R, et al. Epidermal growth factor receptor: A promising therapeutic target for colorectal cancer. Anal Quant Cytol Histol. 2006;28:61–68. [PubMed] [Google Scholar]
- 67.Scartozzi M, Pierantoni C, Berardi R, et al. Anti-EGFR strategies as an incremental step for the treatment of colorectal cancer patients: Moving from scientific evidence to clinical practice. Expert Opin Ther Targets. 2006;10:281–287. doi: 10.1517/14728222.10.2.281. [DOI] [PubMed] [Google Scholar]
- 68.Overman MJ, Hoff PM. EGFR-targeted therapies in colorectal cancer. Dis Colon Rectum. 2007;50:1259–1270. doi: 10.1007/s10350-007-0228-3. [DOI] [PubMed] [Google Scholar]
- 69.Rivera F, Vega-Villegas ME, López-Brea MF. Cetuximab, its clinical use and future perspectives. Anticancer Drugs. 2008;19:99–113. doi: 10.1097/CAD.0b013e3282f23287. [DOI] [PubMed] [Google Scholar]
- 70.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 71.Vallböhmer D, Zhang W, Gordon M, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23:3536–3544. doi: 10.1200/JCO.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 72.Chapusot C, Martin L, Mungra N, et al. Sporadic colorectal cancers with defective mismatch repair display a number of specific morphological characteristics: Relationship between the expression of hMLH1 and hMSH2 proteins and clinicopathological features of 273 adenocarcinomas. Histopathology. 2003;43:40–47. doi: 10.1046/j.1365-2559.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- 73.McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]
- 74.Cunningham MP, Essapen S, Thomas H, et al. Coexpression of the IGF-IR, EGFR and HER-2 is common in colorectal cancer patients. Int J Oncol. 2006;28:329–335. [PubMed] [Google Scholar]
- 75.Spano JP, Lagorce C, Atlan D, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16:102–108. doi: 10.1093/annonc/mdi006. [DOI] [PubMed] [Google Scholar]
- 76.Kluftinger AM, Robinson BW, Quenville NF, et al. Correlation of epidermal growth factor receptor and c-erbB2 oncogene product to known prognostic indicators of colorectal cancer. Surg Oncol. 1992;1:97–105. doi: 10.1016/0960-7404(92)90062-p. [DOI] [PubMed] [Google Scholar]
- 77.Steele RJ, Kelly P, Ellul B, et al. Epidermal growth factor receptor expression in colorectal cancer. Br J Surg. 1990;77:1352–1354. doi: 10.1002/bjs.1800771211. [DOI] [PubMed] [Google Scholar]
- 78.Kountourakis P, Pavlakis K, Psyrri A, et al. Clinicopathologic significance of EGFR and Her-2/neu in colorectal adenocarcinomas. Cancer J. 2006;12:229–236. doi: 10.1097/00130404-200605000-00012. [DOI] [PubMed] [Google Scholar]
- 79.Koretz K, Schlag P, Möller P. Expression of epidermal growth factor receptor in normal colorectal mucosa, adenoma, and carcinoma. Virchows Arch A Pathol Anat Histopathol. 1990;416:343–349. doi: 10.1007/BF01605295. [DOI] [PubMed] [Google Scholar]
- 80.Deng Y, Kurland BF, Wang J, et al. High epidermal growth factor receptor expression in metastatic colorectal cancer lymph nodes may be more prognostic of poor survival than in primary tumor. Am J Clin Oncol. 2009;32:245–252. doi: 10.1097/COC.0b013e3181891326. [DOI] [PubMed] [Google Scholar]
- 81.Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: Implications for a standardized scoring system. Cancer. 2001;92:1331–1346. doi: 10.1002/1097-0142(20010901)92:5<1331::aid-cncr1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 82.Zlobec I, Vuong T, Hayashi S, et al. A simple and reproducible scoring system for EGFR in colorectal cancer: Application to prognosis and prediction of response to preoperative brachytherapy. Br J Cancer. 2007;96:793–800. doi: 10.1038/sj.bjc.6603619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peeters M, Price T, Van Laethem JL. Anti-epidermal growth factor receptor monotherapy in the treatment of metastatic colorectal cancer: Where are we today? The Oncologist. 2009;14:29–39. doi: 10.1634/theoncologist.2008-0167. [DOI] [PubMed] [Google Scholar]
- 84.Ramos FJ, Macarulla T, Capdevila J, et al. Understanding the predictive role of K-ras for epidermal growth factor receptor-targeted therapies in colorectal cancer. Clin Colorectal Cancer. 2008;7(suppl 2):S52–S57. doi: 10.3816/CCC.2008.s.008. [DOI] [PubMed] [Google Scholar]
- 85.Peeters M, Balfour J, Arnold D. Review article: Panitumumab—a fully human anti-EGFR monoclonal antibody for treatment of metastatic colorectal cancer. Aliment Pharmacol Ther. 2008;28:269–281. doi: 10.1111/j.1365-2036.2008.03717.x. [DOI] [PubMed] [Google Scholar]
- 86.Correale P, Cusi MG, Micheli L, et al. Chemo-immunotherapy of colorectal carcinoma: Preclinical rationale and clinical experience. Invest New Drugs. 2006;24:99–110. doi: 10.1007/s10637-006-5932-7. [DOI] [PubMed] [Google Scholar]
- 87.Ma BB, Bristow RG, Kim J, et al. Combined-modality treatment of solid tumors using radiotherapy and molecular targeted agents. J Clin Oncol. 2003;21:2760–2776. doi: 10.1200/JCO.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 88.Williams JP, Stewart T, Li B, et al. The retinoblastoma protein is required for Ras-induced oncogenic transformation. Mol Cell Biol. 2006;26:1170–1182. doi: 10.1128/MCB.26.4.1170-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bendardaf R, Lamlum H, Pyrhönen S. Prognostic and predictive molecular markers in colorectal carcinoma. Anticancer Res. 2004;24:2519–2530. [PubMed] [Google Scholar]
- 90.Midgley RS, Kerr DJ. Ras as a target in cancer therapy. Crit Rev Oncol Hematol. 2002;44:109–120. doi: 10.1016/s1040-8428(01)00189-5. [DOI] [PubMed] [Google Scholar]
- 91.McLeod HL, Church RD. Molecular predictors of prognosis and response to therapy in colorectal cancer. Cancer Chemother Biol Response Modif. 2003;21:791–801. doi: 10.1016/s0921-4410(03)21037-1. [DOI] [PubMed] [Google Scholar]
- 92.Lurje G, Zhang W, Lenz HJ. Molecular prognostic markers in locally advanced colon cancer. Clin Colorectal Cancer. 2007;6:683–690. doi: 10.3816/CCC.2007.n.037. [DOI] [PubMed] [Google Scholar]
- 93.Jiang Y, Kimchi ET, Staveley-O'Carroll KF, et al. Assessment of K-ras mutation: A step toward personalized medicine for patients with colorectal cancer. Cancer. 2009;115:3609–3617. doi: 10.1002/cncr.24434. [DOI] [PubMed] [Google Scholar]
- 94.Westra J, Schaapveld M, Hollema H, et al. Determination of TP53 mutation is more relevant than microsatellite instability status for the prediction of disease-free survival in adjuvant-treated stage III colon cancer patients. J Clin Oncol. 2005;23:5635–5643. doi: 10.1200/JCO.2005.04.096. [DOI] [PubMed] [Google Scholar]
- 95.Neal CP, Garcea G, Doucas H, et al. Molecular prognostic markers in resectable colorectal liver metastases: A systematic review. Eur J Cancer. 2006;42:1728–1743. doi: 10.1016/j.ejca.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 96.Conlin A, Smith G, Carey F, et al. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut. 2005;54:1283–1286. doi: 10.1136/gut.2005.066514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elnatan J, Goh HS, Smith DR. C-KI-RAS activation and the biological behaviour of proximal and distal colonic adenocarcinomas. Eur J Cancer. 1996;32A:491–497. doi: 10.1016/0959-8049(95)00567-6. [DOI] [PubMed] [Google Scholar]
- 98.Samowitz WS, Curtin K, Schaffer D, et al. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: A population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 99.Pajkos G, Kiss I, Sándor J, et al. The prognostic value of the presence of mutations at the codons 12, 13, 61 of K-ras oncogene in colorectal cancer. Anticancer Res. 2000;20:1695–1701. [PubMed] [Google Scholar]
- 100.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 101.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II' study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Russo A, Bazan V, Agnese V, et al. Prognostic and predictive factors in colorectal cancer: Kirsten Ras in CRC (RASCAL) and TP53CRC collaborative studies. Ann Oncol. 2005;16(suppl 4):iv44–iv49. doi: 10.1093/annonc/mdi907. [DOI] [PubMed] [Google Scholar]
- 103.Graziano F, Ruzzo A, Loupakis F, et al. Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J Clin Oncol. 2008;26:1427–1434. doi: 10.1200/JCO.2007.12.4602. [DOI] [PubMed] [Google Scholar]
- 104.Loriot Y, Mordant P, Deutsch E, et al. Are RAS mutations predictive markers of resistance to standard chemotherapy? Nat Rev Clin Oncol. 2009;6:528–534. doi: 10.1038/nrclinonc.2009.106. [DOI] [PubMed] [Google Scholar]
- 105.Tol J, Koopman M, Cats A, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 106.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 107.Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 108.Ciardiello F, Tejpar S, Papamichael D. Implications of KRAS mutation status for the treatment of metastatic colorectal cancer. Target Oncol. 2009;4:311–322. doi: 10.1007/s11523-009-0129-6. [DOI] [PubMed] [Google Scholar]
- 109.Personeni N, Fieuws S, Piessevaux H, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: A fluorescent in situ hybridization study. Clin Cancer Res. 2008;14:5869–5876. doi: 10.1158/1078-0432.CCR-08-0449. [DOI] [PubMed] [Google Scholar]
- 110.Cappuzzo F, Finocchiaro G, Rossi E, et al. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19:717–723. doi: 10.1093/annonc/mdm492. [DOI] [PubMed] [Google Scholar]
- 111.Cappuzzo F, Varella-Garcia M, Finocchiaro G, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–89. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sartore-Bianchi A, Moroni M, Veronese S, et al. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol. 2007;25:3238–3245. doi: 10.1200/JCO.2007.11.5956. [DOI] [PubMed] [Google Scholar]
- 113.Tejpar S, Odze RD. Accomplishments in 2008 in biologic markers for gastrointestinal cancers—focus on colorectal cancer. Gastrointest Cancer Res. 2009;3(5) suppl 2:S73–S78. [PMC free article] [PubMed] [Google Scholar]
- 114.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 115.Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 116.Sauer T, Guren MG, Noren T, et al. Demonstration of EGFR gene copy loss in colorectal carcinomas by fluorescence in situ hybridization (FISH): A surrogate marker for sensitivity to specific anti-EGFR therapy? Histopathology. 2005;47:560–564. doi: 10.1111/j.1365-2559.2005.02252.x. [DOI] [PubMed] [Google Scholar]
- 117.Lenz HJ, Van Cutsem E, Khambata-Ford S, et al. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol. 2006;24:4914–4921. doi: 10.1200/JCO.2006.06.7595. [DOI] [PubMed] [Google Scholar]
- 118.Snyder LC, Astsaturov I, Weiner LM. Overview of monoclonal antibodies and small molecules targeting the epidermal growth factor receptor pathway in colorectal cancer. Clin Colorectal Cancer. 2005;5(suppl 2):S71–S80. doi: 10.3816/ccc.2005.s.010. [DOI] [PubMed] [Google Scholar]
- 119.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 120.Jimeno A, Messersmith WA, Hirsch FR, et al. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: Practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 121.Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol. 2008;8:413–418. doi: 10.1016/j.coph.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 122.Siena S, Sartore-Bianchi A, Di Nicolantonio F, et al. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Siddiqui AD, Piperdi B. KRAS mutation in colon cancer: A marker of resistance to EGFR-I therapy. Ann Surg Oncol. 2010;17:1168–1176. doi: 10.1245/s10434-009-0811-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Normanno N, Tejpar S, Morgillo F, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 125.Bekaii-Saab T. KRAS testing in metastatic colorectal cancer: Implications on the use of biologic agents. Clin Colorectal Cancer. 2009;8:135–140. doi: 10.3816/CCC.2009.n.022. [DOI] [PubMed] [Google Scholar]
- 126.Plesec TP, Hunt JL. KRAS mutation testing in colorectal cancer. Adv Anat Pathol. 2009;16:196–203. doi: 10.1097/PAP.0b013e3181a9d4ed. [DOI] [PubMed] [Google Scholar]
- 127.Chang DZ, Kumar V, Ma Y, et al. Individualized therapies in colorectal cancer: KRAS as a marker for response to EGFR-targeted therapy. J Hematol Oncol. 2009;2:18. doi: 10.1186/1756-8722-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baynes RD, Gansert J. KRAS mutational status as a predictor of epidermal growth factor receptor inhibitor efficacy in colorectal cancer. Am J Ther. 2009;16:554–561. doi: 10.1097/MJT.0b013e318199fa17. [DOI] [PubMed] [Google Scholar]
- 129.Saif MW, Shah M. K-ras mutations in colorectal cancer: A practice changing discovery. Clin Adv Hematol Oncol. 2009;7:45–53. 64. [PubMed] [Google Scholar]
- 130.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 131.Javle M, Hsueh CT. Updates in gastrointestinal oncology—insights from the 2008 44th annual meeting of the American Society of Clinical Oncology. J Hematol Oncol. 2009;2:9. doi: 10.1186/1756-8722-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frattini M, Saletti P, Romagnani E, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 134.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 135.Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 136.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 137.Tejpar S, Peeters M, Humblet Y, et al. Phase I/II study of cetuximab dose-escalation in patients with metastatic colorectal cancer (mCRC) with no or slight skin reactions on cetuximab standard dose treatment (EVEREST): Pharmacokinetic (PK), pharmacodynamic (PD) and efficacy data. J Clin Oncol. 2007;25(18) suppl:4037. [Google Scholar]
- 138.Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 139.Heinemann V, Stintzing S, Kirchner T, et al. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2008;35:262–271. doi: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 140.Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 141.Molinari F, Martin V, Saletti P, et al. Differing deregulation of EGFR and downstream proteins in primary colorectal cancer and related metastatic sites may be clinically relevant. Br J Cancer. 2009;100:1087–1094. doi: 10.1038/sj.bjc.6604848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 144.Meriggi F, Di Biasi B, Abeni C, et al. Anti-EGFR therapy in colorectal cancer: How to choose the right patient. Curr Drug Targets. 2009;10:1033–1040. doi: 10.2174/138945009789577891. [DOI] [PubMed] [Google Scholar]
- 145.Prenen H, De Schutter J, Jacobs B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15:3184–3188. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 146.Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–2629. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 147.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perrone F, Lampis A, Orsenigo M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 149.Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS ONE. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Adjei AA. Ras signaling pathway proteins as therapeutic targets. Curr Pharm Des. 2001;7:1581–1594. doi: 10.2174/1381612013397258. [DOI] [PubMed] [Google Scholar]
- 151.Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochim Biophys Acta. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 152.Halter SA, Webb L, Rose J. Lack of ras mutations and prediction of long-term survival in carcinoma of the colon. Mod Pathol. 1992;5:131–134. [PubMed] [Google Scholar]
- 153.Benhattar J, Losi L, Chaubert P, et al. Prognostic significance of K-ras mutations in colorectal carcinoma. Gastroenterology. 1993;104:1044–1048. doi: 10.1016/0016-5085(93)90272-e. [DOI] [PubMed] [Google Scholar]
- 154.Lee JC, Wang ST, Lai MD, et al. K-ras gene mutation is a useful predictor of the survival of early stage colorectal cancers. Anticancer Res. 1996;16:3839–3844. [PubMed] [Google Scholar]
- 155.Ahnen DJ, Feigl P, Quan G, et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: A Southwest Oncology Group study. Cancer Res. 1998;58:1149–1158. [PubMed] [Google Scholar]
- 156.Belly RT, Rosenblatt JD, Steinmann M, et al. Detection of mutated K12-ras in histologically negative lymph nodes as an indicator of poor prognosis in stage II colorectal cancer. Clin Colorectal Cancer. 2001;1:110–116. doi: 10.3816/CCC.2001.n.011. [DOI] [PubMed] [Google Scholar]
- 157.Font A, Abad A, Monzó M, et al. Prognostic value of K-ras mutations and allelic imbalance on chromosome 18q in patients with resected colorectal cancer. Dis Colon Rectum. 2001;44:549–557. doi: 10.1007/BF02234328. [DOI] [PubMed] [Google Scholar]
- 158.Geido E, Sciutto A, Rubagotti A, et al. Combined DNA flow cytometry and sorting with k-ras2 mutation spectrum analysis and the prognosis of human sporadic colorectal cancer. Cytometry. 2002;50:216–224. doi: 10.1002/cyto.10109. [DOI] [PubMed] [Google Scholar]
- 159.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: Results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 160.Finkelstein SD, Sayegh R, Bakker A, et al. Determination of tumor aggressiveness in colorectal cancer by K-ras-2 analysis. Arch Surg. 1993;128:526–531. doi: 10.1001/archsurg.1993.01420170056008. discussion 531–532. [DOI] [PubMed] [Google Scholar]
- 161.Moerkerk P, Arends JW, van Driel M, et al. Type and number of Ki-ras point mutations relate to stage of human colorectal cancer. Cancer Res. 1994;54:3376–3378. [PubMed] [Google Scholar]
- 162.Span M, Moerkerk PT, de Goeij AF, et al. A detailed analysis of K-ras point mutations in relation to tumor progression and survival in colorectal cancer patients. Int J Cancer. 1996;69:241–245. doi: 10.1002/(SICI)1097-0215(19960621)69:3<241::AID-IJC15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 163.Cerottini JP, Caplin S, Saraga E, et al. The type of K-ras mutation determines prognosis in colorectal cancer. Am J Surg. 1998;175:198–202. doi: 10.1016/s0002-9610(97)00283-3. [DOI] [PubMed] [Google Scholar]
- 164.Bazan V, Migliavacca M, Zanna I, et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13:1438–1446. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 165.Bazan V, Agnese V, Corsale S, et al. Specific TP53 and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: Results of a 5-year Gruppo Oncologico dell'Italia Meridionale (GOIM) prospective study. Ann Oncol. 2005;16(suppl 4):iv50–iv55. doi: 10.1093/annonc/mdi908. [DOI] [PubMed] [Google Scholar]
- 166.Poehlmann A, Kuester D, Meyer F, et al. K-ras mutation detection in colorectal cancer using the Pyrosequencing technique. Pathol Res Pract. 2007;203:489–497. doi: 10.1016/j.prp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 167.Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–445. doi: 10.1002/path.2625. [DOI] [PubMed] [Google Scholar]
- 168.Suchy B, Zietz C, Rabes HM. K-ras point mutations in human colorectal carcinomas: Relation to aneuploidy and metastasis. Int J Cancer. 1992;52:30–33. doi: 10.1002/ijc.2910520107. [DOI] [PubMed] [Google Scholar]
- 169.Linnemann U, Schimanski CC, Gebhardt C, et al. Prognostic value of disseminated colorectal tumor cells in the liver: Results of follow-up examinations. Int J Colorectal Dis. 2004;19:380–386. doi: 10.1007/s00384-003-0555-3. [DOI] [PubMed] [Google Scholar]
- 170.Nash GM, Gimbel M, Shia J, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol. 2010;17:572–578. doi: 10.1245/s10434-009-0605-3. [DOI] [PubMed] [Google Scholar]
- 171.Bell SM, Scott N, Cross D, et al. Prognostic value of p53 overexpression and c-Ki-ras gene mutations in colorectal cancer. Gastroenterology. 1993;104:57–64. doi: 10.1016/0016-5085(93)90835-z. [DOI] [PubMed] [Google Scholar]
- 172.Hardingham JE, Butler WJ, Roder D, et al. Somatic mutations, acetylator status, and prognosis in colorectal cancer. Gut. 1998;42:669–672. doi: 10.1136/gut.42.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liang JT, Cheng YM, Chang KJ, et al. Reappraisal of K-ras and p53 gene mutations in the recurrence of Dukes' B2 rectal cancer after curative resection. Hepatogastroenterology. 1999;46:830–837. [PubMed] [Google Scholar]
- 174.Tortola S, Marcuello E, González I, et al. p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. J Clin Oncol. 1999;17:1375–1381. doi: 10.1200/JCO.1999.17.5.1375. [DOI] [PubMed] [Google Scholar]
- 175.Esteller M, González S, Risques RA, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001;19:299–304. doi: 10.1200/JCO.2001.19.2.299. [DOI] [PubMed] [Google Scholar]
- 176.González-Aguilera JJ, Oliart S, Azcoita MM, et al. Simultaneous mutations in K-ras and TP53 are indicative of poor prognosis in sporadic colorectal cancer. Am J Clin Oncol. 2004;27:39–45. doi: 10.1097/01.coc.0000045920.49210.7a. [DOI] [PubMed] [Google Scholar]
- 177.Janssen KP, Abal M, El Marjou F, et al. Mouse models of K-ras-initiated carcinogenesis. Biochim Biophys Acta. 2005;1756:145–154. doi: 10.1016/j.bbcan.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 178.Lee S, Cho NY, Choi M, et al. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 2008;58:104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 179.Nash GM, Gimbel M, Cohen AM, et al. KRAS mutation and microsatellite instability: Two genetic markers of early tumor development that influence the prognosis of colorectal cancer. Ann Surg Oncol. 2010;17:416–424. doi: 10.1245/s10434-009-0713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kern SE, Fearon ER, Tersmette KW, et al. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] JAMA. 1989;261:3099–3103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- 181.Laurent-Puig P, Olschwang S, Delattre O, et al. Survival and acquired genetic alterations in colorectal cancer. Gastroenterology. 1992;102:1136–1141. [PubMed] [Google Scholar]
- 182.Rochlitz CF, Heide I, de Kant E, et al. Position specificity of Ki-ras oncogene mutations during the progression of colorectal carcinoma. Oncology. 1993;50:70–76. doi: 10.1159/000227150. [DOI] [PubMed] [Google Scholar]
- 183.Morrin M, Kelly M, Barrett N, et al. Mutations of Ki-ras and p53 genes in colorectal cancer and their prognostic significance. Gut. 1994;35:1627–1631. doi: 10.1136/gut.35.11.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dix BR, Robbins P, Soong R, et al. The common molecular genetic alterations in Dukes' B and C colorectal carcinomas are not short-term prognostic indicators of survival. Int J Cancer. 1994;59:747–751. doi: 10.1002/ijc.2910590606. [DOI] [PubMed] [Google Scholar]
- 185.Markowitz S, Hines JD, Lutterbaugh J, et al. Mutant K-ras oncogenes in colon cancers do not predict patient's chemotherapy response or survival. Clin Cancer Res. 1995;1:441–445. [PubMed] [Google Scholar]
- 186.Kastrinakis WV, Ramchurren N, Maggard M, et al. K-ras status does not predict successful hepatic resection of colorectal cancer metastasis. Arch Surg. 1995;130:9–14. doi: 10.1001/archsurg.1995.01430010011001. [DOI] [PubMed] [Google Scholar]
- 187.Pricolo VE, Finkelstein SD, Wu TT, et al. Prognostic value of TP53 and K-ras-2 mutational analysis in stage III carcinoma of the colon. Am J Surg. 1996;171:41–46. doi: 10.1016/S0002-9610(99)80071-3. [DOI] [PubMed] [Google Scholar]
- 188.Wadler S, Bajaj R, Neuberg D, et al. Prognostic implications of c-Ki-ras2 mutations in patients with advanced colorectal cancer treated with 5-fluorouracil and interferon: A study of the Eastern Cooperative Oncology Group (EST 2292) Cancer J Sci Am. 1997;3:284–288. [PubMed] [Google Scholar]
- 189.Andersen SN, Løvig T, Breivik J, et al. K-ras mutations and prognosis in large-bowel carcinomas. Scand J Gastroenterol. 1997;32:62–69. doi: 10.3109/00365529709025065. [DOI] [PubMed] [Google Scholar]
- 190.Kressner U, Bjorheim J, Westring S, et al. Ki-ras mutations and prognosis in colorectal cancer. Eur J Cancer. 1998;34:518–521. doi: 10.1016/s0959-8049(97)10111-3. [DOI] [PubMed] [Google Scholar]
- 191.Liang JT, Chang KJ, Chen JC, et al. Clinicopathologic and carcinogenetic appraisal of DNA replication error in sporadic T3N0M0 stage colorectal cancer after curative resection. Hepatogastroenterology. 1999;46:883–890. [PubMed] [Google Scholar]
- 192.Bouzourene H, Gervaz P, Cerottini JP, et al. p53 and Ki-ras as prognostic factors for Dukes' stage B colorectal cancer. Eur J Cancer. 2000;36:1008–1015. doi: 10.1016/s0959-8049(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 193.Bleeker WA, Hayes VM, Karrenbeld A, et al. Prognostic significance of K-ras and TP53 mutations in the role of adjuvant chemotherapy on survival in patients with Dukes C colon cancer. Dis Colon Rectum. 2001;44:358–363. doi: 10.1007/BF02234733. [DOI] [PubMed] [Google Scholar]
- 194.Bleeker WA, Hayes VM, Karrenbeld A, et al. Impact of KRAS and TP53 mutations on survival in patients with left- and right-sided Dukes' C colon cancer. Am J Gastroenterol. 2000;95:2953–2957. doi: 10.1111/j.1572-0241.2000.02327.x. [DOI] [PubMed] [Google Scholar]
- 195.Rosty C, Chazal M, Etienne MC, et al. Determination of microsatellite instability, p53 and K-ras mutations in hepatic metastases from patients with colorectal cancer: Relationship with response to 5-fluorouracil and survival. Int J Cancer. 2001;95:162–167. doi: 10.1002/1097-0215(20010520)95:3<162::aid-ijc1028>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 196.Petrowsky H, Sturm I, Graubitz O, et al. Relevance of Ki-67 antigen expression and K-ras mutation in colorectal liver metastases. Eur J Surg Oncol. 2001;27:80–87. doi: 10.1053/ejso.2000.1029. [DOI] [PubMed] [Google Scholar]
- 197.Gervaz P, Bouzourene H, Cerottini JP, et al. Dukes B colorectal cancer: Distinct genetic categories and clinical outcome based on proximal or distal tumor location. Dis Colon Rectum. 2001;44:364–372. doi: 10.1007/BF02234734. [DOI] [PubMed] [Google Scholar]
- 198.Clarke GA, Ryan E, Crowe JP, et al. Tumour-derived mutated K-ras codon 12 expression in regional lymph nodes of stage II colorectal cancer patients is not associated with increased risk of cancer-related death. Int J Colorectal Dis. 2001;16:108–111. doi: 10.1007/s003840100291. [DOI] [PubMed] [Google Scholar]
- 199.Okulczyk B, Piotrowski Z, Kovalchuk O, et al. Evaluation of K-RAS gene in colorectal cancer. Folia Histochem Cytobiol. 2003;41:97–100. [PubMed] [Google Scholar]
- 200.Losi L, Luppi G, Benhattar J. Assessment of K-ras, Smad4 and p53 gene alterations in colorectal metastases and their role in the metastatic process. Oncol Rep. 2004;12:1221–1225. [PubMed] [Google Scholar]
- 201.Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 203.Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Finocchiaro G, Cappuzzo F, Janne PA. EGFR, HER2 and Kras as predictive factors for cetuximab sensitivity in colorectal cancer. J Clin Oncol. 2007;25:168s. [Google Scholar]
- 205.De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 206.Etienne-Grimaldi MC, Formento JL, Francoual M, et al. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008;14:4830–4835. doi: 10.1158/1078-0432.CCR-07-4906. [DOI] [PubMed] [Google Scholar]
- 207.Lurje G, Nagashima F, Zhang W, et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 208.Freeman DJ, Juan T, Reiner M, et al. Association of K-ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer. 2008;7:184–190. doi: 10.3816/CCC.2008.n.024. [DOI] [PubMed] [Google Scholar]
- 209.Tol J, Koopman M, Rodenburg CJ, et al. A randomised phase III study on capecitabine, oxaliplatin and bevacizumab with or without cetuximab in first-line advanced colorectal cancer, the CAIRO2 study of the Dutch Colorectal Cancer Group (DCCG). An interim analysis of toxicity. Ann Oncol. 2008;19:734–738. doi: 10.1093/annonc/mdm607. [DOI] [PubMed] [Google Scholar]
- 210.Garm Spindler KL, Pallisgaard N, Rasmussen AA, et al. The importance of KRAS mutations and EGF61A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol. 2009;20:879–884. doi: 10.1093/annonc/mdn712. [DOI] [PubMed] [Google Scholar]
- 211.Sohn BS, Kim TW, Lee JL, et al. The role of KRAS mutations in predicting the efficacy of cetuximab-plus-irinotecan therapy in irinotecan-refractory Korean metastatic colorectal cancer patients. Oncology. 2009;77:224–230. doi: 10.1159/000236046. [DOI] [PubMed] [Google Scholar]
- 212.Maestro ML, Vidaurreta M, Sanz-Casla MT, et al. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14:1229–1236. doi: 10.1245/s10434-006-9111-z. [DOI] [PubMed] [Google Scholar]
- 213.Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–2259. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 214.Oliveira C, Westra JL, Arango D, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–2311. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- 215.Deng G, Bell I, Crawley S, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 216.Wang L, Cunningham JM, Winters JL, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–5212. [PubMed] [Google Scholar]
- 217.Zlobec I, Bihl MP, Schwarb H, et al. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367–380. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 218.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 219.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 220.Seth R, Crook S, Ibrahem S, et al. Concomitant mutations and splice variants in KRAS and BRAF demonstrate complex perturbation of the Ras/Raf signalling pathway in advanced colorectal cancer. Gut. 2009;58:1234–1241. doi: 10.1136/gut.2008.159137. [DOI] [PubMed] [Google Scholar]
- 221.Oliveira C, Velho S, Moutinho C, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- 222.Kim JH, Shin SH, Kwon HJ, et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 223.Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 224.Rashid A, Shen L, Morris JS, et al. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Qi J, Zhu YQ, Huang MF, et al. Hypermethylation of CpG island in O6-methylguanine-DNA methyltransferase gene was associated with K-ras G to A mutation in colorectal tumor. World J Gastroenterol. 2005;11:2022–2025. doi: 10.3748/wjg.v11.i13.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Whitehall VL, Walsh MD, Young J, et al. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 227.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 228.Ward RL, Cheong K, Ku SL, et al. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 229.Shen L, Catalano PJ, Benson AB, 3rd, et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13:6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: Study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 231.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 232.Hawkins N, Norrie M, Cheong K, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 233.van Rijnsoever M, Elsaleh H, Joseph D, et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 234.Ratto C, Sofo L, Ippoliti M, et al. Prognostic factors in colorectal cancer. Literature review for clinical application. Dis Colon Rectum. 1998;41:1033–1049. doi: 10.1007/BF02237397. [DOI] [PubMed] [Google Scholar]
- 235.Shankaran V, Wisinski KB, Mulcahy MF, et al. The role of molecular markers in predicting response to therapy in patients with colorectal cancer. Mol Diagn Ther. 2008;12:87–98. doi: 10.1007/BF03256274. [DOI] [PubMed] [Google Scholar]
- 236.O'Connell MJ, Schaid DJ, Ganju V, et al. Current status of adjuvant chemotherapy for colorectal cancer. Can molecular markers play a role in predicting prognosis? Cancer. 1992;70(6) suppl:1732–1739. doi: 10.1002/1097-0142(19920915)70:4+<1732::aid-cncr2820701614>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 237.Jen J, Kim H, Piantadosi S, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–221. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 238.Graziano F, Catalano V, Baldelli AM, et al. Prognostic biomarkers in resected colorectal cancer: Implications for adjuvant chemotherapy. Expert Rev Anticancer Ther. 2001;1:247–257. doi: 10.1586/14737140.1.2.247. [DOI] [PubMed] [Google Scholar]
- 239.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060–2070. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 240.Popat S, Zhao D, Chen Z, et al. Relationship between chromosome 18q status and colorectal cancer prognosis: A prospective, blinded analysis of 280 patients. Anticancer Res. 2007;27:627–633. [PubMed] [Google Scholar]
- 241.Ogino S, Nosho K, Irahara N, et al. Prognostic significance and molecular associations of 18q loss of heterozygosity: A cohort study of microsatellite stable colorectal cancers. J Clin Oncol. 2009;27:4591–4598. doi: 10.1200/JCO.2009.22.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 243.Benson AB., 3rd New approaches to assessing and treating early-stage colon and rectal cancers: Cooperative group strategies for assessing optimal approaches in early-stage disease. Clin Cancer Res. 2007;13:6913s–6920s. doi: 10.1158/1078-0432.CCR-07-1188. [DOI] [PubMed] [Google Scholar]
- Benson AB., 3rd Present and future role of prognostic and predictive markers for patients with colorectal cancer. Am Soc Clin Oncol Educ Book. 2006:187–190. [Google Scholar]
- 245.Baddi L, Benson A., 3rd Adjuvant therapy in stage II colon cancer: Current approaches. The Oncologist. 2005;10:325–331. doi: 10.1634/theoncologist.10-5-325. [DOI] [PubMed] [Google Scholar]
- 246.Ohtani H. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 247.Atreya I, Neurath MF. Immune cells in colorectal cancer: Prognostic relevance and therapeutic strategies. Expert Rev Anticancer Ther. 2008;8:561–572. doi: 10.1586/14737140.8.4.561. [DOI] [PubMed] [Google Scholar]
- 248.Shunyakov L, Ryan CK, Sahasrabudhe DM, et al. The influence of host response on colorectal cancer prognosis. Clin Colorectal Cancer. 2004;4:38–45. doi: 10.3816/ccc.2004.n.008. [DOI] [PubMed] [Google Scholar]
- 249.Titu LV, Monson JR, Greenman J. The role of CD8(+) T cells in immune responses to colorectal cancer. Cancer Immunol Immunother. 2002;51:235–247. doi: 10.1007/s00262-002-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dalerba P, Maccalli C, Casati C, et al. Immunology and immunotherapy of colorectal cancer. Crit Rev Oncol Hematol. 2003;46:33–57. doi: 10.1016/s1040-8428(02)00159-2. [DOI] [PubMed] [Google Scholar]
- 251.Koch M, Beckhove P, Op den Winkel J, et al. Tumor infiltrating T lymphocytes in colorectal cancer: Tumor-selective activation and cytotoxic activity in situ. Ann Surg. 2006;244:986–992. doi: 10.1097/01.sla.0000247058.43243.7b. discussion 992–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Loose D, Van de Wiele C. The immune system and cancer. Cancer Biother Radiopharm. 2009;24:369–376. doi: 10.1089/cbr.2008.0593. [DOI] [PubMed] [Google Scholar]
- 253.Okada K, Komuta K, Hashimoto S, et al. Frequency of apoptosis of tumor-infiltrating lymphocytes induced by Fas counterattack in human colorectal carcinoma and its correlation with prognosis. Clin Cancer Res. 2000;6:3560–3564. [PubMed] [Google Scholar]
- 254.Evans C, Dalgleish AG, Kumar D. Review article: Immune suppression and colorectal cancer. Aliment Pharmacol Ther. 2006;24:1163–1177. doi: 10.1111/j.1365-2036.2006.03075.x. [DOI] [PubMed] [Google Scholar]
- 255.Houston AM, Michael-Robinson JM, Walsh MD, et al. The “Fas counterattack” is not an active mode of tumor immune evasion in colorectal cancer with high-level microsatellite instability. Hum Pathol. 2008;39:243–250. doi: 10.1016/j.humpath.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 256.Vermorken JB, Claessen AM, van Tinteren H, et al. Active specific immunotherapy for stage II and stage III human colon cancer: A randomised trial. Lancet. 1999;353:345–350. doi: 10.1016/S0140-6736(98)07186-4. [DOI] [PubMed] [Google Scholar]
- 257.Brandacher G, Perathoner A, Ladurner R, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: Effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 258.Jass JR. Lymphocytic infiltration and survival in rectal cancer. J Clin Pathol. 1986;39:585–589. doi: 10.1136/jcp.39.6.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ropponen KM, Eskelinen MJ, Lipponen PK, et al. Prognostic value of tumour-infiltrating lymphocytes (TILs) in colorectal cancer. J Pathol. 1997;182:318–324. doi: 10.1002/(SICI)1096-9896(199707)182:3<318::AID-PATH862>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 260.Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 261.Guidoboni M, Gafa R, Viel A, et al. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am J Pathol. 2001;159:297–304. doi: 10.1016/S0002-9440(10)61695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Diederichsen AC, Hjelmborg JB, Christensen PB, et al. Prognostic value of the CD4+/CD8+ ratio of tumour infiltrating lymphocytes in colorectal cancer and HLA-DR expression on tumour cells. Cancer Immunol Immunother. 2003;52:423–428. doi: 10.1007/s00262-003-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Funada Y, Noguchi T, Kikuchi R, et al. Prognostic significance of CD8+ T cell and macrophage peritumoral infiltration in colorectal cancer. Oncol Rep. 2003;10:309–313. [PubMed] [Google Scholar]
- 264.Menon AG, Janssen-van Rhijn CM, Morreau H, et al. Immune system and prognosis in colorectal cancer: A detailed immunohistochemical analysis. Lab Invest. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 265.Pages F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 266.Qiu H, Xiao-Jun W, Zhi-Wei Z, et al. The prognostic significance of peripheral T-lymphocyte subsets and natural killer cells in patients with colorectal cancer. Hepatogastroenterology. 2009;56:1310–1315. [PubMed] [Google Scholar]
- 267.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lugli A, Karamitopoulou E, Panayiotides I, et al. CD8+ lymphocytes/tumour-budding index: An independent prognostic factor representing a 'pro-/anti-tumour' approach to tumour host interaction in colorectal cancer. Br J Cancer. 2009;101:1382–1392. doi: 10.1038/sj.bjc.6605318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 270.Atreya I, Schimanski CC, Becker C, et al. The T-box transcription factor eomesodermin controls CD8 T cell activity and lymph node metastasis in human colorectal cancer. Gut. 2007;56:1572–1578. doi: 10.1136/gut.2006.117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Phillips SM, Banerjea A, Feakins R, et al. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg. 2004;91:469–475. doi: 10.1002/bjs.4472. [DOI] [PubMed] [Google Scholar]
- 272.Banerjea A, Bustin SA, Dorudi S. The immunogenicity of colorectal cancers with high-degree microsatellite instability. World J Surg Oncol. 2005;3:26. doi: 10.1186/1477-7819-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Saeterdal I, Bjørheim J, Lislerud K, et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:13255–13260. doi: 10.1073/pnas.231326898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Linnebacher M, Gebert J, Rudy W, et al. Frameshift peptide-derived T-cell epitopes: A source of novel tumor-specific antigens. Int J Cancer. 2001;93:6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 275.Ishikawa T, Fujita T, Suzuki Y, et al. Tumor-specific immunological recognition of frameshift-mutated peptides in colon cancer with microsatellite instability. Cancer Res. 2003;63:5564–5572. [PubMed] [Google Scholar]
- 276.Tougeron D, Fauquembergue E, Rouquette A, et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod Pathol. 2009;22:1186–1195. doi: 10.1038/modpathol.2009.80. [DOI] [PubMed] [Google Scholar]
- 277.Dolcetti R, Viel A, Doglioni C, et al. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154:1805–1813. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Takemoto N, Konishi F, Yamashita K, et al. The correlation of microsatellite instability and tumor-infiltrating lymphocytes in hereditary non-polyposis colorectal cancer (HNPCC) and sporadic colorectal cancers: The significance of different types of lymphocyte infiltration. Jpn J Clin Oncol. 2004;34:90–98. doi: 10.1093/jjco/hyh018. [DOI] [PubMed] [Google Scholar]
- 279.Michael-Robinson JM, Biemer-Hüttmann A, Purdie DM, et al. Tumour infiltrating lymphocytes and apoptosis are independent features in colorectal cancer stratified according to microsatellite instability status. Gut. 2001;48:360–366. doi: 10.1136/gut.48.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Drescher KM, Sharma P, Watson P, et al. Lymphocyte recruitment into the tumor site is altered in patients with MSI-H colon cancer. Fam Cancer. 2009;8:231–239. doi: 10.1007/s10689-009-9233-0. [DOI] [PubMed] [Google Scholar]
- 281.Buckowitz A, Knaebel HP, Benner A, et al. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer. 2005;92:1746–1753. doi: 10.1038/sj.bjc.6602534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Baker K, Zlobec I, Tornillo L, et al. Differential significance of tumour infiltrating lymphocytes in sporadic mismatch repair deficient versus proficient colorectal cancers: A potential role for dysregulation of the transforming growth factor-beta pathway. Eur J Cancer. 2007;43:624–631. doi: 10.1016/j.ejca.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 283.Prall F, Duhrkop T, Weirich V, et al. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Human Pathol. 2004;35:808–816. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 284.Clarke SL, Betts GJ, Plant A, et al. CD4+CD25+FOXP3+ regulatory T cells suppress anti-tumor immune responses in patients with colorectal cancer. PLoS ONE. 2006;1:e129. doi: 10.1371/journal.pone.0000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Yaqub S, Henjum K, Mahic M, et al. Regulatory T cells in colorectal cancer patients suppress anti-tumor immune activity in a COX-2 dependent manner. Cancer Immunol Immunother. 2008;57:813–821. doi: 10.1007/s00262-007-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Loddenkemper C, Schernus M, Noutsias M, et al. In situ analysis of FOXP3+ regulatory T cells in human colorectal cancer. J Transl Med. 2006;4:52. doi: 10.1186/1479-5876-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ling KL, Pratap SE, Bates GJ, et al. Increased frequency of regulatory T cells in peripheral blood and tumour infiltrating lymphocytes in colorectal cancer patients. Cancer Immun. 2007;7:7. [PMC free article] [PubMed] [Google Scholar]
- 288.Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 289.Suzuki H, Chikazawa N, Tasaka T, et al. Intratumoral CD8(+) T/FOXP3 (+) cell ratio is a predictive marker for survival in patients with colorectal cancer. Cancer Immunol Immunother. 2010;59:653–661. doi: 10.1007/s00262-009-0781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Le Gouvello S, Bastuji-Garin S, Aloulou N, et al. High prevalence of Foxp3 and IL17 in MMR-proficient colorectal carcinomas. Gut. 2008;57:772–779. doi: 10.1136/gut.2007.123794. [DOI] [PubMed] [Google Scholar]
- 291.Frey DM, Droese RA, Viehl CT, et al. High frequency of tumor-infiltrating FOXP3+ regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int J Cancer. 2010;126:2635–2643. doi: 10.1002/ijc.24989. [DOI] [PubMed] [Google Scholar]
- 292.Michel S, Benner A, Tariverdian M, et al. High density of FOXP3-positive T cells infiltrating colorectal cancers with microsatellite instability. Br J Cancer. 2008;99:1867–1873. doi: 10.1038/sj.bjc.6604756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Assmann V, Jenkinson D, Marshall JF, et al. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J Cell Sci. 1999;112:3943–3954. doi: 10.1242/jcs.112.22.3943. [DOI] [PubMed] [Google Scholar]
- 294.Yang B, Zhang L, Turley EA. Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J Biol Chem. 1993;268:8617–8623. [PubMed] [Google Scholar]
- 295.Assmann V, Gillett CE, Poulsom R, et al. The pattern of expression of the microtubule-binding protein RHAMM/IHABP in mammary carcinoma suggests a role in the invasive behaviour of tumour cells. J Pathol. 2001;195:191–196. doi: 10.1002/path.941. [DOI] [PubMed] [Google Scholar]
- 296.Maxwell CA, Keats JJ, Belch AR, et al. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res. 2005;65:850–860. [PubMed] [Google Scholar]
- 297.Turley EA, Noble PW, Bourguignon LY. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–4592. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 298.Lugli A, Zlobec I, Günthert U, et al. Overexpression of the receptor for hyaluronic acid mediated motility is an independent adverse prognostic factor in colorectal cancer. Mod Pathol. 2006;19:1302–1309. doi: 10.1038/modpathol.3800648. [DOI] [PubMed] [Google Scholar]
- 299.Hall CL, Yang B, Yang X, et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell. 1995;82:19–26. doi: 10.1016/0092-8674(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 300.Zlobec I, Minoo P, Baumhoer D, et al. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112:495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]
- 301.Zlobec I, Baker K, Terracciano LM, et al. RHAMM, p21 combined phenotype identifies microsatellite instability-high colorectal cancers with a highly adverse prognosis. Clin Cancer Res. 2008;14:3798–3806. doi: 10.1158/1078-0432.CCR-07-5103. [DOI] [PubMed] [Google Scholar]
- 302.Jehan Z, Bavi P, Sultana M, et al. Frequent PIK3CA gene amplification and its clinical significance in colorectal cancer. J Pathol. 2009;219:337–346. doi: 10.1002/path.2601. [DOI] [PubMed] [Google Scholar]
- 303.Kato S, Iida S, Higuchi T, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer. 2007;121:1771–1778. doi: 10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 304.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Jang KS, Song YS, Jang SH, et al. Clinicopathological significance of nuclear PTEN expression in colorectal adenocarcinoma. Histopathology. 2010;56:229–239. doi: 10.1111/j.1365-2559.2009.03468.x. [DOI] [PubMed] [Google Scholar]
- 306.Colakoglu T, Yildirim S, Kayaselcuk F, et al. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: Is PTEN loss predictor of local recurrence? Am J Surg. 2008;195:719–725. doi: 10.1016/j.amjsurg.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 307.Sawai H, Yasuda A, Ochi N, et al. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol. 2008;8:56. doi: 10.1186/1471-230X-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 309.Zlobec I, Molinari F, Kovac M, et al. Prognostic and predictive value of TOPK stratified by KRAS and BRAF gene alterations in sporadic, hereditary and metastatic colorectal cancer patients. Br J Cancer. 2010;102:151–161. doi: 10.1038/sj.bjc.6605452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zhu F, Zykova TA, Kang BS, et al. Bidirectional signals transduced by TOPK-ERK interaction increase tumorigenesis of HCT116 colorectal cancer cells. Gastroenterology. 2007;133:219–231. doi: 10.1053/j.gastro.2007.04.048. [DOI] [PubMed] [Google Scholar]