The study investigates the use of seven different micro-RNAs as novel tumor markers in breast cancer, prostate cancer, colon cancer, renal cancer, and melanoma patients and finds miR-195 to be useful in differentiating breast cancer from the other cancer types.
Keywords: miRNAs, Tumor markers, Noninvasive biomarkers, Breast cancer, Cancer diagnostics
Learning Objectives
After completing this course, the reader will be able to:
Describe the site-specific dysregulation of some miRNAs and explain their potential as non-invasive biomarkers for diagnosis, prognostication, or markers of treatment response.
Cite the characteristics reflected by circulating miR-195 and explain how elevated levels could be used as a breast tumor marker.
This article is available for continuing medical education credit at CME.TheOncologist.com
Abstract
Purpose.
The potential of microRNAs (miRNAs) as novel tumor markers has been the focus of recent scrutiny because of their tissue specificity, stability, and association with clinicopathological parameters. Data have emerged documenting altered systemic miRNA expression across a spectrum of cancers; however, it remains uncertain as to whether circulating miRNAs are tumor specific. Our aim was to assess a panel of cancer-associated miRNAs in the circulation of patients with various malignancies, to determine whether these “oncomirs” were tumor specific, and thus to establish whether systemic miRNA analysis has utility in cancer diagnosis.
Patients and Methods.
Whole blood samples were prospectively collected from preoperative cancer patients (breast, prostate, colon, and renal cancer and melanoma; n = 163) and healthy age- and sex-matched controls (n = 63). Total RNA was isolated, and a panel of seven miRNAs was quantified by real-time quantitative polymerase chain reaction in each sample.
Results.
Differential expression of the general oncomirs let 7a, miR-10b, and miR-155, was observed in the majority of cancer patients in a nonspecific manner. Significantly, elevated circulating miR-195 was found to be breast cancer specific and could differentiate breast cancer from other cancers and from controls with a sensitivity of 88% at a specificity of 91%. A combination of three circulating miRNAs, including miR-195, further enhanced the discriminative power of this test for breast cancer to 94%.
Conclusion.
These findings suggest that individual cancers display specific systemic miRNA profiles, which could aid in discriminating among cancer types. This finding is of notable clinical consequence because it illustrates the potential of systemic miRNAs as sensitive, specific, noninvasive cancer biomarkers.
Introduction
Early diagnosis of cancer remains a compelling challenge for clinicians; it is the ultimate goal in order to minimize treatment-associated morbidity and mortality and achieve maximal long-term survival. Currently, the concept of individualized therapeutic regimens for cancer patients is in vogue, as clinicians and translational researchers attempt to tailor treatment regimens in order that each patient receives maximal benefit in the neoadjuvant and adjuvant settings. The discovery of novel classes of molecular markers in cancer has provided exciting, potentially viable biomarkers that may have utility in early cancer detection. These biomarkers may facilitate accurate tumor stratification, predict response to treatments, predict the risk for disease recurrence or progression, or even represent novel therapeutic targets. One notable example in recent years was the exciting discovery by Slamon and colleagues, in 1985, that the human epidermal growth factor receptor (HER)-2/neu oncogene was overexpressed in 20%–30% of human breast cancers, and that the prognosis for patients whose tumors overexpress HER-2/neu is poor [1, 2]. This finding led to the development of a specific recombinant monoclonal antibody against HER-2/neu, trastuzumab, to treat patients whose tumors overexpress this oncogene. Subsequent trials evaluating this antibody, involving >10,000 women, established that adjuvant trastuzumab therapy halves the recurrence rate and reduces mortality by 30% [3–5]. Currently, HER-2/neu expression in breast tumor tissue is routinely evaluated, and when overexpressed, trastuzumab is considered for inclusion in individual adjuvant therapy regimens.
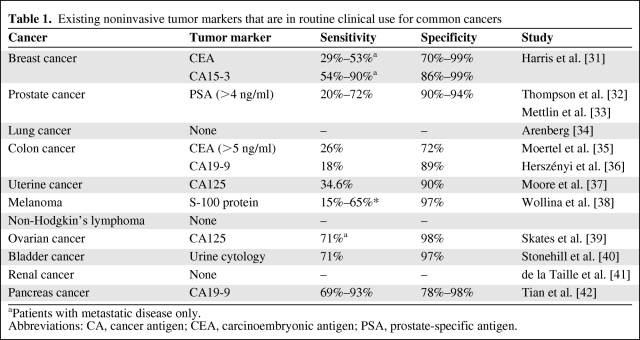
Despite stellar efforts in probing the molecular biology of common cancers, progress similar to that observed in breast cancer has not been mirrored; there remains an enormous dearth of knowledge regarding the molecular taxonomy and complex pathways of other malignancies. Currently, there are few known or validated biomarkers for early detection, treatment planning, follow-up, or targeted therapy of cancer. The utility of currently available tumor markers is limited by disappointing sensitivities and specificities, even in the case of prostate-specific antigen (PSA), which is widely used in routine clinical practice for the screening and management of prostate cancer patients (Table 1).
Table 1.
Existing noninvasive tumor markers that are in routine clinical use for common cancers
aPatients with metastatic disease only.
Abbreviations: CA, cancer antigen; CEA, carcinoembryonic antigen; PSA, prostate-specific antigen.
MicroRNAs (miRNAs) are a contemporary class of very short RNAs that control gene expression by targeting messenger RNAs and triggering either translational repression or RNA degradation [6, 7]. Aberrant miRNA expression underpins a variety of pathological processes, including carcinogenesis, and a number of miRNAs are known to be dysregulated in tumor tissues [8, 9] (supplemental online Table S1). The potential of miRNAs as novel tumor markers has been the focus of much recent attention because of their tissue specificity and unique ability to predict clinicopathological parameters with accuracy superior to that of mRNA expression profiling [10]. This recognition led to the exploration of these tiny molecules in the circulation in the hope that, if present, systemic miRNA analysis could herald a breakthrough in clinical practice, where the quest for sensitive and specific noninvasive cancer biomarkers persists. Recent reports have documented altered serum or plasma miRNAs in a variety of cancers, including prostate cancer, colon cancer, lung cancer, and, most recently, breast cancer [11–16]. However, it is unknown whether the specific miRNAs reported to be altered in these studies are disease specific or a global cancer phenomenon. The aim of this study was to investigate the utility of a panel of circulating miRNAs (miR-10b, miR-21, miR-145, miR-155, miR-195, and let 7a) as potential cancer biomarkers, in particular for early-stage disease. These target miRNAs have previously been reported to be dysregulated in various malignancies and have been identified to play key regulatory roles through their functional interactions with critical cancer-associated genes. Our recently reported findings that circulating miR-195 and let-7a are significantly elevated in breast cancer patients raised the obvious question as to whether elevated levels of these markers are specific to breast cancer patients or a general cancer phenomenon.
Materials and Methods
Study Cohort
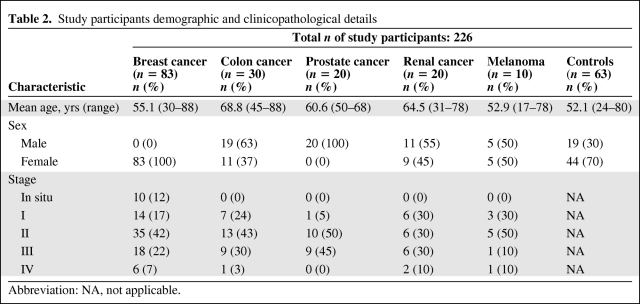
Following ethical approval and written informed consent, whole blood samples were collected prospectively from 226 participants, including 83 consecutive breast cancer patients, 30 colon cancer patients, 20 prostate cancer patients, 20 renal cell carcinoma patients, and 10 individuals with malignant melanoma. The control group comprised 63 healthy age-matched individuals from the community (female, 44; male, 19). All 163 cancer patients presented to the tertiary referral cancer center in the west of Ireland for management of their malignancy. Each case had a histologically confirmed diagnosis and the histological tumor profiles reflect those of typical cohorts for the respective malignancies (Table 2). Breast cancer cases were predominantly of the ductal type (71%), luminal A epithelial subtype (63%), and almost three quarters of the cohort had early-stage disease (71%), inclusive of a 12% proportion with in situ disease. The prostate cancer cohort comprised 20 males with adenocarcinoma of the prostate, 55% of whom had early, stage I and stage II, disease; the average Gleason score was 7 (n = 13), with a range of 6–9, and the mean PSA level at diagnosis was 7.67 (range, 3.1–17.5). Colon cancer cases (n = 30; male, 19) were all infiltrating adenocarcinomas, were predominantly left sided (73%), and 67% were early, stage I and stage II disease. The renal cell carcinoma cohort (n = 20; male, 11) had an average tumor size of 5.9 cm (range, 2–12 cm); 65% had Fuhrman nuclear grade II disease, 23% had extracapsular invasion, and 29% had node-positive disease. Of the 10 malignant melanoma cases (male, 5), seven patients presented with Clarkes level IV or V lesions; the mean Breslow's thickness was 2.3 mm (range, 0.9–5.0) and two patients had a positive sentinel node at presentation. All patients' demographic and clinicopathological details were entered in a prospectively maintained cancer database. The control blood samples were collected from age-matched healthy men and women residing in the same catchment area from which cases originated, and were collected on a contemporaneous basis with cases so as to minimize potential bias because of differential seasonal or environmental exposures. Control individuals were interviewed by a clinician prior to being enrolled in this study to ensure they had no current or previous malignancy, or concurrent inflammatory condition.
Table 2.
Study participants demographic and clinicopathological details
Abbreviation: NA, not applicable.
Blood Collection
We recently reported that systemic miRNA analysis is optimally performed on unclotted whole blood samples, versus serum or plasma [16]. Venous blood samples (nonfasting) were collected from each participant as follows: whole blood was collected in a Vacuette EDTA K3E blood bottle (Grenier Bio-One International AG, Kremsmünster, Austria). Unprocessed whole blood samples were stored at 4°C until required.
RNA Isolation: Copurification of Total RNA Using Trizol
Total RNA was extracted from 1 ml of whole blood using an adaptation of the TRI Reagent® BD technique (Molecular Research Center, Inc., Cincinnati, OH), as previously described. The RNA concentration was determined using a NanoDrop® spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). The wavelength-dependent extinction coefficient “33” was taken to represent the microcomponent of all RNA in solution. In general, concentrations in the range of 30–300 ng/μl of miRNA were obtained per sample. Integrity was assessed using RNA 6000 Nano LabChip Series II Assays (for small RNA) on a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
Each 1 ml of whole blood yielded 60 μl of total RNA, with yields in the range of 30–300 ng/μl of small RNA, which was then transferred to storage tubes prior to storage at −80°C.
Analysis of miRNA Gene Expression
We chose to study a panel of seven miRNAs in the circulation of all cancer patients (miR-10b, miR-21, miR-145, miR-155, miR-195, let 7a, and miR-16). These were chosen based on their previously documented associations with malignancies [17, 18] or for their potential as endogenous controls in the circulation [14–16].
Real-time quantitative polymerase chain reaction (RQ-PCR) quantification of miRNA expression was performed using TaqMan MicroRNA® Assays (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Total RNA was reverse transcribed using the MultiScribe™-based High-Capacity cDNA Archive kit (Applied Biosystems). RT− controls were included in each batch of reactions. PCR reactions were carried out in final volumes of 10 μl using an ABI 7900 HT Fast Real-Time PCR System (Applied Biosystems). Briefly, reactions consisted of 0.7 μl cDNA, 5 μl TaqMan® Universal PCR Fast Master Mix, and 0.2 μM TaqMan® primer–probe mix (Applied Biosystems). Reactions were initiated with a 10-minute incubation at 95°C followed by 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds. An interassay control derived from a breast cancer cell line (ZR-75-1) was included on each plate and all reactions were performed in triplicate. miR-16 was used as an endogenous control to standardize miRNA expression. The threshold standard deviation (SD) for intra-assay and interassay replicates was 0.3. The percentage PCR amplification efficiencies (E) for each assay were calculated using the slope of the semilog regression plot of cycle threshold versus log input of cDNA (10-fold dilution series of five points), with the following equation, and a threshold of 10% above or below 100% efficiency was applied: E = (10−1/slope − 1) × 100.
The relative quantity of miRNA expression was calculated using the comparative cycle threshold (ΔΔCt) method [19], normalized to miR-16 levels, and the lowest expressed sample was used as a calibrator.
Statistical Analysis
Data were analyzed using the software package SPSS 17.0 for Windows (SPSS Inc., Chicago, IL). Because of the magnitude and range of relative miRNA expression levels observed, results data were log transformed for analysis. There was no evidence against normality for the log-transformed data as confirmed using the Kolmogorov–Smirnov test and so data are presented as the mean ± SD. Analysis of variance (ANOVA), followed by the Tukey honestly significant difference post hoc test, was used to compare the mean response between the levels of the between-subject factors of interest whereas the two-sample t-test was used for any two-sample comparisons. All tests were two tailed and results with a p < .05 were considered statistically significant. Receiver operating characteristic (ROC) curves were constructed and the area under the curve (AUC) was calculated to assess the ability of each miRNA to differentiate between cancer cases and controls, by computing sensitivity and specificity for each possible cutoff point of the individual miRNAs. This was performed univariately for each individual miRNA, and multivariately for combinations of the six target miRNAs in our panel via logistic regression analysis.
Results
Dysregulated Expression of miRNAs in the Circulation of Cancer Patients
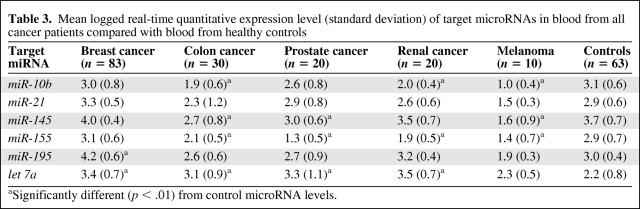
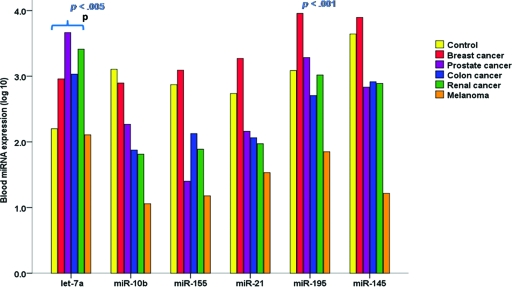
Expression of seven miRNAs, selected for their established relevance to cancer (miR-10b, miR-21, miR-145, miR-155, miR-195, miR-16, and let-7a), was detectable at variable levels in the circulation of all 226 study participants (163 cancer patients and 63 healthy age-matched controls) (Table 3). miR-16 expression was stable and reproducible across all 226 participants' peripheral blood samples (supplemental online Fig. S1) and was therefore used to normalize RQ-PCR data. To explore the potential of using this panel of miRNAs as specific biomarkers for breast cancer, we compared levels of the six target miRNAs in the circulation of 83 consecutive breast cancer patients with those of 80 other cancer patients and 63 healthy control subjects. Circulating levels of five cancer-associated miRNAs (let-7a, miR-10b, miR-145, miR-155, and miR-21) were generally dysregulated in the presence of several cancers, with no specific one of these five markers denoting a particular malignancy. By contrast, an elevated level of systemic miR-195 was unique to the breast cancer cohort (p < .001), indicating that it may be a breast cancer–specific marker. This expression pattern held true for cancer patients with early-stage disease specifically (TNM stage, in situ, I, and II) (Fig. 1).
Table 3.
Mean logged real-time quantitative expression level (standard deviation) of target microRNAs in blood from all cancer patients compared with blood from healthy controls
aSignificantly different (p < .01) from control microRNA levels.
Figure 1.
Circulating micro-RNA (miRNA) expression in early-stage cancers. Comparing early-stage cancers (TNM stages, in situ, I, and II; n = 110) with controls (n = 63), miR-195 expression was observed to be significantly elevated only in breast cancer patients (p < .001). Circulating let-7a levels were significantly elevated in patients with several early-stage visceral malignancies.
Abbreviation: TNM, tumor–node–metastasis.
Generic “Oncomirs”
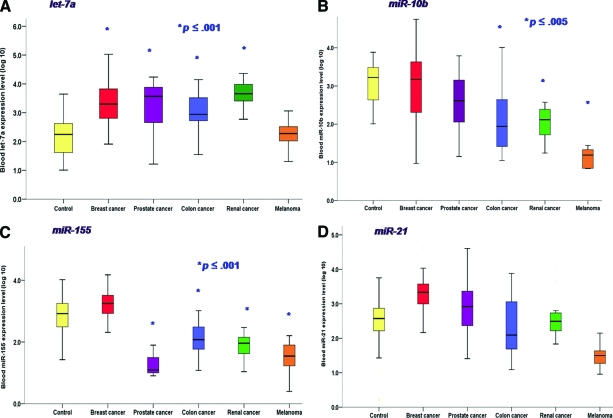
Let-7a levels were observed to be significantly higher in the circulation of patients with several visceral malignancies (breast, prostate, colon, and renal cancers) than in controls (ANOVA p <0.001). Systemic let-7a levels were similarly higher in patients with these malignancies, with no difference among the various cancer patient types (Fig. 2A). Patients with malignant melanoma were not observed to have altered circulating let-7a levels (p = 1.00).
Figure 2.
Nonspecific dysregulation of common “oncomirs” in the circulation. (A): Let-7a levels were significantly elevated in patients with a variety of visceral malignancies. (B): Circulating miR-10b expression levels in cancer patients. (C): Circulating miR-155 expression levels in cancer patients. (D): miR-21 expression was not significantly different among any of the cancer subgroups or when compared with controls.
miR-10b, a prometastatic miRNA [20], was found to be significantly lower, on average, in blood from colon and renal cancer patients as well as melanoma patients (all stages of disease). Levels of circulating miR-10b did not differ significantly according to stage of disease in these patients. Systemic miR-10b levels were within the normal range in patients with breast and prostate cancers (Fig. 2B).
The level of miR-145, a tumor suppressor miRNA [21], was significantly lower in blood from colon cancer, prostate cancer, and melanoma patients than in control patients (p = .001), although levels in breast cancer patients did not differ from those of the control group (p = .162).
The level of miR-155, a miRNA associated with a variety of malignant tumors [22], was observed to be significantly lower systemically in patients with all malignancies (p < .001) except breast cancer, for whom levels were similar to those of the control group (p = .38) (Fig. 2C).
miR-21, a well-described oncogenic miRNA [23], was observed to be expressed at generally higher levels in the circulation of cancer patients, with the exception of melanoma cases. However this difference did not reach statistical significance in this study cohort (Fig. 2D).
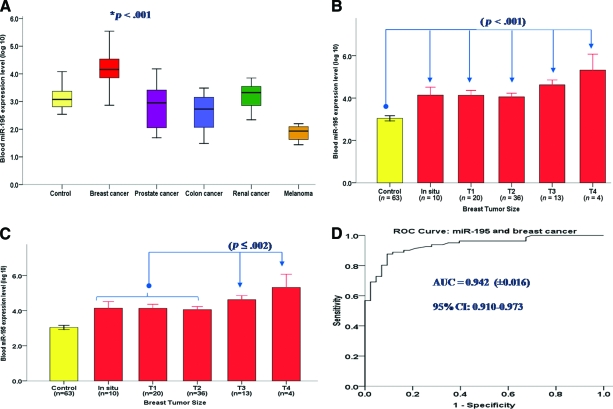
Breast Cancer–Specific Tumor Marker
Only one of our panel of miRNAs, miR-195, was exclusively overexpressed in a specific cancer population. miR-195 was observed to be significantly overexpressed only in blood from breast cancer patients (p < .001), with levels in other cancer patients largely comparable with those of healthy controls (Fig. 3A). On average, levels of miR-195 in breast cancer patients were 25-fold (unlogged fold change) higher than levels in control subjects. Circulating miR-195 levels correlated significantly with tumor size (Pearson's correlation coefficient, 0.446; p < .001), and patients with all types of tumors, irrespective of size, expressed significantly higher levels of miR-195 in the circulation than did the control cohort (Fig. 3B). We observed significant incrementally higher systemic miR-195 levels between small tumors (T1 and T2) and both the T3 and T4 tumor sizes (p = .002 and p < .001, respectively; ANOVA and Tukey post-hoc analysis) (Fig. 3C). miR-195 was also detectable in patients with noninvasive disease, at levels significantly higher than in the control group (Fig. 3B, 3C). ROC analysis determined the optimal cutoff value for miR-195 to differentiate breast cancer cases from controls, and from this analysis the sensitivity of circulating miR-195 alone was determined to be 87.7%, at a specificity of 91%, with an area under the ROC curve of 0.937 (Fig. 3D). A panel of three circulating miRNAs further increased the sensitivity over individual markers in isolation. The combination of circulating levels of miR-195, let-7a, and miR-155 increased the sensitivity for differentiating breast cancer cases from controls to 94% (logistic regression analysis, p < .001). Circulating miR-195 levels did not correlate with levels of the existing breast tumor marker cancer antigen 15.3 (Pearson's correlation coefficient, 0.138; p = .346).
Figure 3.
Circulating miR-195: A breast cancer–specific tumor marker. (A): Circulating miR-195 was significantly higher in patients with breast cancer than in control patients and patients with other cancers. (B, C): Circulating miR-195 according to breast tumor size. (D): ROC curve of the breast cancer sample set analyzed for systemic miR-195 expression.
Tumor size is documented as per the AJCC tumor–node–metastasis system: Tis, carcinoma in situ; T1, tumor ≤2 cm; T2, tumor 2.1–4.9 cm; T3, tumor ≥5 cm; T4, tumor of any size penetrating the skin or chest wall.
Abbreviations: AJCC, American Joint Committee on Cancer; AUC, area under the curve; CI, confidence interval; ROC, receiver operating characteristic.
Discussion
Our results demonstrate that cancer-associated miRNAs are generally dysregulated in the circulation of patients with visceral malignancy; this dysregulation appears to be relatively nonspecific. This is somewhat predictable, given that the majority of miRNAs investigated in this study have been associated with a variety of common cancers and are known to be involved in multiple critical stages of carcinogenesis. However, a growing body of evidence, based on high-throughput miRNA microarray studies in many cancer types, has identified other miRNAs that are specific for a given tissue type [10]. Our data for circulating miR-195, which was observed to be significantly elevated only in breast cancer patients, support the hypothesis that certain miRNAs are site specific. miR-195 had previously been demonstrated to be overexpressed in primary breast cancer tissue, which prompted its inclusion in the panel of miRNAs investigated in this study. Mattie et al. [24] identified miR-195 levels to be significantly higher in HER-2/neu+ than in HER-2/neu− breast cancers. Subsequently, Zhang et al. [25] identified miR-195 to be significantly elevated in breast tumor compared with normal breast tissue, a finding that was reproducible in our breast cancer cohort, thus validating these results [16].
Several findings from this, and previously published data from our group [16], provide convincing evidence to support circulating miR-195 as a breast cancer–specific tumor marker. miR-195 expression is higher in breast tumors than in normal breast tissue, a finding mirrored in the circulation, wherein miR-195 levels are considerably higher (19-fold) in breast cancer patients than in healthy controls. Two weeks following curative resection, systemic levels decreased to a basal level, comparable with those of the control group. Furthermore, both tumor and circulating levels correlated with disease burden (tumor size and stage of disease). Finally, miR-195 was not elevated in blood from patients with other malignancies (prostate, colon, and renal cancer or melanoma).
let-7a, one of the well-established cancer-associated miRNAs, was observed to be significantly elevated in almost all of our cancer patients, with the exception of those with malignant melanoma. Our findings support published data that implicate let-7a as a protagonist in many cancers, particularly lung, breast, colon, gastric, and ovarian cancer. However, we observed a paradoxical effect in the circulation (i.e., significantly higher systemic let-7a levels in cancer patients than in controls), compared with that described previously at the tumor tissue level, where let-7a is most commonly found to be underexpressed in tumors, compared with normal tissue, for each specific cancer [26]. Although unexpected, this finding may be explained by the interaction of let-7a with its target mRNA, the KRAS oncogene, at the cellular level. Recent evidence proposes that dysfunctional interaction between let-7 and KRAS, resulting from a single nucleotide polymorphism in the let-7 complementary site in the KRAS 3′ untranslated region, prevents let-7 from binding and exerting its tumor suppressor effect, resulting in overexpression of the oncogene [27]. A plausible hypothesis is that this particular failure of miRNA and mRNA to bind could lead to lower expression levels of let-7a in tumor tissues [28], and a reciprocal increase in free let-7a sequences entering the tumor microenvironment, and subsequently, the circulation. This hypothesis warrants further elucidation in order to define the precise mechanism by which miRNAs enter the circulation.
miR-155 levels were observed to be significantly lower in the circulation of all cancer patients, with the exception of breast cancer patients, in contrast to expression levels observed in primary breast tumor tissue [22]. Volinia et al. [29] identified miR-155 as an oncogenic miRNA overexpressed in several solid tumors, including colon cancer, lung cancer, lymphoma, and breast cancer. Subsequent functional studies have further defined its critical role in carcinogenesis [30]; it is known to promote cell migration and invasion by targeting RhoA, a gene involved in cell junction formation and stabilization. miR-155 also mediates transforming growth factor β–induced epithelial-to-mesenchymal transition—a remarkable process central to the development of tumor invasion and metastasis that involves the dissolution of epithelial tight junctions, intonation of adherens junctions, remodeling of the cytoskeleton, and loss of apical–basal polarity. Our cohort of prostate cancer, renal cancer, colon cancer, and malignant melanoma patients had a greater proportion of advanced cancers (TNM stages 2–4) than our breast cancer subgroup, 29% of whom had in situ or stage I disease. This may have contributed to the disparity in circulating miR-155 expression levels between breast cancer patients and patients with other cancers. However, our data suggest that miR-155 does have specific value as a biomarker for breast cancer. In our breast cancer subgroup, circulating levels of miR-155 in combination with miR-195 and let-7a did increase the ability of these miRNAs to discriminate cancer cases from controls, above the sensitivity of either miRNA alone. The analysis of this panel of miRNAs in blood could achieve a sensitivity of 94%.
Although our findings demonstrate the potential utility of circulating miRNAs as cancer-specific biomarkers, it is important to acknowledge that the sample size of cancers evaluated in this study is relatively small and that the panel of miRNAs selected for evaluation was biased toward our search for breast cancer–specific markers. Nonetheless, these data suggest that sustained effort toward developing circulating miRNAs as cancer-specific biomarkers is warranted.
Conclusion
The unique properties of miRNAs, including their remarkable stability, tissue-specific expression profiles, and the ease with which they are quantified, herald these molecules as ideal cancer biomarkers. Our data demonstrate the specificity of elevated circulating miR-195 for breast cancer, and the remarkably high sensitivity of miR-195 in combination with the general oncomirs let-7a and miR-155 for discriminating breast cancer cases from controls, thus prompting their potential utility as unique, noninvasive breast tumor markers. Further evaluation of blood-based miRNAs in larger cancer cohorts is necessary to validate these findings, and to further elucidate the feasibility of developing circulating miRNA assays specific for individual cancers as clinically useful tools to detect even early-stage malignancy.
Supplementary Material
Acknowledgments
Funding was provided by a Health Research Board fellowship award (H.M.H.) and the National Breast Cancer Research Institute (NBCRI), Ireland. We gratefully acknowledge Ms. Emer Hennessy for technical assistance and curation of the BioBank. We also thank Ms. Catherine Curran for collation of clinicopathological data.
This work was presented in part at the San Antonio Breast Cancer Symposium, December 2009.
Author Contributions
Conception/Design: Helen M. Heneghan, Nicola Miller, Michael J. Kerin
Provision of study material or patients: Helen M. Heneghan, Michael J. Kerin
Collection and/or assembly of data: Helen M. Heneghan, Ronan Kelly
Data analysis and interpretation: Helen M. Heneghan, Nicola Miller, John Newell
Manuscript writing: Helen M. Heneghan, Michael J. Kerin
Final approval of manuscript: Helen M. Heneghan, Nicola Miller, Ronan Kelly, John Newell, Michael J. Kerin
References
- 1.King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 1985;229:974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 3.Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med. 2006;354:809–820. doi: 10.1056/NEJMoa053028. [DOI] [PubMed] [Google Scholar]
- 4.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 5.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michael MZ, O'Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resnick KE, Alder H, Hagan JP, et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 16.Heneghan HM, Miller N, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 17.Heneghan HM, Miller N, Lowery AJ, et al. MicroRNAs as novel biomarkers for breast cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowery AJ, Miller N, McNeill RE, et al. MicroRNAs as prognostic indicators and therapeutic targets: Potential effect on breast cancer management. Clin Cancer Res. 2008;14:360–365. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 19.Davoren PA, McNeill RE, Lowery AJ, et al. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol. 2008;9:76. doi: 10.1186/1471-2199-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–572. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki HI, Yamagata K, Sugimoto K, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 22.Faraoni I, Antonetti FR, Cardone J, et al. miR-155 gene: A typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Talotta F, Cimmino A, Matarazzo MR, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 24.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Su B, Zhou QM, et al. [Differential expression profiles of microRNAs between breast cancer cells and mammary epithelial cells.] Ai Zheng. 2009;28:493–499. In Chinese. [PubMed] [Google Scholar]
- 26.Zhang B, Pan X, Cobb GP, et al. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 27.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong W, Yang H, He L, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 32.Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level ≤4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 33.Mettlin C, Littrup PJ, Kane RA, et al. Relative sensitivity and specificity of serum prostate specific antigen (PSA) level compared with age-referenced PSA, PSA density, and PSA change. Data from the American Cancer Society National Prostate Cancer Detection Project. Cancer. 1994;74:1615–1620. doi: 10.1002/1097-0142(19940901)74:5<1615::aid-cncr2820740520>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 34.Arenberg D. In search of the holy grail: Lung cancer biomarkers. Chest. 2004;126:325–326. doi: 10.1378/chest.126.2.325. [DOI] [PubMed] [Google Scholar]
- 35.Moertel CG, O'Fallon JR, Go VL, et al. The preoperative carcinoembryonic antigen test in the diagnosis, staging, and prognosis of colorectal cancer. Cancer. 1986;58:603–610. doi: 10.1002/1097-0142(19860801)58:3<603::aid-cncr2820580302>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.Herszényi L, Farinati F, Cardin R, et al. Tumor marker utility and prognostic relevance of cathepsin B, cathepsin L, urokinase-type plasminogen activator, plasminogen activator inhibitor type-1, CEA and CA 19–9 in colorectal cancer. BMC Cancer. 2008;8:194. doi: 10.1186/1471-2407-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore RG, Brown AK, Miller MC, et al. Utility of a novel serum tumor biomarker HE4 in patients with endometrioid adenocarcinoma of the uterus. Gynecol Oncol. 2008;110:196–201. doi: 10.1016/j.ygyno.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wollina U, Karte K, Hipler UC, et al. Serum protein s100β in patients with malignant melanoma detected by an immunoluminometric assay. J Cancer Res Clin Oncol. 2000;126:107–110. doi: 10.1007/s004320050018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skates SJ, Horick N, Yu Y, et al. Preoperative sensitivity and specificity for early-stage ovarian cancer when combining cancer antigen CA-125II, CA 15-3, CA 72-4, and macrophage colony-stimulating factor using mixtures of multivariate normal distributions. J Clin Oncol. 2004;22:4059–4066. doi: 10.1200/JCO.2004.03.091. [DOI] [PubMed] [Google Scholar]
- 40.Stonehill WH, Goldman HB, Dmochowski RR. The use of urine cytology for diagnosing bladder cancer in spinal cord injured patients. J Urol. 1997;157:2112–2114. [PubMed] [Google Scholar]
- 41.de la Taille A, Buttyan R, Katz AE, et al. Biomarkers of renal cell carcinoma. Past and future considerations. Urol Oncol. 2000;5:139–148. doi: 10.1016/s1078-1439(00)00064-8. [DOI] [PubMed] [Google Scholar]
- 42.Tian F, Appert HE, Myles J, et al. Prognostic value of serum CA 19-9 levels in pancreatic adenocarcinoma. Ann Surg. 1992;215:350–355. doi: 10.1097/00000658-199204000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.