Abstract
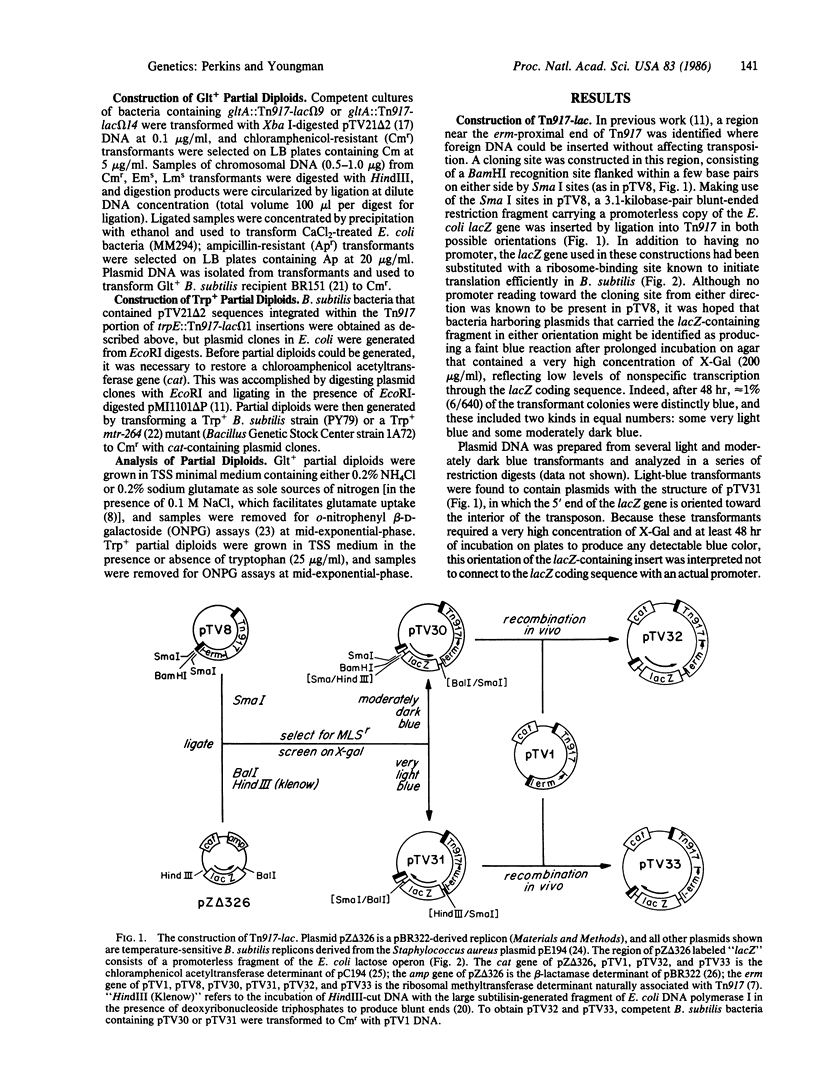
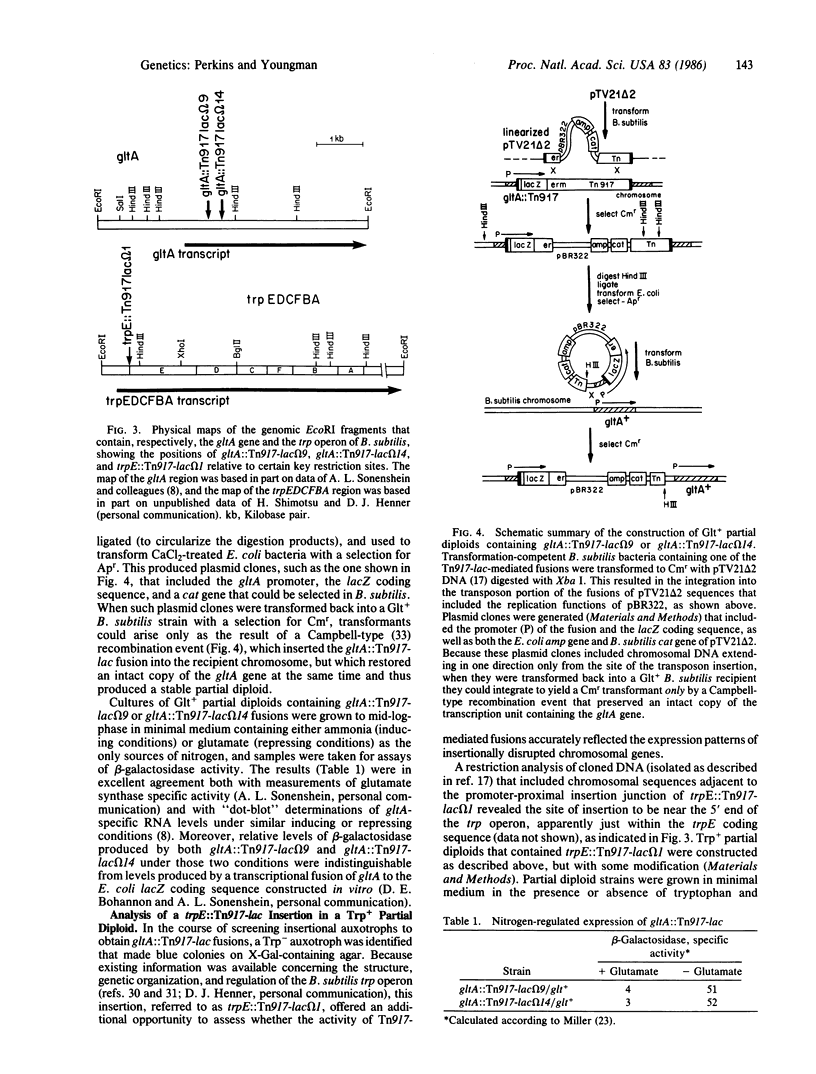
A derivative of Tn917 was constructed, referred to as Tn917-lac, which is capable of generating fusions that connect the transcripts of Bacillus subtilis chromosomal genes to the coding sequence of the lacZ gene of Escherichia coli. Two independent insertions of Tn917-lac into the gltA gene and one insertion into the trpE gene (in the trpEDCFBA operon) of B. subtilis were studied in detail, and the results confirmed that Tn917-lac-mediated transcriptional fusions produce levels of beta-galactosidase that reflect accurately the regulated expression of interrupted genes. To facilitate these studies, a procedure was developed that permits the analysis of Tn917-lac-mediated fusions in partial diploids where insertional mutations are complemented by an intact copy of the interrupted genes. Tn917 is known to function efficiently in bacteria representing three quite different Gram-positive genera (Streptococcus, Bacillus, and Staphylococcus) and is known to display a relatively high degree of randomness in its insertions into bacterial genomes, making it likely that Tn917-lac will be useful for the identification and study of many kinds of regulated genes in a wide range of Gram-positive species.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellofatto V., Shapiro L., Hodgson D. A. Generation of a Tn5 promoter probe and its use in the study of gene expression in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1035–1039. doi: 10.1073/pnas.81.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannon D. E., Rosenkrantz M. S., Sonenshein A. L. Regulation of Bacillus subtilis glutamate synthase genes by the nitrogen source. J Bacteriol. 1985 Sep;163(3):957–964. doi: 10.1128/jb.163.3.957-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. R., Hoch J. A., Aronson A. I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977 Sep;131(3):981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R., Smith I. Transformation and transduction in Bacillus subtilis: evidence for separate modes of recombinant formation. J Mol Biol. 1969 Oct 28;45(2):155–179. doi: 10.1016/0022-2836(69)90097-7. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Hahn J., Contente S., Dubnau D. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J Bacteriol. 1982 Nov;152(2):722–735. doi: 10.1128/jb.152.2.722-735.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O. Mapping of the 5-methyltryptophan resistance locus in Bacillus subtilis. J Bacteriol. 1974 Jan;117(1):315–317. doi: 10.1128/jb.117.1.315-317.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O., Roth C. W., Crawford I. P., Nester E. W. Control of tryptophan biosynthesis by the methyltryptophan resistance gene in Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):38–45. doi: 10.1128/jb.105.1.38-45.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love P. E., Lyle M. J., Yasbin R. E. DNA-damage-inducible (din) loci are transcriptionally activated in competent Bacillus subtilis. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6201–6205. doi: 10.1073/pnas.82.18.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Reznikoff W. S., Silverstone A. E., Ippen K., Signer E. R., Beckwith J. R. Fusions of the lac and trp Regions of the Escherichia coli Chromosome. J Bacteriol. 1970 Dec;104(3):1273–1279. doi: 10.1128/jb.104.3.1273-1279.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. J. A physical and functional analysis of Tn917, a Streptococcus transposon in the Tn3 family that functions in Bacillus. Plasmid. 1984 Sep;12(2):119–138. doi: 10.1016/0147-619x(84)90058-1. [DOI] [PubMed] [Google Scholar]
- Schreier H. J., Smith T. M., Bernlohr R. W. Regulation of nitrogen catabolic enzymes in Bacillus spp. J Bacteriol. 1982 Aug;151(2):971–975. doi: 10.1128/jb.151.2.971-975.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Characterization of the Bacillus subtilis tryptophan promoter region. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6315–6319. doi: 10.1073/pnas.81.20.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Clewell D. B. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1980 Mar;141(3):1366–1374. doi: 10.1128/jb.141.3.1366-1374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Reznikoff W. S. Cloning DNA restriction endonuclease fragments with protruding single-stranded ends. Gene. 1980 May;9(3-4):307–319. doi: 10.1016/0378-1119(90)90329-p. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Young F. E., Smith C., Reilly B. E. Chromosomal location of genes regulating resistance to bacteriophage in Bacillus subtilis. J Bacteriol. 1969 Jun;98(3):1087–1097. doi: 10.1128/jb.98.3.1087-1097.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins J. B., Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195(3):424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- Youngman P., Perkins J. B., Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984 Jul;12(1):1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Youngman P., Zuber P., Perkins J. B., Sandman K., Igo M., Losick R. New ways to study developmental genes in spore-forming bacteria. Science. 1985 Apr 19;228(4697):285–291. doi: 10.1126/science.228.4697.285. [DOI] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]



