The validity of a recently developed microRNA-based quantitative reverse transcriptase polymerase chain reaction test for identifying the tumor tissue of origin, first in a consecutive cohort of metastatic tumors of known origin and then in a cohort of cancer of unknown primary cases resected from the central nervous system, was evaluated.
Keywords: MicroRNA, qRT-PCR, Carcinoma of unknown primary (CUP), Tumor of unknown origin
Abstract
Background.
Identification of the tissue of origin of a brain metastatic tumor is vital to its management. Carcinoma of unknown primary (CUP) is common in oncology, representing 3%–5% of all invasive malignancies. We aimed to validate a recently developed microRNA-based quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) test for identifying the tumor tissue of origin, first in a consecutive cohort of metastatic tumors of known origin and then in a cohort of CUP cases resected from the central nervous system (CNS).
Patients and Methods.
One hundred two resected CNS metastatic tumors with known origin, previously classified based on the patient's clinical history and pathological data, as well as a second cohort of resected CNS tumors from 57 patients originally diagnosed as CUP were studied. A qRT-PCR diagnostic assay that measures the expression level of 48 microRNAs was used to classify the tissue of origin of these metastatic tumors.
Results.
In this blinded study, the test predictions correctly identified the reference diagnosis of the samples of known origin, excluding samples from prostate origin, in 84% of cases. In the second CUP patient cohort, the test prediction was in agreement with the diagnosis that was later confirmed clinically or with pathological evaluation in 80% of cases.
Conclusion.
In a cohort of brain and spinal metastases, a previously developed test based on the expression of 48 microRNAs allowed accurate identification of the tumor tissue of origin in the majority of cases. The high accuracy of this test in identifying the tissue of origin of metastases of unknown primary is demonstrated for the first time and may have broad clinical application.
Introduction
Metastatic tumors are the most common central nervous system (CNS) neoplasms. The reported incidence of up to 11 per 100,000 population per year is probably an underestimate resulting from underdiagnosis and inaccurate reporting [1]. Autopsy studies have revealed that CNS metastases occur in about 25% of patients who die from cancer [2]. The most common sources of brain metastases are, in descending order, lung cancer (especially small cell lung cancer [SCLC] and adenocarcinoma), breast cancer, melanoma, renal cancer, and colon cancer [1]. The histological, ultrastructural, and immunohistochemical (IHC) features of secondary CNS tumors are as diverse as in the primary tumors from which they arise. In the majority of cases, secondary CNS tumors show IHC characteristics similar to those of their respective primary tumors. IHC analyses are therefore often helpful in cases of unknown primary tumors for assessment of the exact nature and origin of the metastatic neoplasm [3, 4]. However, in an estimated 11% of patients with brain metastases, the primary tumor site remains unknown despite extensive workup by IHC and molecular biology [5], resulting in a time-consuming, labor-intensive, and expensive systematic search for a primary tumor site in these patients. In another 10% of patients with brain metastases, no primary tumor is found at presentation despite an extensive clinical workup [6], even though IHC or molecular data derived from the metastatic tissue seem to have reliably identified the probable primary tumor site. And in yet another small subset of patients, even though IHC results on metastatic tumor tissue seem to have reliably identified the primary tumor site, the individual clinical course may reveal an unsuspected primary tumor in later stages of disease or at autopsy [7], highlighting the limits of IHC approaches.
The prognosis of patients with CNS metastases is strongly influenced by the location of the CNS metastases, primary tumor control, and sensitivity to therapy. In cancer of unknown primary (CUP) patients, therapeutic regimens are chosen based only on the metastatic tissue properties. When the metastatic tissue doesn't hold suggestive features of a probable primary tumor, the oncologist lacks essential information and therefore may miss promising therapeutic opportunities for the patient.
In the past, to further minimize the number of patients with secondary CNS tumors and unidentified primary tumor sites, diagnosticians explored the feasibility of diverse molecular tests comparing genetic or chromosomal features of primary tumors with those of their respective metastases. Data from comparative genomic hybridization (CGH) revealed a high degree of conformity between brain metastases and corresponding primary tumors [8]. However, the CGH technique is not suitable for routine diagnostic workup of tumor specimens, and is further restricted in its feasibility by routine fixation procedures in pathology departments. A test that reliably identifies the primary tumor site should therefore use formalin-fixed paraffin-embedded (FFPE) tissue, similarly to all other standard pathological techniques, and it should ideally be superior to the IHC markers available to date.
MicroRNAs are a group of short (21–23 nucleotides), noncoding genes that play an important role in regulating gene expression [9]. Mature microRNA is incorporated into a large protein complex, the RNA-induced silencing complex, where it binds its corresponding mRNA and inhibits protein translation. MicroRNAs have an important role in tissue differentiation [10, 11] and tumorigenesis [12, 13] and have highly tissue-specific expression patterns [14–16]. MicroRNAs are stable in tissue samples, stored frozen or as FFPE samples, as well as in body fluids like serum [17–19]. In previous studies, we demonstrated that a biologically motivated classifier based on expression of a relatively small number of microRNAs can accurately identify tissue of origin [20]. Based on these results, a quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assay was developed that measures the expression of 48 microRNAs and predicts tissue of origin in FFPE metastatic tumor samples [21]. The goal of this retrospective study was to assess the performance characteristics of this assay by applying it to unselected, archival brain and spinal metastases, from known origin as well as from real CUP cases.
Materials and Methods
Patients
In phase I, we examined 102 consecutive resection specimens of brain and spinal metastases identified between 1995 and 2008 from the database of the neuropathology department, Institute of Pathology at Ruprecht-Karls University, Heidelberg, Germany, after approval by the institutional review board. The set included 83 brain and 19 spine metastases, with clinically known primaries consistent with one of the 25 tumor classes the assay had been trained to recognize [22]. Phase II consisted of 60 FFPE blocks obtained from 57 different patients whose original diagnosis was defined as CUP, including three different metastases for one patient and two samples from an additional patient. Forty-seven patients had brain metastases and 10 had spinal metastases. For 22 patients, the primary tumor was later identified. For 31 patients, a suspected origin was suggested by the pathological data collected at the time of surgery or after the test results were obtained. The primary tumor for four patients remained unknown even after extensive clinicopathological workup, and no suggested origin existed for comparison. The analyses were performed blinded without knowledge of or access to the clinical information, original diagnosis, or hematoxylin and eosin (H&E) slide.
Sample Preparation
For each case, five 10-μm sections were cut into a microcentrifuge tube. A control H&E slide was reviewed by a pathologist (W.C.M.) to ensure sufficient tumor cellular content. In four cases for which the tumor cellular content was <50%, an additional test was performed on microdissected samples from five 10-μm sections mounted onto glass slides and scraped by a size 11 scalpel blade. Microdissection enriched the tumor cellular content to 80%–95%.
RNA Extraction
Total RNA was extracted as previously described [22]. Briefly, FFPE sections were deparaffinized with xylene, washed in ethanol, and digested with proteinase K. RNA was extracted using acid phenol–chloroform followed by ethanol precipitation and DNase digestion. Following a second acid phenol–chloroform extraction, the pellet was resuspended in diethylpyrocarbonate-treated water and analyzed for its concentration and purity by spectrophotometry (Nanodrop ND-1000, PEQLAB, Erlangen, Germany).
qRT-PCR Test
Forty-eight microRNAs were quantified using a qRT-PCR method in duplicate on 96-well plates as recently described [21]. RNA was polyadenylated, reverse transcribed, and measured by qPCR.
Samples were processed in batches starting with the extraction. Each batch harbored a positive control sample with a defined cycle threshold (CT) for all microRNAs. A “no sample,” which underwent RNA extraction, and a “no RNA” sample were used as negative controls to detect potential contamination.
Quality assessment of each well and the CTmiR were calculated and rescaled as previously described [21]. These values were used as input to two classifiers [21]—a binary decision tree and a K nearest neighbor (KNN). In short, the decision tree uses logistic regression on combinations of one to three microRNAs in each node to make binary decisions. The KNN is based on comparing the expression of all 48 microRNAs in each sample with all samples in the training database. Each classifier, the decision tree and KNN, returns a predicted tissue of origin and histological type when applicable. The test returns the two different predictions or a single consensus prediction if the predictions concur. When the decision tree and KNN predict different histological types for the same tissue of origin, the tissue of origin is returned as a consensus prediction with no histological type indicated.
IHC
IHC was performed semiautomatically on 5-μm FFPE sections mounted on coated slides (Dako REAL™, catalogue number S2024; Dako, Berlin, Germany) using the Ventana Benchmark XT (Ventana Medical Systems Inc., Strasbourg, France). Antigen retrieval procedures prior to primary antibody exposure and antibody dilutions were applied as determined for each antibody. Primary antibody-binding sites were visualized using the ultraView Universal DAB Detection Kit (catalogue number 760–500; Ventana/Roche, Mannheim, Germany). Nuclei were automatically counterstained using hematoxylin (catalogue number 760-2021; Ventana/Roche) and bluing reagent (catalogue number 760-2037; Ventana/Roche).
Results
MicroRNA-Based Results for Brain Metastatic Tumors of Known Origin (Phase I)
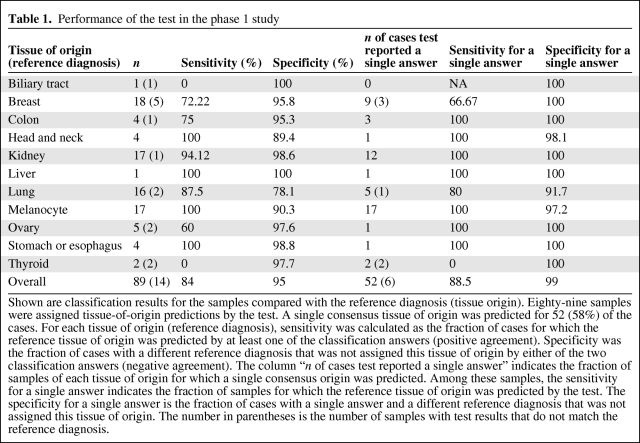
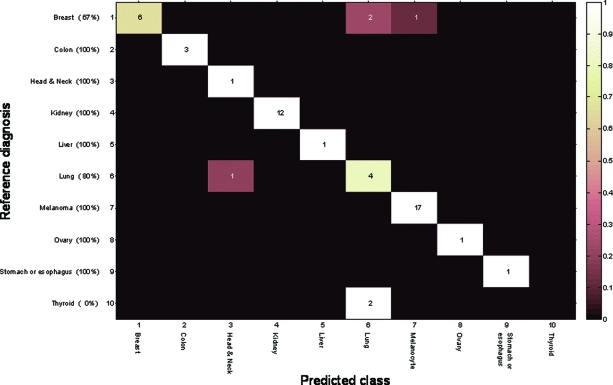
One of the 102 samples failed QA criteria (insufficient RNA amount to run the assay). For the remaining 101 samples, the test was completed successfully and produced tissue-of-origin predictions. Nine of the 12 prostate cancer metastases (75%) were classified incorrectly and were subsequently analyzed separately; therefore, we calculated this set to include 89 samples. For 75 of the 89 samples (84%), the reference diagnosis for tissue of origin was predicted by at least one of the classifiers (Table 1). For 52 of the 89 samples (58%), the two classifiers agreed, generating a consensus tissue-of-origin prediction. In the 37 cases for which two answers were provided, one by each classifier, the accuracy of the results was comparable between the two classifiers. For these 52 single-prediction cases, the sensitivity (positive agreement) was 88.5% (46 of 52 cases) and it was >90% for most tissue types (Fig. 1, Table 1). Specificity (negative agreement) in this group was in the range of 97%–100% and averaged >99%. Reassuringly, although the vast majority of samples with which the test was developed were primaries and metastases not from the CNS, the pattern and overall performance values were similar to the results obtained in the test validation study [21]. Furthermore, there was no statistically significant difference among poorly, moderately, or well-differentiated tumors in terms of classification results (data not shown).
Table 1.
Performance of the test in the phase 1 study
Shown are classification results for the samples compared with the reference diagnosis (tissue origin). Eighty-nine samples were assigned tissue-of-origin predictions by the test. A single consensus tissue of origin was predicted for 52 (58%) of the cases. For each tissue of origin (reference diagnosis), sensitivity was calculated as the fraction of cases for which the reference tissue of origin was predicted by at least one of the classification answers (positive agreement). Specificity was the fraction of cases with a different reference diagnosis that was not assigned this tissue of origin by either of the two classification answers (negative agreement). The column “n of cases test reported a single answer” indicates the fraction of samples of each tissue of origin for which a single consensus origin was predicted. Among these samples, the sensitivity for a single answer indicates the fraction of samples for which the reference tissue of origin was predicted by the test. The specificity for a single answer is the fraction of cases with a single answer and a different reference diagnosis that was not assigned this tissue of origin. The number in parentheses is the number of samples with test results that do not match the reference diagnosis.
Figure 1.
Confusion matrix for the 52 phase 1 samples resulting in a single answer. Rows indicate the reference diagnosis tissue and columns indicate the tissue determined by the microRNA test (shown are only the origins relevant for the single-answer cases). Numbers in parentheses (along the y-axis) denote the overall sensitivity per tissue of origin among the single-answer cases.
MicroRNA-Based Results for CNS Metastatic Tumors of Prostate Origin
When developing the test, because of the difficulty of obtaining sufficient numbers of metastatic samples, a comparison of microRNA expression in primary and metastatic tumor samples from the same origin was made to determine whether primaries can be used to augment the training set. In many cases, no significant difference was observed [20] (online supplemental data). In other cases, expression levels of all microRNAs were found to be very similar, with a small set differentially expressed, probably because of their expression in the surrounding tissue [23]. In order to avoid such bias, we trained the test on primary and metastatic tissues whenever possible. However, for prostate cancer, we were able to obtain only two metastases for training.
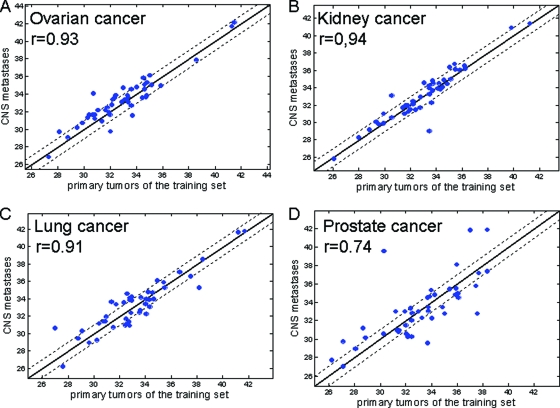
The brain metastatic set described in this paper included 12 metastases from prostate origin, nine classified incorrectly. We hypothesized that, in prostate cancer, microRNA expression between primaries and metastases is uncharacteristically different. To check this hypothesis, we compared the microRNA expression level in primary tissues from the training set with that of the metastatic samples from the present cohort. We found no significant differences and >0.9 correlation for ovarian, kidney, and lung cancer (Fig. 2A–2C), whereas for prostate cancer the correlation was only 0.74 (Fig. 2D).
Figure 2.
MicroRNA median CT signals in central nervous system metastases from phase 1 compared with primaries from the set used to develop the assay. Signals are normalized CT values. Dotted lines indicate factor 2. Analysis excluded three brain-specific microRNAs (miR-124, miR-9*, and miR-138), because we assume that their expression was derived from the surrounding tissue.
Abbreviation: CT, cycle threshold.
Classification Example
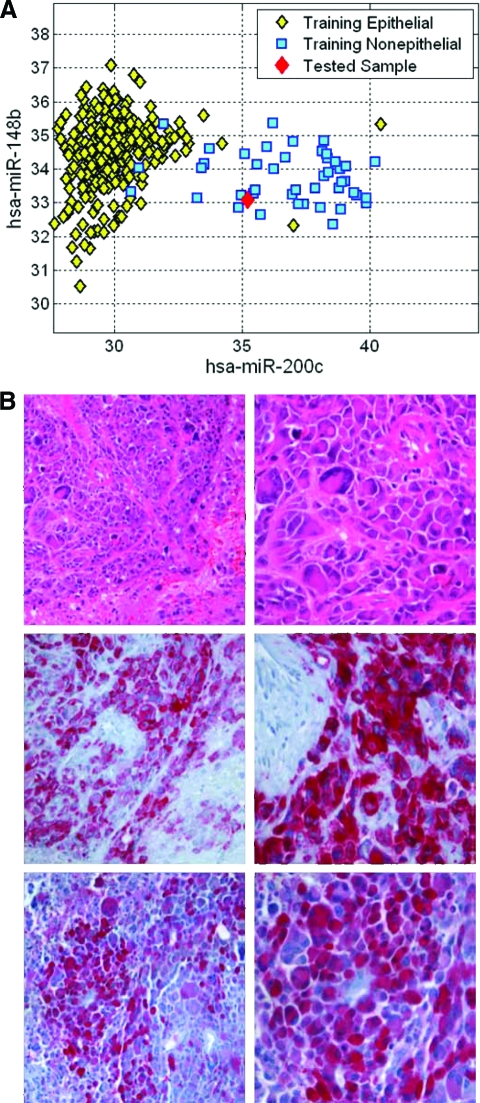
One of the samples originally diagnosed in the clinical setting as a brain metastatic tumor of breast cancer was classified by the test as a metastatic melanoma. Figure 3A presents the separation of epithelial and nonepithelial samples of the training set in the relevant decision tree node [21]. The tested sample plotted on this space clearly demonstrates nonepithelial features, suggesting that the breast is unlikely to be the correct tissue of origin. Upon re-examination of the clinical record, we found that this sample was originally classified as a breast metastasis based on the medical history of the patient, who was identified as having a breast lesion several months earlier, diagnosed as a poorly differentiated adenocarcinoma with tumor giant cells, adenoid formations, and IHC negative for estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2/neu. The brain metastasis showed the same morphology and IHC profile. IHC of the brain metastases was performed following the qRT-PCR test results (Fig. 3B) and supported a diagnosis of melanoma, with HMB45 being positive and S-100 being focally positive, even though MELAN A was found to be negative. To further explore this case, a sample derived from the breast lesion, considered the primary tumor, was also studied with the microRNA-based test and was identified as melanoma by both the tree and the KNN algorithm. IHC of the breast lesion was later found to be positive for three melanoma markers (HMB45, S-100, and MELAN A).
Figure 3.
Classification example. (A): Measured levels (normalized CT, inversely proportional to log(abundance)) of hsa-miR-200c and hsa-miR-148b are compared for all training set samples and the tested sample. (B): Hematoxylin and eosin staining (upper panel) showed that the metastasis was composed of undifferentiated pleomorphic tumor cells. These reveal focally strong and specific immunopositivity for S100 (lower panel) and HMB45 (middle panel).
Abbreviation: CT, cycle threshold.
MicroRNA-Based Results for CUP Tumors (Phase II)
The set included 60 brain and spinal metastatic archival samples, obtained from 57 patients whose original clinical diagnosis was defined as CUP. Three of the 60 samples (5%) failed QA criteria. For 57 samples obtained from 54 different individuals, 47 brain and 10 intraspinal, the test was completed successfully and produced tissue-of-origin predictions (supplemental online Table 1). Three different brain metastases from one patient (ID21), later found to have lung adenocarcinoma, were correctly identified by the test. For another patient (ID39), two metachronous metastases were studied and resulted in a prediction of lung carcinoid origin, whereas the patient's final clinical evaluation determined a neuroendocrine tumor of unknown primary. The results are presented for 54 patients to avoid duplication in reporting agreement. For 27 of the 54 samples (50%), the two classifiers agreed, generating a consensus tissue-of-origin prediction.
To evaluate the performance of the test, we developed a concordance score based on the clinicopathological data available at the time of diagnosis and additional information gathered after the test result had been obtained. The score divides the results into four main categories: type 1, clinical match, diagnosis obtained with the assay is clinically confirmed and pathological findings are compatible; type 2, pathology match (no clinically verified primary tumor), subdivided into type 2a, pathology findings are “consistent with” the test results, and type 2b, pathology findings “cannot rule out” the test results; type 3, pathology mismatch (no clinically verified primary tumor), pathology workup is not typical for the test diagnosis (when the test predicts two possible origins, the pathology workup is not typical of both); and type 4, clinical mismatch, the clinical diagnosis is discordant with the test results. For four samples, no suggested origin existed and an “unknown” score was given.
We found that the test result predicted a convincing suggested origin in 40 (80%) of the 50 samples that had a suggested origin based on clinical and/or pathological data. Twenty-two samples (44%) were scored as type 1 and 18 (36%) were scored as type 2 (14 as type 2a and four as type 2b). Ten (20%) of the 50 samples were discordant, four as type 3 (pathology discordance) and six as type 4 (clinical discordance).
Microdissection
In four FFPE blocks, the tumor cellular content was ≤50%. These samples were processed twice, using five 10-μm sections and after microdissection, comparing the test predictions. In two cases, the classification was the same and agreed with the suggested origin. In the additional two cases, the predicted origins after microdissection were different from the nondissected ones. The first case, clinically suspected to originate from kidney cancer, was from a spine metastasis block containing 50% tumor cells. In the nonmicrodissected sample, the test resulted in colon or bile duct carcinoma, whereas the classification after microdissection changed to renal cell carcinoma or melanoma, correctly predicting the suspected origin. The second metastasis, suspected to originate from stomach adenocarcinoma, was of a brain metastasis block containing only 20% tumor cells. The nondissected sample rendered a renal cell carcinoma result, whereas after microdissection lung carcinoid or stomach adenocarcinoma were suggested, again successfully predicting the suspected origin.
Discussion
The current work describes the first validation of a newly developed microRNA-based qRT-PCR test that uses microRNA biomarkers for the identification of tumor tissue of origin. Here, we validated the test in a set of metastatic samples, which were derived from the CNS, of known origin as well as samples from true CUP cases.
Recently, a few papers describing gene expression–based assays to determine the tumor tissue of origin for clinical use were published [22–25]. Most studies aimed at identifying tissue of origin used mRNA profiling of tumor samples [26–29] and presented the development of the classifier based on a training set and validation results for an independent test set composed of samples that were either not relevant in clinical settings, like primary tumors, or samples that were more clinically relevant, taken from metastatic tissue with known origin. Ma et al. [28] described the development of a 92-gene qPCR test to differentiate 32 tumor classes that was validated on a small set of 119 independent FFPE tumor samples and resulted in an overall accuracy of 82%. In another study [25], the tissue of origin was determined based on 1,550 mRNAs measured on microarrays, resulting in an overall sensitivity of 87.8% on a validation set of 547 frozen samples.
Only a few attempts have been made to study the ability of a classifier to determine the tumor tissue of origin in a setting of metastatic tissue derived from real CUP patients. Varadhachary et al. [23] studied a large cohort of 120 CUP patients using a test that distinguished only six primary sites. The test predicted results for 63 patients, that is, it did not yield a meaningful result in 48% of patients. In another study, Horlings et al. [22] used an mRNA microarray–based assay in 38 CUP patients and compared the results with tentative suggestions derived from clinicopathological investigation, resulting in consistency with the clinical data for most samples. Here, for the first time, we studied the performance of a molecular test on a consecutive large prospective cohort of metastatic samples from a specific site, the CNS.
In the first phase, we studied test performance in a cohort of samples with “known” tumor of origin. Interestingly, although the test was developed using a training set that contained only a small number of brain metastatic samples, representing only five of the 25 different tumor classes in the assay, the assay performance was very similar to the validation set [21]. While analyzing the results, we realized that the performance for prostate cancer CNS metastases was much lower than expected. As this manuscript was prepared, a few papers [30, 31] demonstrated the role of microRNAs in prostate cancer, specifically in antiandrogen therapy and resistance [30]. Leite et al. [31] compared the expression of only 14 microRNAs between localized high-grade and metastatic androgen-independent prostate carcinoma and found half of them to be differentially expressed. When we compared microRNA expression, we found that the correlation between the primary and metastatic prostate tissue was significantly lower than seen in other tissues. Because prostate cancer is not considered a common tissue of origin, representing only 2% of CUP cases, as confirmed by autopsy studies [32], we decided to exclude this origin from further analyses.
Because in CUP, by definition, the primary site in many cases remains unknown, there is inherent difficulty in validating a CUP assay. To tackle this problem, we developed a scoring system that compares the test result with all available clinicopathological data. Based on this scoring system, we demonstrated that, for the vast majority of the true CUP cases, namely, 40 (80%) of 50 cases for which a suggested origin existed, the microRNA-based test was in agreement with the available clinicopathological data.
In some cases, pathological data alone correctly identified the primary tumor site. For such cases, the microRNA test is a confirmative tool. In others, the clinical workup of a patient or the pathology of a metastatic lesion indicated more than one potential primary tumor site, and the microRNA test was helpful in identifying the true primary lesion, as in case ID136. In that case, clinical workup prior to metastatic tumor surgery had detected a lesion suspicious for thyroid tumor. The pathology, however, was in favor of lung cancer as the underlying primary tumor. The microRNA test identified lung as the primary tumor site upfront, resolving the clinical and pathology conflict. Imaging analyses following metastatic surgery identified a lung lesion, histopathologically verified after resection.
Despite extensive workup, including IHC and all currently available clinicodiagnostic procedures, <20% of patients with CUP have a primary site of their cancer identified ante mortem [33]. Autopsy studies have reported that 70% of cases remain undiagnosed [33].
These data impressively show the futility of unguided clinicodiagnostic procedures, which are often time-consuming and expensive. In addition, even though the number of primary tumors likely to metastasize to the brain is limited and a growing number of organ-specific antibodies has helped to elucidate the provenance of metastatic lesions in the majority of cases, 2%–18% of all metastatic lesions to the brain remain tumors of unknown primary site [3, 5, 34, 35]. This holds true even if taking into account that very practical algorithms for the IHC evaluation of common CNS metastatic neoplasms, including poorly differentiated neoplasms, have been developed. These are common in the way they implement a combination of antibodies to verify or exclude potential primary tumor sites [3, 5, 33, 36]. These data underscore the necessity of developing diagnostic tests that go beyond IHC.
In a considerable number of cases, the microRNA-based CUP test will be able to do both—identify the potential primary tumor lesion despite ambiguous IHC results and guide clinicodiagnostic procedures—thereby influencing not only diagnostic but also therapeutic decisions in individual patients.
These findings also imply that the primary lesions of CUP tumors that metastasize to the brain are either miniscule at the time of diagnosis and evade diagnostic testing, even at autopsy of the patient, or that CUP tumors may be a distinct group of tumors without true primary lesions but featuring the same metastatic potential as primary tumor lesions outside the CNS. The latter issue has not been extensively investigated on a molecular basis, and the limited information available is still controversial and inconclusive.
There is consent, however, that CUP is a heterogeneous group of tumors that is not a distinct biological entity involving specific genetic and phenotypic alterations. This argues for CUP lesions being derived from miniscule primary tumor sites and argues for the development of more refined molecular tests to identify the true primary tumor location. MicroRNAs are expressed in a highly tissue-specific manner. Because of their small size, microRNAs are robust and stable even in FFPE tissue samples. Thus, microRNAs constitute ideal diagnostic targets for the identification of tissue of origin in otherwise completely dedifferentiated tumors. In our view and experience, investigation of the microRNA profile of a metastatic lesion does not aim to replace either the clinicodiagnostic workup or IHC in surgical pathology or neuropathology but rather significantly extends the diagnostic repertoire of the investigating pathologist to guide clinicians in their search for a primary tumor lesion in true CUP cases lacking organ-specific IHC results. Thus, even when the test results in two possible tissues of origin, it helps the physician to focus the clinicodiagnostic workup to come up with a diagnosis and treatment decision.
Finally, because microRNA profiling is an unbiased analysis investigating a multitude of single independent parameters, it is objective, is investigator independent, and may, in the future, potentially be able to elucidate and define further subsets of metastatic CUP lesions that may follow a better clinical course and may require distinct therapies. Also, with the help of suitable training sets of tumors, the system is open to be extended to tumor types that belong to more favorable subgroups of metastatic CUP lesions that are currently known [37] and to any favorable subgroup to be identified in the future, to ensure their recognition in clinical practice and allow for specific and individualized treatment and patient management, especially in patients with CUP lesions with a potentially favorable outcome.
As pointed out earlier, a major advantage of the microRNA test is its unbiased nature. In patients who have a history of cancer, the investigating pathologist might be tempted to consider the clinically confirmed primary tumor as the most likely source of the metastatic lesion. In these cases, IHC investigations are aimed at confirming this notion, rather than at identifying other possibilities. This might lead to misinterpretation of the metastases, more so if the primary tumor has already been misinterpreted. Here, a case in the first cohort provides an excellent example. The microRNA test suggested melanoma as the origin, in contrast to the clinicopathological diagnosis of a breast cancer metastasis. This prompted melanoma-specific IHC that was confirmed, not only for the metastatic lesion in the brain but also for the supposedly primary breast tumor.
To date, many anatomic pathology tests, including IHC, have not been extensively studied for their cost-effectiveness [38]. This lack of information makes an upfront comparison of IHC and any other newly developed diagnostic test in pathology (e.g., the microRNA CUP test presented in this report and mRNA tests) regarding cost-effectiveness difficult, if not impossible. A commonly used matrix to determine the cost-effectiveness of any given diagnostic test is to look at its “cost per increase in patient life expectancy.” If one assumes that all patients with metastatic cancer have a universally poor prognosis and thus a similar life expectancy, then the “cost per increase in patient life expectancy” for any medical procedure in this cohort of patients would likely not be highly cost-effective. However, this assumption is flawed because not all patients with metastatic cancer have a universally negative outcome, nor is life expectancy the most important variable in many cases in which customized palliative treatments based on tumor type would have a profoundly positive impact on quality of life. In addition, increases in patient life expectancy are also likely not the most appropriate measure of an anatomic or molecular pathology test that imparts information rather than a tangible procedure [38]. Despite all these limitations, it is fair to say that IHC remains, by far, the most inexpensive test in pathology when compared with mRNA- or microRNA-based diagnostic tests. However, this reflects the cost per test only and does not reflect the potential impact of the test on patient management, disease outcome, or patient quality of life. Despite a low cost per test, extensive IHC staining does fail to identify the primary tumor site in metastatic lesions in 3%–10% of cases [36, 39]. Relative to the costs of other medical tests and procedures in patients with undefined primary malignancies, the microRNA-based CUP test is still rather inexpensive, follows strong and valid biological reasoning, and successfully identifies primary tumor sites in cases with ambiguous IHC results, thereby providing benefits other than and beyond cost-effectiveness, and it may avert even more costly procedures that otherwise would be performed [38].
Finally, tumor therapies have become more individualized based on the histomorphological or molecular features of the primary tumor. However, distinct histomorphological subtypes are often clouded by tumor dedifferentiation. Furthermore, clinicians often base their therapeutic strategies on findings in metastatic lesions and refrain from removing additional tissue for tumor classification, even if a primary tumor is identified later. Thus, in practice, the exact classification of the underlying primary tumor rests on the metastatic lesion. Here, refined molecular profiling might prove to be superior to conventional workup in characterizing the actual subtype of the primary tumor. In case ID151, the test succeeded in identifying the true morphological subtype of that particular lung cancer; here, SCLC was validated and confirmed by additional IHC performed following the test results. The small cell nature of the metastatic lesion was missed on conventional pathology.
In conclusion, we demonstrated that this microRNA expression–based assay provides an important objective tool for the diagnosis of tumor tissue of origin in CNS metastases, especially in CUP patients.
Supplementary Material
Author Contributions
Conception/Design: Wolf C. Mueller, Ayelet Chajut, Ranit Aharonov
Provision of study material or patients: Wolf C. Mueller, Ulrike Lass, Diana Jaeger
Collection and/or assembly of data: Wolf C. Mueller, Tina Bocker Edmonston, Brianna St. Cyr, Ulrike Lass, Yael Spector
Data analysis and interpretation: Wolf C. Mueller, Shai Rosenwald, Tina Bocker Edmonston, Yael Spector, Ayelet Chajut, Ranit Aharonov
Manuscript writing: Wolf C. Mueller, Tina Bocker Edmonston, Ayelet Chajut, Ranit Aharonov
Final approval of manuscript: Wolf C. Mueller, Shai Rosenwald, Tina Bocker Edmonston, Brianna St. Cyr, Ulrike Lass, Yael Spector, Ayelet Chajut, Ranit Aharonov, Diana Jaeger
References
- 1.Suki D. The epidemiology of brain metastases. In: Sawaya R, editor. Intracranial Metastases: Current Management Strategies. Malden, MA: Blackwell Futura Publishing; 2004. pp. 20–34. [Google Scholar]
- 2.Gavrilovic IT, Posner JB. Brain metastases: Epidemiology and pathophysiology. J Neurooncol. 2005;75:5–14. doi: 10.1007/s11060-004-8093-6. [DOI] [PubMed] [Google Scholar]
- 3.Becher MW, Abel TW, Thompson RC, et al. Immunohistochemical analysis of metastatic neoplasms of the central nervous system. J Neuropathol Exp Neurol. 2006;65:935–944. doi: 10.1097/01.jnen.0000235124.82805.2b. [DOI] [PubMed] [Google Scholar]
- 4.Drlicek M, Bodenteich A, Urbanits S, et al. Immunohistochemical panel of antibodies in the diagnosis of brain metastases of the unknown primary. Pathol Res Pract. 2004;200:727–734. doi: 10.1016/j.prp.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan RB, DeAngelis LM. Brain metastases. In: Schiff D, Wen PY, editors. Cancer Neurology in Clinical Practice. Totowa, NJ: Humana Press; 2003. pp. 73–86. [Google Scholar]
- 7.Hill HC. Challenges of utilizing immunostains to facilitate the diagnosis and management of metastatic adenocarcinoma. J Natl Med Assoc. 2008;100:1469–1473. doi: 10.1016/s0027-9684(15)31549-2. [DOI] [PubMed] [Google Scholar]
- 8.Petersen I, Hidalgo A, Petersen S, et al. Chromosomal imbalances in brain metastases of solid tumors. Brain Pathol. 2000;10:395–401. doi: 10.1111/j.1750-3639.2000.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 11.Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu A, Tetzlaff MT, Vanbelle P, et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 21.Rosenwald S, Gilad S, Benjamin S, et al. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol. 2010;23:814–823. doi: 10.1038/modpathol.2010.57. [DOI] [PubMed] [Google Scholar]
- 22.Horlings HM, van Laar RK, Kerst JM, et al. Gene expression profiling to identify the histogenetic origin of metastatic adenocarcinomas of unknown primary. J Clin Oncol. 2008;26:4435–4441. doi: 10.1200/JCO.2007.14.6969. [DOI] [PubMed] [Google Scholar]
- 23.Varadhachary GR, Talantov D, Raber MN, et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J Clin Oncol. 2008;26:4442–4448. doi: 10.1200/JCO.2007.14.4378. [DOI] [PubMed] [Google Scholar]
- 24.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 25.Monzon FA, Medeiros F, Lyons-Weiler M, et al. Identification of tissue of origin in carcinoma of unknown primary with a microarray-based gene expression test. Diagn Pathol. 2010;5:3. doi: 10.1186/1746-1596-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender RA, Erlander MG. Molecular classification of unknown primary cancer. Semin Oncol. 2009;36:38–43. doi: 10.1053/j.seminoncol.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Bloom G, Yang IV, Boulware D, et al. Multi-platform, multi-site, microarray-based human tumor classification. Am J Pathol. 2004;164:9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma XJ, Patel R, Wang X, et al. Molecular classification of human cancers using a 92-gene real-time quantitative polymerase chain reaction assay. Arch Pathol Lab Med. 2006;130:465–473. doi: 10.5858/2006-130-465-MCOHCU. [DOI] [PubMed] [Google Scholar]
- 29.Tothill RW, Kowalczyk A, Rischin D, et al. An expression-based site of origin diagnostic method designed for clinical application to cancer of unknown origin. Cancer Res. 2005;65:4031–4040. doi: 10.1158/0008-5472.CAN-04-3617. [DOI] [PubMed] [Google Scholar]
- 30.Gandellini P, Folini M, Zaffaroni N. Emerging role of microRNAs in prostate cancer: Implications for personalized medicine. Discov Med. 2010;9:212–218. [PubMed] [Google Scholar]
- 31.Leite KR, Sousa-Canavez JM, Reis ST, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2009 Apr 15; doi: 10.1016/j.urolonc.2009.02.002. [Epub ahead of print], doi: 10.1016/j.urolonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Pentheroudakis G, Greco FA, Pavlidis N. Molecular assignment of tissue of origin in cancer of unknown primary may not predict response to therapy or outcome: A systematic literature review. Cancer Treat Rev. 2009;35:221–227. doi: 10.1016/j.ctrv.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Pavlidis N, Fizazi K. Carcinoma of unknown primary (CUP) Crit Rev Oncol Hematol. 2009;69:271–278. doi: 10.1016/j.critrevonc.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Barnholtz-Sloan JS, Sloan AE, Davis FG, et al. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004;22:2865–2872. doi: 10.1200/JCO.2004.12.149. [DOI] [PubMed] [Google Scholar]
- 35.Lassman AB, DeAngelis LM. Brain metastases. Neurol Clin. 2003;21:1–23, vii. doi: 10.1016/s0733-8619(02)00035-x. [DOI] [PubMed] [Google Scholar]
- 36.Pavlidis N, Briasoulis E, Hainsworth J, et al. Diagnostic and therapeutic management of cancer of an unknown primary. Eur J Cancer. 2003;39:1990–2005. doi: 10.1016/s0959-8049(03)00547-1. [DOI] [PubMed] [Google Scholar]
- 37.Pavlidis N, Fizazi K. Cancer of unknown primary (CUP) Crit Rev Oncol Hematol. 2005;54:243–250. doi: 10.1016/j.critrevonc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Raab SS. Cost-Effectiveness of Immunohistochemistry. Philadelphia: Churchill Livingstone; 2002. pp. 45–56. [Google Scholar]
- 39.van de Wouw AJ, Janssen-Heijnen ML, Coebergh JW, et al. Epidemiology of unknown primary tumours; incidence and population-based survival of 1285 patients in Southeast Netherlands, 1984–1992. Eur J Cancer. 2002;38:409–413. doi: 10.1016/s0959-8049(01)00378-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.