The phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) network plays a key regulatory function in cell survival, proliferation, migration, metabolism, angiogenesis, and apoptosis. Genetic aberrations found at different levels make this pathway one of the most commonly disrupted in human breast cancer. Because the PI3K pathway has divergent downstream effects, the identification of the key effectors of the pathway and their presence in the different subtypes of breast tumors will allow the development of ideal targeted therapies with meaningful clinical efficacy.
Learning Objectives
After completing this course, the reader will be able to:
Describe how PTEN loss, PIK3CA mutations, and AKT dysregulation affect the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling network in human breast cancer.
Review the current state of AKT and mTOR inhibitor development, and describe its potential for clinical applications.
This article is available for continuing medical education credit at CME.TheOncologist.com
Abstract
The phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) network plays a key regulatory function in cell survival, proliferation, migration, metabolism, angiogenesis, and apoptosis. Genetic aberrations found at different levels, either with activation of oncogenes or inactivation of tumor suppressors, make this pathway one of the most commonly disrupted in human breast cancer. The PI3K-dependent phosphorylation and activation of the serine/threonine kinase AKT is a key activator of cell survival mechanisms. The activation of the oncogene PIK3CA and the loss of regulators of AKT including the tumor suppressor gene PTEN are mutations commonly found in breast tumors. AKT relieves the negative regulation of mTOR to activate protein synthesis and cell proliferation through S6K and 4EBP1. The common activation of the PI3K pathway in breast cancer has led to the development of compounds targeting the effector mechanisms of the pathway including selective and pan-PI3K/pan-AKT inhibitors, rapamycin analogs for mTOR inhibition, and TOR-catalytic subunit inhibitors. The influences of other oncogenic pathways such as Ras-Raf-Mek on the PI3K pathway and the known feedback mechanisms of activation have prompted the use of compounds with broader effect at multiple levels and rational combination strategies to obtain a more potent antitumor activity and possibly a meaningful clinical effect. Here, we review the biology of the network, its role in the development and progression of breast cancer, and the evaluation of targeted therapies in clinical trials.
Introduction
The transformation of normal mammary epithelial cells into cancer cells involves a multistep process with alterations in signal transduction pathways that confer important survival and growth advantages to malignant cells [1]. As part of the growth factor receptor (GFR) signaling, the phosphatidylinositol 3-kinase (PI3K) pathway is a key mediator of cell metabolism and cell growth that is affected by genetic aberrancies at different levels, becoming a crucial pathway for cancer development and representing a therapeutic target against breast cancer [2–5]. Understanding the principal effector mechanisms of the PI3Ks and the cross talk with other oncogenic signaling pathways has been the focus of extensive research to develop drugs with clinical efficacy [6].
PI3K Signaling Pathway
Phosphatidylinositol is a component of eukaryotic cell membranes. The inositol head of the phospholipid can be phosphorylated at multiple sites by phosphoinositide kinases (PIKs), which act as signal transducers involved in the regulation of multiple cell functions [7]. The PI3K superfamily has been studied profoundly since the discovery of PI3K activity associated with viral oncoproteins and its role in growth regulation and prevention of apoptosis and other cellular responses [7]. PI3Ks are grouped into classes I, II or III, depending on their subunit structure, regulation, and substrate selectivity. Each class contains various isoforms, class IA being the most studied in cancer [5]. Class IA PI3Ks (PIK3Cα, PIK3Cβ, and PIK3Cδ) are heterodimeric proteins with a regulatory subunit (p85) and a catalytic subunit (p110), that phosphorylate 4,5-phosphoinositide (4,5-PIP2) and generate the second messenger 3,4,5-phosphoinosite trisphosphate (PIP3) [7, 8]. The p110s are encoded by the PI3KCA gene and are regulated upstream by growth factor binding to tyrosine kinases receptors and G protein-coupled receptors. Activating mutations in the PI3KCA gene and the regulator p85 have been identified in breast cancer [9]. Activated RAS protein can interact with p110 and also activate class IA PI3Ks.
The generation of the second messenger 3,4,5-PIP3 by class IA PI3Ks plays a key role in downstream signaling by several effector proteins including the serine/threonine kinase AKT and PDK1 (phosphoinositide-dependent kinase 1) [10]. The membrane colocalization of both PDK1 and AKT through their pleckstrin homology domains results in phosphorylation at Thr308 and partial activation of AKT kinase. The phosphorylation of Ser473 by PDK2 generates complete activation of AKT [11]. AKT and its isoforms AKT-1, AKT-2, and AKT-3 have cell-transforming properties through the phosphorylation of multiple protein targets including mTOR (mammalian target of rapamycin), Bad, Caspase 9, Tuberin, GSK3b, and forkhead transcription factors involved in cell survival and apoptosis. Signaling through the PI3K/AKT pathway is negatively regulated by the tumor-suppressor gene PTEN (phosphatase and tensin homolog) localized in chromosome 10 [12–14].
AKT Downstream Signaling
AKT is a key regulator of a variety of proteins involved in cell proliferation, metabolism, survival, invasion, migration, apoptosis, and DNA repair. To execute this variety of actions, AKT relieves the negative regulation of mTOR mediated by the tumor-suppressor proteins: TSC1 and TSC2 (tuberous sclerosis complex proteins) [15–17]. Activation of mTOR plays a key role in the activation of protein synthesis contributing to the pathogenesis of multiple tumor types. Phosphorylation of TSC2 by AKT inactivates the GTP hydrolysis of the small GTP-binding protein Rheb (ras homologue enriched in the brain), permitting Rheb to remain in the GTP-bound state. Rheb-GTP binds and activates the mTOR kinase domain [18]. The proline-rich AKT substrate (PRAS40) is also a negative regulator of mTOR and it is inactivated by AKT phosphorylation [19, 20]. These findings expose the fundamental role of AKT in the mTOR activation by growth factors in that AKT inactivates two negative regulators of mTOR [21]. The TSC1/2 complex is also regulated by the LKB1-AMPK (AMP-dependent kinase) and MAPK pathways. These pathways are activated based on the nutritional (amino acids) and energy status of the cell. The convergence of these signals through the TSC1/2 complex allows mTOR to control cell growth and proliferation based on the availability of nutrients and energy sources [22].
mTOR exists in two multiprotein complexes: mTOR complexes 1 and 2 (mTORC1 and mTORC2). mTORC1 complex is composed of mTOR, raptor, mammalian LST8 (mLST-8/GβL), and PRAS40. mTORC1 activation controls protein synthesis by phosphorylating two translational regulatory proteins: eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4EBP1) and p70 ribosomal protein S6 kinase (S6K1). Raptor binds to S6K and 4EBP1 substrates and presents them to mTOR for phosphorylation [23]. The activation of S6K and 4EBP1 promotes translation initiation for protein synthesis [24–28]. Important proteins for cell cycle control like D-type cyclins, c-myc, and ornithine decarboxylase are also regulated by this complex [28]. mTOR also decreases ribosome biogenesis regulating transcription of ribosomal RNA and the eukaryotic elongation factor 2 kinase. In regulating the initiation and elongation steps, mTOR controls the overall rate of protein synthesis. The capacity of mTOR to regulate protein synthesis explains in part how the tumor-promoting functions of deregulated mTOR may be distributed among multiple targets [29, 30].
mTORC2 complex consists of mTOR, mSIN-1, mLST-8, PRR5 (proctor), and a different scaffolding protein called rictor (rapamycin-insensitive companion of mTOR) [31]. The activation of this complex remains poorly understood; it appears to be through growth factors in an AKT-independent manner [32]. mTORC2 phosphorylates AKT at Ser473 [33], leading AKT activation toward the Forkhead transcription factor FOXO and the apoptosis regulator BAD. mTORC2 also regulates the cell cytoskeleton and cell polarity through the phosphorylation of protein kinase C (PKCα) [31]. Recent studies in cell lines of colorectal cancer have shown that mTOR-associated proteins, Raptor and Rictor, are overexpressed in colorectal cancer cells [34]. The rapamycin-like drugs directly inhibit mTORC1 but not mTORC2. The Rictor protein makes the FRB domain of mTOR inaccessible to the rapamycin–FKBP-12 complex [35] (Figure 1). In some tumor cells, the inhibition of mTORC1 can enhance PI3K/AKT activation. Under normal conditions, the mTORC1 substrate S6K1 delivers a negative feedback signal by phosphorylating insulin receptor substrate 1 (IRS-1), preventing IRS-1 from recruiting PI3K to the receptor for activation [36, 37]. The inhibition of mTORC1 blocks the S6K-mediated negative feedback, resulting in enhanced PI3K/AKT activation that could activate survival pathways as possible means of resistance [38]. Therapeutic inhibition of mTORC2 may therefore potentiate the effect of mTORC1 inhibitors by preventing AKT activation.
Figure 1.
The PI3K/AKT/mTOR signaling network regulates cell survival, proliferation, migration, metabolism, and apoptosis, integrating the growth factor signaling pathway, nutrient status, and other oncogenic pathways. Aberrations at different levels of the network are implicated in breast cancer development and progression. Different therapies targeting the pathway are being developed and included in clinical trials. Arrows represent activation; bars represent inhibition. Abbreviations: 4EBP1, 4E-binding protein 1; AMPK, adenosine monophosphate-activated protein kinase; Bad, BCL2-associated agonist of cell death; FOXO, forkhead box O1; GPCR, G protein-coupled receptor; GSK3, glycogen synthase kinase 3; IRS1, insulin receptor substrate 1; mLST8, mTOR associated protein, LST8 homolog; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; PI3K, phosphatidylinositol 3-kinase; PIP2, phosphatidylinositol (4,5) biphosphate; PIP3, phosphatidylinositol (3,4,5) triphosphate; PRAS40, proline-rich Akt substrate 40; PTEN, phosphatase and tensin homolog; Rheb, Ras homolog enriched in brain; S6K, ribosomal protein S6 kinase; TKR, tyrosine kinase receptors; TSC1, tuberous sclerosis complex 1; TSC2, tuberous sclerosis complex 2.
PI3K Pathway Aberrations in Breast Cancer
The PI3K pathway has shown to be activated in a diversity of malignancies including breast, colorectal, ovarian, pancreas, brain, endometrium, and other tumor types. After p53, this pathway is considered to be more affected by genetic alterations than any other pathway in cancer [39]. The role of PI3Ks proteins in oncogenesis has been validated by multiple studies [40] showing that aberrations in this pathway are potential causes of cell transformation and, more significant, that PI3K pathway inhibition causes tumor regression [41, 42].
The PI3K signaling network is known to be affected at different levels in human breast cancer [43]. More than 70% of breast tumors have molecular alterations in at least one component of the pathway [44]. Loss of PTEN, PIK3CA mutations, and mutations or other aberrations at the level of PDK1, AKT1, AKT2, and p70S6kinase are some of the known mechanisms that activate the pathway [12]. The identification of genomic alterations and their frequency in the different subtypes of breast cancer may predict responsiveness to targeted therapies. Mouse models and in vitro experiments have shown that tumors with PTEN loss or PIK3CA mutations are predicted to be more sensitive to PI3K pathway inhibitors [45, 46]. Both PIK3CA mutations and loss of the regulatory actions of PTEN enhance AKT-dependent [47, 48] and AKT-independent [45] downstream pathways and are frequently found in breast cancer.
PTEN Loss
PTEN is a tumor-suppressor gene that inhibits the PI3K/AKT/mTOR pathway by cleaving a phosphate group from the PI3K-activated second messenger PIP-3 [49–51]. The lack of its negative regulatory action causes the activation of the PI3K pathway through the phosphorylation of AKT [52]. PTEN loss has been found in many cancers including breast, endometrial, prostate, and thyroid, among others. Initial studies demonstrated a decreased expression or loss of PTEN in up to 33% of breast tumors and a direct relation of this aberrancy with progression of breast cancer [53, 54]. The loss of PTEN occurs through different ways including somatic mutations, loss of heterozygosity, epigenetic modifications, and protein instability and leads to activation of Akt/mTOR-dependent cell proliferation. Cell lines with PTEN deficiency are mainly inhibited by agents targeting mTOR [55–57]. The association of PTEN loss with clinicopathologic markers and prognosis remains unclear; however, some studies have shown an association of PTEN loss with high tumor grade, tumor size, and negative hormone receptor status [44, 58, 59].
PIK3CA Activation
PIK3CA oncogene mutations are particularly common in breast cancer. The PIK3CA gene encodes the p110α catalytic subunit, which plays a key role in the activation of AKT downstream signaling and mammary tumor progression. Activating mutations clustered in the “hot spots” of exons 9 and 20, which correspond to the helical and catalytic domains of p110α, have been reported in up to 26% of breast tumor samples and in 30% of cell lines [60, 61]. In these studies, mutations in exon 20 (catalytic domain) are the most common in breast cancer, in contrast to colorectal cancer where exon 9 mutations are predominant [62]. Several analyses have revealed a direct relation of PI3K activation with lymph node involvement, estrogen receptor (ER), progesterone receptor (PR) positivity, and HER2 overexpression [61, 63]. However, the association with pathologic markers and clinical outcomes is still controversial [44, 64, 65]. Interestingly, there is an inverse relationship between PIK3CA-activating mutations and PTEN loss. A recent analysis by Saal et al. reported that, in tumors with PIK3CA mutation, only 13% had PTEN loss, whereas 34% expressed PTEN normally [61]. PIK3CA and PTEN mutations seem to be mutually exclusive and the identification of the driving mutation of each tumor may direct efficacious targeted therapies. Recent data show evidence that PIK3CA mutations may contribute to carcinogenesis through both AKT-dependent and AKT-independent mechanisms. In the absence of AKT activation, PDK1 may transmit an alternative signal that engages downstream substrates such as SGK3 in PIK3CA mutant cancer cells [45]. The exact substrates of SGK3 remain to be elucidated [66]. When the AKT-dependent signal is compromised, such as in normal PTEN levels, PIK3CA mutations may transduce an AKT-independent signal that engages PDK1 and SGK3 [45].
AKT Dysregulation
The AKT pathway has been found to be dysregulated in a variety of ways in human breast cancer. AKT plays a central role in the pathway and represents an attractive therapeutic target since multiple upstream signaling components converge in AKT. Different studies have demonstrated different roles for the subclasses of AKT in the biological behavior of breast cancer cells. AKT2 activation promotes transition from epithelial to mesenchymal cells, induces secretion of matrix metalloproteinases, and upregulates 1-integrins contributing to tumor invasion and metastasis [66–69]; furthermore, germline deletion of AKT2 in MMTV-ErbB2 mice was shown to decrease lung metastases [70]. Other studies with cell culture systems have shown that the overexpression of AKT1 in breast cancer cell lines results in a decrease in migration and invasion [71, 72]. In transgenic mouse models these two AKT family members have shown to achieve opposing functions in terms of breast tumor metastasis. The hypotheses for the various roles of AKT subclasses include differences in activation levels, interacting partners, downstream substrates, or subcellular localization. The identification of the substrates unique to each isoform of AKT will lead to the therapeutic targeting of specific aspects of tumorigenesis [73–76]. Lopez-Knowles et al. found AKT positivity in 24% of 292 invasive breast cancer patients and a positive association of AKT with high tumor grade, ER and PR negativity, HER2 positivity, and breast cancer–specific death [44].
PI3K Pathway and Breast Cancer Subtypes
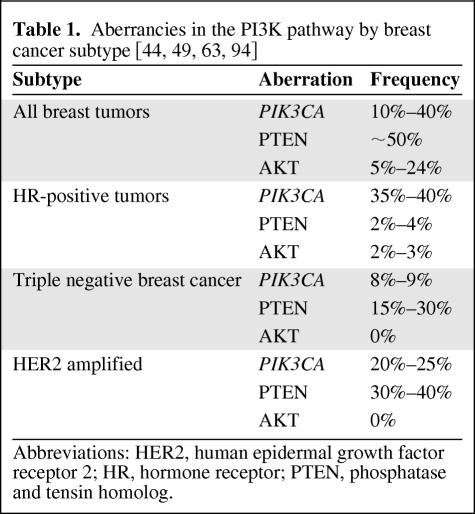
Gene expression profiles have classified breast cancer in luminal A, luminal B, HER2-enriched, and basal-like tumors [77] with each subtype reflecting different biology and clinical outcome. A surrogate classification using immunohistochemistry classifies patients based on ER/PR status, HER2, cytokeratin 5/6 (CK5/6), and EGFR status. The frequency and type of PI3K pathway aberrations vary among the different breast cancer subtypes (Table 1) [44, 63]. Each molecular alteration may have a different clinical impact depending on the breast cancer molecular background, the presence of other aberrations, and the treatments received. The genetic heterogeneity of breast cancer and likely different cell origin for each tumor subtype make necessary an independent analysis of the PI3K pathway aberrations by tumor subtype.
Table 1.
Abbreviations: HER2, human epidermal growth factor receptor 2; HR, hormone receptor; PTEN, phosphatase and tensin homolog.
Hormone Receptor–Positive Tumors
PIK3CA mutations have been found in up to 40% of hormone receptor–positive breast cancer and it is the most frequent aberration of the PI3K pathway found in these tumors [44]. In this subtype, PIK3CA mutations have been associated with low mTORC1 signaling and better clinical outcomes in patients treated with tamoxifen monotherapy [78]. The underlying mechanism and downstream signaling pathways to support this favorable association are under investigation. Negative regulatory genes, mTORC1 downregulating feedback mechanisms, and alternative stronger activators are some of the hypotheses postulated [78]. Gene expression profiling analyses are studying the downstream target genes and signaling pathways activated by PIK3CA mutations in ERα-positive tumors. Cizkova et al. [79] reported an overexpression of genes involved in the human Wnt signaling pathway, which plays a major role in tumor invasion, metastasis, angiogenesis, and cancer stem cell self-renewal. Some of the genes identified have been linked to tumors with less aggressive features and favorable outcomes. AKT1 mutations seem to be restricted to hormone receptor–positive tumors. AKT1-activating mutations have been linked to initial tumorigenesis with posterior inhibition of invasion and metastasis. In fact, AKT1 may prevent tumor progression and may be associated with good outcomes in this subtype of breast cancer [63].
The critical importance of ER and PR in the development and progression of breast cancer [80], and the association of reduced expression of these receptors with poor response to antiestrogen therapy and worse prognosis [81], are well known. The PI3K pathway influences the levels and activity of ER/PR for which this cross talk is a major determinant of both breast cancer progression and response to therapy [82]. The endogenous membrane ER can activate GFRs and PI3K/AKT [83]. The bidirectional cross talk promotes phosphorylation and genomic activation of ER on gene transcription [84–86]. In the presence of hyperactive GFR signaling, as often occurs in breast cancer (e.g., HER2 overexpression), an excessive phosphorylation of ER may diminish the inhibitory effects of the endocrine therapies and lead to endocrine resistance. Cumulative clinical data have shown that patients with HER2- and EGFR-overexpressing tumors have a poorer outcome and are less responsive to tamoxifen [87, 88]. In fact, recent studies using molecular signatures have reported that in ER-positive breast cancer, the GFR/PI3K pathway is associated with lower ER levels and, more importantly, that these levels could be increased by inhibiting the PI3K pathway. The authors suggested that some tumors may rely more on the PI3K signaling than on estrogen for growth and that by blocking the GRF/PI3K pathway these tumors would resort to the estrogen-signaling pathway for survival and restore hormonal sensitivity. Combining the ER blockage and PI3K inhibition might be a more potent treatment strategy [89–91].
Triple Negative Breast Cancer
The basal-like tumors (triple negative for ER, PR, and HER2 and positive for CK5/6 or EGFR) have also shown enhanced PI3K activity mainly through PTEN loss. In gene expression analyses of the main regulators of the pathway, PTEN loss was associated with the basal-like phenotype whereas high PTEN levels were more frequent in luminal A cancers [44]. The loss of PTEN has been reported in approximately 30% of basal-like breast cancers [46] and may play a major role in the pathogenesis of these tumors and poor clinical outcomes of the patients. The aggressive nature and lack of directed therapies against these cancers have promoted a promising growth in the investigation and discovery of potential targets with clinical efficacy. From pharmacogenomic analysis of breast cancer cell lines, genes that constitute the RAS/RAF/MEK signature are the identifiers of the basal-like tumors sensitive to MEK inhibitors. In these studies, loss of the PTEN markedly attenuated the response to MEK inhibition in basal-like tumors. The compensatory upregulation of PI3K/AKT as a survival pathway as a result of PTEN loss is likely the most important mechanism of resistance. The combined treatment with PI3K and MEK inhibitors generated a synergistic effect inhibiting basal-like cell lines. The design of clinical trials with combination therapies including MEK and PI3K inhibitors for this patient population might be a more efficacious approach than single-pathway inhibition therapy [4, 92].
HER2-Amplified Tumors
The HER2/neu gene is amplified in 20%–25% of human breast cancer and is associated with aggressive phenotypes and poor outcomes. Despite the major advances in the treatment of HER2-amplified breast cancer with trastuzumab, the development of therapeutic resistance is a current challenge. Only about 30% of HER2-amplified breast cancers respond to trastuzumab therapy. The HER2-positive tumors have shown enhanced PI3K activity mainly through PTEN loss [93]. A combined signature of PTEN loss and PIK3CA mutation in HER2-positive breast cancer is a strong predictor of trastuzumab resistance [63]. Recent studies in breast cancer cultured cells have shown that the loss of PTEN or activating mutations in PI3K determine resistance of these cells to trastuzumab, but not to lapatinib [94]. In addition, the identification of tumors with the PIK3CA mutation and ER+/HER+ as a group with likely normal PTEN is important since the therapeutic response to trastuzumab is dependent on an intact PTEN [61, 93].
The mechanisms of resistance remain under investigation. In a recent analysis published by Junttila et al., trastuzumab significantly reduced the level of phosphorylation of HER3 and AKT, causing a potent inhibition of the HER3/PI3K/AKT pathway. In these studies, the inhibition of proliferation strongly correlated with the degree of pAKT inhibition and suggested that activators of the PI3K pathway are an important cause of trastuzumab resistance. In cell lines treated with GDC-0941 (agent inhibiting p110α, p110β, and p110δ subunits of PI3K), there was a 40%–85% inhibition of pAKT in all cell lines including trastuzumab-sensitive and -insensitive cells, suggesting a direct correlation between the PI3K/AKT pathway and HER2-positive cells. When these cells were treated with both trastuzumab and GDC-0941, there was a synergistic effect in the inhibition of AKT and downstream targets. The addition of GDC-0941 resulted in inhibition of proliferation in breast cancer cells resistant to trastuzumab because of PTEN loss and activating PIK3CA mutations. The combination of the agents was more efficient in the inhibition of the tumors than either of the single agents [95].
In a study correlating the status of multiple components of the PI3K pathway with trastuzumab resistance, Esteva et al. found that in 137 patients with HER2-positive breast cancer treated with trastuzumab, those who had PTEN-deficient tumors were more likely to be resistant to trastuzumab-based therapy and had decreased overall survival. The combination of other components of the pathway with PTEN loss showed that patients with PTEN−/AKT+ and PTEN−/70S6K+ tumors had more trastuzumab resistance and less overall survival than patients with PTEN loss alone [96]. These data highlight the clinical implications of the PI3K pathway in the mechanisms of resistance to trastuzumab and its potential as a biomarker of prognosis.
Targeting the PI3K Pathway
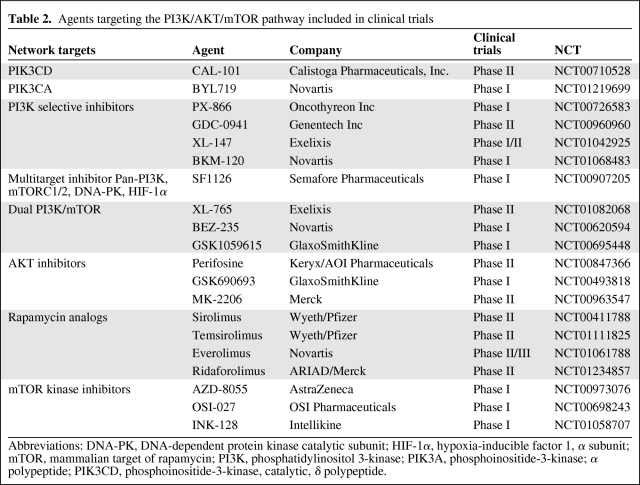
The PI3K pathway and its upstream and downstream effectors comprise many potential targets for drug development in breast cancer. Agents inhibiting the network at different levels used alone or in combination with chemotherapy, radiation, or other targeted therapies are being evaluated in constantly emerging preclinical and clinical trials (Table 2) [97]. The complexity of the PI3K/AKT/mTOR pathway and the influence of activating alternative cascades and feedback loops have prompted the study of combination therapies and the identification of predictive factors.
Table 2.
Agents targeting the PI3K/AKT/mTOR pathway included in clinical trials
Abbreviations: DNA-PK, DNA-dependent protein kinase catalytic subunit; HIF-1α, hypoxia-inducible factor 1, α subunit; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; PIK3A, phosphoinositide-3-kinase; α polypeptide; PIK3CD, phosphoinositide-3-kinase, catalytic, δ polypeptide.
PI3K Inhibitors
In the early 1990s the first synthetic PI3K inhibitor LY-294002 was developed [97]. The conjugation of LY-294002 with Arg-Gly-Asp peptides is derived in a prodrug SF-1126 that is in phase I clinical trials as a multimodal Pan-PI3K inhibitor. Phase I clinical trials of BKM120 (a PI3K inhibitor) or BEZ235 (a PI3K/mTOR inhibitor) in combination with endocrine therapy are in progress for postmenopausal patients with hormone receptor–positive metastatic breast cancer. The new generation of PI3K inhibitors is focused on enhancing the potency and the specificity of the compound for particular PI3K isoforms [98]. There are multiple isoform-specific PI3K inhibitors under investigation. CAL-101 (Calistoga Pharmaceuticals Inc.) is a PIK3Cδ selective inhibitor in phase II studies.
As reviewed above, the complexity and multiple interactions of the PI3K pathway make difficult a homogeneous response to targeted treatments. For example, the activation of the pathway by activated RAS mutations limits the effects of single PI3K inhibitors. A more efficacious approach to these tumors is using medications with pan-PI3K inhibitory actions. Pan-PI3K inhibitors include GDC-0941 (Genentech Inc.), which is in a phase I clinical trial, in combination with paclitaxel and bevacizumab for metastatic breast cancer and XL-147 (Exelixis/Sanofi-Aventis) in phase I/II clinical trials alone or combined with trastuzumab and paclitaxel. Pan-PI3K inhibitors with dual PI3K/mTOR inhibitory activity such as XL-765, SF-1126, BEZ-235, GDC-0941, and GSK1059615 are currently in clinical trials for the treatment of breast cancer and other solid tumors. They are thought to work better overcoming the reactivation of the pathway by feedback loops.
AKT Inhibitors
Drug targeting the AKT family has focused on the development of subunit selective inhibiting molecules including ATP competitors, PIP3 analogs, allosteric inhibitors, pseudosubstrate peptides, and other mechanisms. Preclinical studies have shown that dual AKT-1 and AKT-2 inhibition might be more effective than single inhibition [71]. AKT-3 blockage is more important in tumors like melanoma. The side-effect profile is also isoform-specific and is mainly related to hyperglycemia caused by AKT-2 inhibition [92]. Pan-Akt inhibitors with ATP-competitive properties like AT-13148 and A-443654 are under investigation for clinical development [99, 100].
Allosteric AKT inhibitors disrupt access to the PDK1-dependent AKT phosphorylation site. Compared with ATP-competitive inhibitors, this strategy is more specific and provides better isoform selectivity [101]. The compound MK-2206 (Merck & Co., Inc.) is included in a phase II clinical trial for advanced breast cancer. GSK690693 have entered clinical trials for advanced solid tumors. These compounds are allosteric inhibitors with activity against all three AKT isoforms [102].
mTOR Inhibitors
Since the discovery of rapamycin by Sehgal and colleagues in 1975, extensive work has been done with these compounds as potential agents against cancer [103]. Rapamycin inhibits mTOR [104] regulating the phosphorylation of S6K and 4EBP1/EBP2 for which its direct effect on protein synthesis was elucidated [105]. The mTOR pathway has become an attractive target for drug development against cancer since temsirolimus, a generic analog of rapamycin, was approved for renal cell carcinoma [21, 106]. Preclinical studies in breast cancer have suggested that rapamycin may enhance chemotherapy-induced apoptosis acting in synergisms when combined with standard agents such as paclitaxel, carboplatin, and vinorelbine [106]. The combination of temsirolimus with the aromatase inhibitor letrozole showed some biological and clinical activity in a phase II clinical trial in breast cancer; however, the phase III trial was terminated because of lack of efficacy [107]. Rapamycin analogs temsirolimus (CCI-779) and everolimus (RAD-001) are being used for metastatic breast cancer in combinations with drugs such as capecitabine and exemestane. Ridaforolimus (MK-8669) is in early clinical trials for ER-positive breast cancer.
The majority of preclinical and clinical efforts to target mTOR have involved rapamycin analogs that suppress mTORC1 and do not acutely inhibit mTORC2. The feedback activation of PI3K and AKT limits the efficacy of these compounds [108]. New agents that block this feedback loop by the inhibition of the catalytic activity of TOR with both TORC1 and TORC2 inhibition cause broader suppression of the PI3K/AKT/TOR signaling pathway.
The use of mTOR inhibitors in combination with other targeted agents and chemotherapy may be limited by side effects like myelosuppression, mucositis. and bowel perforation. A new challenge of research is the identification and validation of biologic markers to predict response and to select the high-risk patients that will benefit the most from these therapies [106].
Conclusion
A large amount of clinical data exploring new single and combined therapies to inhibit the PI3K pathway is constantly emerging. Because the PI3K pathway has divergent downstream effects, the identification of the key effectors of the pathway and their presence in the different subtypes of breast tumors will allow the development of ideal targeted therapies with meaningful clinical efficacy. The development of medications with multitarget properties and the identification of potent drug combinations are expected to generate results in the management of breast tumors driven by multiple oncogenic pathways and to overcome resistance by feedback mechanisms. In addition, the heterogeneity of breast cancer makes imperative the identification of biological markers that define molecular profiles for a rational use of PI3K inhibitors.
Acknowledgments
This work was supported in part by the National Cancer Institute 1K23CA121994 (A.M.G.), AACR SU2C Dream Team (A.M.G.), and National Cancer Institute through The University of Texas MD Anderson's Cancer Center Support Grant (P30 CA016672).
Author Contributions
Conception/Design: Leonel F. Hernandez-Aya, Ana Gonzalez-Angulo
Provision of study material or patients: Leonel F. Hernandez-Aya, Ana Gonzalez-Angulo
Collection and/or assembly of data: Leonel F. Hernandez-Aya, Ana Gonzalez- Angulo
Data analysis and interpretation: Leonel F. Hernandez-Aya, Ana Gonzalez- Angulo
Manuscript writing: Leonel F. Hernandez-Aya, Ana Gonzalez-Angulo
Final approval of manuscript: Leonel F. Hernandez-Aya, Ana Gonzalez- Angulo
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 3.Chang HW, Aoki M, Fruman D, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 4.Hoeflich KP, O'Brien C, Boyd Z, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 5.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal R, Carey M, Hennessy B, et al. PI3K pathway-directed therapeutic strategies in cancer. Curr Opin Investig Drugs. 2010;11:615–628. [PubMed] [Google Scholar]
- 7.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 8.Guillermet-Guibert J, Bjorklof K, Salpekar A, et al. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci U S A. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J, Cantley LC. The negative regulation of phosphoinositide 3-kinase signaling by p85 and its implication in cancer. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- 10.Bellacosa A, Testa JR, Staal SP, et al. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 11.Wang DS, Ching TT, St Pyrek J, et al. Biotinylated phosphatidylinositol 3,4,5-trisphosphate as affinity ligand. Anal Biochem. 2000;280:301–307. doi: 10.1006/abio.2000.4525. [DOI] [PubMed] [Google Scholar]
- 12.Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 13.Tokunaga E, Oki E, Egashira A, et al. Deregulation of the Akt pathway in human cancer. Curr Cancer Drug Targets. 2008;8:27–36. doi: 10.2174/156800908783497140. [DOI] [PubMed] [Google Scholar]
- 14.Katso R, Okkenhaug K, Ahmadi K, et al. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 15.Inoki K, Li Y, Zhu T, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 16.Manning BD, Tee AR, Logsdon MN, et al. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 17.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Corradetti MN, Inoki K, et al. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Oshiro N, Takahashi R, Yoshino K, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Harris TE, Roth RA, et al. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons JJ, Abraham RT, Yu K. Mammalian target of rapamycin: discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin Oncol. 2009;36(Suppl 3):S3–S17. doi: 10.1053/j.seminoncol.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim DH, Sabatini DM. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol. 2004;279:259–270. doi: 10.1007/978-3-642-18930-2_15. [DOI] [PubMed] [Google Scholar]
- 24.Barbet NC, Schneider U, Helliwell SB, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunn GJ, Hudson CC, Sekulic A, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 26.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 27.Dorrello NV, Peschiaroli A, Guardavaccaro D, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 28.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proud CG. mTORC1 signalling and mRNA translation. Biochem Soc Trans. 2009;37:227–231. doi: 10.1042/BST0370227. [DOI] [PubMed] [Google Scholar]
- 31.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, Dibble CC, Matsuzaki M, et al. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarbassov DD, Guertin DA, Ali SM, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 34.Gulhati P, Cai Q, Li J, et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Ozes ON, Akca H, Mayo LD, et al. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc Natl Acad Sci U S A. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tremblay F, Brule S, Hee Um S, et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007;12:104–107. doi: 10.1016/j.ccr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Krasilnikov MA. Phosphatidylinositol-3 kinase dependent pathways: the role in control of cell growth, survival, and malignant transformation. Biochemistry (Mosc) 2000;65:59–67. [PubMed] [Google Scholar]
- 41.Bayascas JR, Leslie NR, Parsons R, et al. Hypomorphic mutation of PDK1 suppresses tumorigenesis in PTEN(+/−) mice. Curr Biol. 2005;15:1839–1846. doi: 10.1016/j.cub.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 42.Chen ML, Xu PZ, Peng XD, et al. The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/− mice. Genes Dev. 2006;20:1569–1574. doi: 10.1101/gad.1395006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills GB, Kohn E, Lu Y, et al. Linking molecular diagnostics to molecular therapeutics: targeting the PI3K pathway in breast cancer. Semin Oncol. 2003;30:93–104. doi: 10.1053/j.seminoncol.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Lopez-Knowles E, O'Toole SA, McNeil CM, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer. 2010;126:1121–1131. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 45.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura N, Ramaswamy S, Vazquez F, et al. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol Cell Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 50.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 51.Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim Biophys Acta. 2000;1470:M21–M35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 52.Carnero A, Blanco-Aparicio C, Renner O, et al. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 53.Perren A, Weng LP, Boag AH, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155:1253–1260. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bose S, Wang SI, Terry MB, et al. Allelic loss of chromosome 10q23 is associated with tumor progression in breast carcinomas. Oncogene. 1998;17:123–127. doi: 10.1038/sj.onc.1201940. [DOI] [PubMed] [Google Scholar]
- 55.Neshat MS, Mellinghoff IK, Tran C, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi Y, Gera J, Hu L, et al. Enhanced sensitivity of multiple myeloma cells containing PTEN mutations to CCI-779. Cancer Res. 2002;62:5027–5034. [PubMed] [Google Scholar]
- 57.Steelman LS, Navolanic PM, Sokolosky ML, et al. Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene. 2008;27:4086–4095. doi: 10.1038/onc.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bose S, Chandran S, Mirocha JM, et al. The Akt pathway in human breast cancer: a tissue-array-based analysis. Mod Pathol. 2006;19:238–245. doi: 10.1038/modpathol.3800525. [DOI] [PubMed] [Google Scholar]
- 59.Lee JS, Kim HS, Kim YB, et al. Reduced PTEN expression is associated with poor outcome and angiogenesis in invasive ductal carcinoma of the breast. Appl Immunohistochem Mol Morphol. 2004;12:205–210. doi: 10.1097/00129039-200409000-00004. [DOI] [PubMed] [Google Scholar]
- 60.Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 61.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 62.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 63.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li SY, Rong M, Grieu F, et al. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- 65.Maruyama N, Miyoshi Y, Taguchi T, et al. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13:408–414. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 66.Slagsvold T, Marchese A, Brech A, et al. CISK attenuates degradation of the chemokine receptor CXCR4 via the ubiquitin ligase AIP4. EMBO J. 2006;25:3738–3749. doi: 10.1038/sj.emboj.7601267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 68.Thant AA, Nawa A, Kikkawa F, et al. Fibronectin activates matrix metalloproteinase-9 secretion via the MEK1-MAPK and the PI3K-Akt pathways in ovarian cancer cells. Clin Exp Metastasis. 2000;18:423–428. doi: 10.1023/a:1010921730952. [DOI] [PubMed] [Google Scholar]
- 69.Arboleda MJ, Lyons JF, Kabbinavar FF, et al. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- 70.Maroulakou IG, Oemler W, Naber SP, et al. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle T transgenic mice. Cancer Res. 2007;67:167–177. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]
- 71.Irie HY, Pearline RV, Grueneberg D, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoeli-Lerner M, Yiu GK, Rabinovitz I, et al. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 73.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 74.Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 75.Nakatani K, Thompson DA, Barthel A, et al. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J Biol Chem. 1999;274:21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- 76.Dillon RL, Muller WJ. Distinct biological roles for the akt family in mammary tumor progression. Cancer Res. 2010;70:4260–4264. doi: 10.1158/0008-5472.CAN-10-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loi S, Haibe-Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A. 2010;107:10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cizkova M, Cizeron-Clairac G, Vacher S, et al. Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: major implication of the Wnt signaling pathway. PLoS One. 2010;5:e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui X, Schiff R, Arpino G, et al. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–7735. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Bardou VJ, Arpino G, Elledge RM, et al. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21:1973–1979. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 82.Arpino G, Wiechmann L, Osborne CK, et al. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee AV, Guler BL, Sun X, et al. Oestrogen receptor is a critical component required for insulin-like growth factor (IGF)-mediated signalling and growth in MCF-7 cells. Eur J Cancer. 2000;36(Suppl 4):109–110. doi: 10.1016/s0959-8049(00)00264-1. [DOI] [PubMed] [Google Scholar]
- 84.Martin MB, Franke TF, Stoica GE, et al. A role for Akt in mediating the estrogenic functions of epidermal growth factor and insulin-like growth factor I. Endocrinology. 2000;141:4503–4511. doi: 10.1210/endo.141.12.7836. [DOI] [PubMed] [Google Scholar]
- 85.Kato S, Endoh H, Masuhiro Y, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 86.Razandi M, Pedram A, Park ST, et al. Proximal events in signaling by plasma membrane estrogen receptors. J Biol Chem. 2003;278:2701–2712. doi: 10.1074/jbc.M205692200. [DOI] [PubMed] [Google Scholar]
- 87.De Laurentiis M, Arpino G, Massarelli E, et al. A meta-analysis on the interaction between HER-2 expression and response to endocrine treatment in advanced breast cancer. Clin Cancer Res. 2005;11:4741–4748. doi: 10.1158/1078-0432.CCR-04-2569. [DOI] [PubMed] [Google Scholar]
- 88.Arpino G, Green SJ, Allred DC, et al. HER-2 amplification, HER-1 expression, and tamoxifen response in estrogen receptor-positive metastatic breast cancer: a southwest oncology group study. Clin Cancer Res. 2004;10:5670–5676. doi: 10.1158/1078-0432.CCR-04-0110. [DOI] [PubMed] [Google Scholar]
- 89.Martin LA, Head JE, Pancholi S, et al. The farnesyltransferase inhibitor R115777 (tipifarnib) in combination with tamoxifen acts synergistically to inhibit MCF-7 breast cancer cell proliferation and cell cycle progression in vitro and in vivo. Mol Cancer Ther. 2007;6:2458–2467. doi: 10.1158/1535-7163.MCT-06-0452. [DOI] [PubMed] [Google Scholar]
- 90.deGraffenried LA, Friedrichs WE, Russell DH, et al. Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant Akt Activity. Clin Cancer Res. 2004;10:8059–8067. doi: 10.1158/1078-0432.CCR-04-0035. [DOI] [PubMed] [Google Scholar]
- 91.Creighton CJ, Fu X, Hennessy BT, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rexer BN, Ghosh R, Arteaga CL. Inhibition of PI3K and MEK: it is all about combinations and biomarkers. Clin Cancer Res. 2009;15:4518–4520. doi: 10.1158/1078-0432.CCR-09-0872. [DOI] [PubMed] [Google Scholar]
- 93.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 94.O'Brien NA, Browne BC, Chow L, et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9:1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. [DOI] [PubMed] [Google Scholar]
- 95.Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 96.Esteva FJ, Guo H, Zhang S, et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:1647–1656. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marone R, Cmiljanovic V, Giese B, et al. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 98.Knight ZA, Gonzalez B, Feldman ME, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yap TA, Garrett MD, Walton MI, et al. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 100.Luo Y, Shoemaker AR, Liu X, et al. Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Mol Cancer Ther. 2005;4:977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Z, Leister WH, Robinson RG, et al. Discovery of 2,3,5-trisubstituted pyridine derivatives as potent Akt1 and Akt2 dual inhibitors. Bioorg Med Chem Lett. 2005;15:905–909. doi: 10.1016/j.bmcl.2004.12.062. [DOI] [PubMed] [Google Scholar]
- 102.Lindsley CW. The Akt/PKB family of protein kinases: a review of small molecule inhibitors and progress towards target validation: a 2009 update. Curr Top Med Chem. 2010;10:458–477. doi: 10.2174/156802610790980602. [DOI] [PubMed] [Google Scholar]
- 103.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 104.Choi J, Chen J, Schreiber SL, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 105.Kuo CJ, Chung J, Fiorentino DF, et al. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 106.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leary A, Dowsett M. Combination therapy with aromatase inhibitors: the next era of breast cancer treatment? Br J Cancer. 2006;95:661–666. doi: 10.1038/sj.bjc.6603316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun SY, Rosenberg LM, Wang X, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]