Abstract
Three different pathways lead to the synthesis of phosphatidylethanolamine (PtdEtn) in yeast, one of which is localized to the inner mitochondrial membrane. To study the contribution of each of these pathways, we constructed a series of deletion mutants in which different combinations of the pathways are blocked. Analysis of their growth phenotypes revealed that a minimal level of PtdEtn is essential for growth. On fermentable carbon sources such as glucose, endogenous ethanolaminephosphate provided by sphingolipid catabolism is sufficient to allow synthesis of the essential amount of PtdEtn through the cytidyldiphosphate (CDP)-ethanolamine pathway. On nonfermentable carbon sources, however, a higher level of PtdEtn is required for growth, and the amounts of PtdEtn produced through the CDP-ethanolamine pathway and by extramitochondrial phosphatidylserine decarboxylase 2 are not sufficient to maintain growth unless the action of the former pathway is enhanced by supplementing the growth medium with ethanolamine. Thus, in the absence of such supplementation, production of PtdEtn by mitochondrial phosphatidylserine decarboxylase 1 becomes essential. In psd1Δ strains or cho1Δ strains (defective in phosphatidylserine synthesis), which contain decreased amounts of PtdEtn, the growth rate on nonfermentable carbon sources correlates with the content of PtdEtn in mitochondria, suggesting that import of PtdEtn into this organelle becomes growth limiting. Although morphological and biochemical analysis revealed no obvious defects of PtdEtn-depleted mitochondria, the mutants exhibited an enhanced formation of respiration-deficient cells. Synthesis of glycosylphosphatidylinositol-anchored proteins is also impaired in PtdEtn-depleted cells, as demonstrated by delayed maturation of Gas1p. Carboxypeptidase Y and invertase, on the other hand, were processed with wild-type kinetics. Thus, PtdEtn depletion does not affect protein secretion in general, suggesting that high levels of nonbilayer-forming lipids such as PtdEtn are not essential for membrane vesicle fusion processes in vivo.
INTRODUCTION
The zwitterionic phospholipid phosphatidylethanolamine (PtdEtn) has a strong tendency to form nonbilayer structures and is the most abundant phospholipid of this type in eukaryotic cells (reviewed by de Kruijff, 1997). The potential of membranes with high PtdEtn content to undergo laminar-hexagonal phase transition has been proposed to affect membrane-membrane contact and bilayer fusion during processes of vesicle formation and vesicle-mediated protein trafficking. In addition, nonbilayer lipids may affect integration of proteins into membranes, their lateral movement within the membrane, and folding and stabilization of certain membrane protein complexes.
The most prominent biological system that has provided both genetic and biochemical evidence for specific roles of PtdEtn in cell function is Escherichia coli (reviewed by Dowhan, 1997). In this prokaryote, lack of PtdEtn can be compensated by elevated levels of cardiolipin (CL) in the presence of divalent cations, thereby maintaining the potential of bilayer-to-nonbilayer phase transition of membranes (Morein et al., 1996). A PtdEtn-deficient E. coli mutant displays complex phenotypic changes, including filamentous growth (Mileykovskaya et al., 1998) and decreased activity of lactose permease. The latter observation was ascribed to misfolding of the permease due to lack of PtdEtn that acts as a molecular chaperone for this transporter (Bogdanov et al., 1999). In vitro, nonbilayer lipids stimulate the activity of the reconstituted bacterial protein translocase (van der Does et al., 2000).
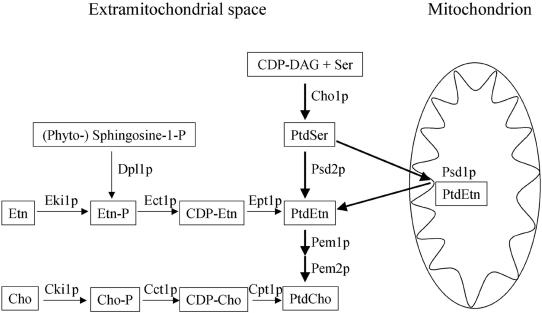
Biosynthesis of PtdEtn in Saccharomyces cerevisiae can be accomplished by two de novo pathways of phosphatidylserine (PtdSer) formation and decarboxylation and by the cytidyldiphosphate (CDP)-ethanolamine branch of the Kennedy pathway (Figure 1). In this organism, PtdEtn is synthesized primarily by the two de novo pathways (reviewed by Daum et al., 1998). Decarboxylation of PtdSer by phosphatidylserine decarboxylase 1 (Psd1p) occurs in the inner mitochondrial membrane (Zinser et al., 1991), whereas phosphatidylserine decarboxylase 2 (Psd2p) was localized to a Golgi/vacuolar compartment (Trotter and Voelker, 1995). Methylation of PtdEtn by PtdEtn methyltransferases 1 (Pem1p) and 2 (Pem2p) yields phosphatidylcholine (PtdCho), the final product of the de novo route of aminoglycerophospholipid synthesis.
Figure 1.
Pathways of PtdEtn biosynthesis. Biosynthesis of PtdEtn in S. cerevisiae occurs by three pathways, namely, the de novo or CDP-DAG pathway (thick arrows) via either 1) mitochondrial Psd1p or 2) Psd2p, and 3) the CDP-ethanolamine branch of the Kennedy pathway (thin arrows).
Ethanolamine (Etn) or choline (Cho) exogenously added to a yeast culture or endogenously formed through lipolytic processes is used for PtdEtn or phosphatidylcholine (PtdCho) synthesis via the Kennedy pathway. The initial enzymes of this branched pathway, ethanolamine kinase (Eki1p) and choline kinase (Cki1p), have overlapping substrate specificities with Eki1p being primarily responsible for Etn phosphorylation and Cki1p for Cho phosphorylation (Kim et al., 1999). Both gene products together represent the total cellular ethanolamine and choline kinase activities in S. cerevisiae. Ethanolamine phosphate (Etn-P) and choline phosphate (Cho-P) are activated by reaction with cytidyltriphosphate (CTP), and cytidyldiphosphate ethanolamine (CDP-Etn) and cytidyldiphosphate choline (CDP-Cho) are finally linked to diacylglycerol to yield PtdEtn and PtdCho. A cpt1Δ ept1Δ double mutant, which is defective in the final steps of this pathway, is viable, suggesting that in yeast the Kennedy pathway is not essential under standard growth conditions (McGee et al., 1994). The Kennedy pathway is linked to sphingolipid catabolism through a reaction catalyzed by dihydrosphingosine-phosphate lyase (Dpl1p). This enzyme cleaves phosphorylated sphingoid base to long chain aldehyde and ethanolaminephosphate (Etn-P) (Saba et al., 1997) allowing incorporation of the latter component into PtdEtn through the Kennedy pathway (Mandala et al., 1998). This finding is consistent with the observation of Hikiji et al. (1988) that cho1Δ cells, which are defective in phosphatidylserine synthase, accumulated some PtdEtn on choline-supplemented media.
Yeast PtdSer is synthesized from cytidyldiphosphate diacylglycerol (CDP-DAG) and serine (Ser) by the action of PtdSer synthase Cho1p (Figure 1), which is localized to the endoplasmic reticulum (reviewed by Daum et al., 1998). Mutants deleted of CHO1 do not contain detectable amounts of PtdSer and are auxotrophic for Cho or Etn, indicating that Cho1p is the only PtdSer synthase in yeast and that PtdSer is not essential (Atkinson et al., 1980). Although PtdSer-deficient yeast cells are viable, they exhibit a number of defects such as decreased tryptophan transport activity (Nakamura et al., 2000) and abnormal vacuolar function and morphogenesis (Hamamatsu et al., 1994). Mutants defective in either of the PtdSer decarboxylases, Psd1p or Psd2p, grow like wild-type on glucose medium, but psd1 psd2 double mutants are auxotrophic for Etn or Cho (Trotter and Voelker, 1995). The fact that cho1 and psd1 psd2 mutants can be rescued by Cho alone suggested that PtdCho is an essential lipid, and that PtdEtn is either nonessential or can be synthesized in adequate amounts from the Etn-P provided by sphingolipid breakdown. The essentiality of PtdCho was supported by the observation that strains defective in both methyltransferases, Pem1p and Pem2p, are auxotrophic for Cho (Summers et al., 1988; Kodaki and Yamashita, 1989); thus, PtdEtn alone does not fully substitute for the methylated phospholipids.
The high level of PtdEtn in mitochondria (Tuller et al., 1999) and the presence of Psd1p in the inner mitochondrial membrane suggest a specific requirement of this organelle for PtdEtn. To investigate the contributions and relative efficiencies of the three pathways of PtdEtn synthesis described above, we 1) genetically dissected each of these pathways, 2) analyzed the specific role of mitochondrial PtdEtn production, and 3) studied the efficiency of PtdEtn import into mitochondria. We demonstrate that the requirement for PtdEtn is more stringent on nonfermentable than on fermentable carbon sources and that PtdEtn is imported into mitochondria only with moderate efficiency.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Culture Conditions
Strains and plasmids used in this study are listed in Table 1. The open reading frames of PSD1, PSD2, and CHO1 were replaced by the KanMX4 marker by using a polymerase chain reaction (PCR)-mediated one-step (PSD1 and CHO1) or two-step (PSD2) gene replacement strategy (Wach et al., 1994). Positions 4 to 1162 of PSD1 (total length 1503 bp), positions −1 to 3427 of PSD2 (total length 3416 bp), and positions 4 to 828 of CHO1 (total length 831 bp) were replaced by using primers listed in Table 2. These constructs were used for transformation of the diploid wild-type strain FY1679 (Table 1). Upon tetrad dissection, the deletions showed 2:2 segregation as monitored by kanamycin resistance. Diploid and haploid deletion strains were tested for proper insertion of the KanMX4 marker by colony PCR with appropriate primers (Table 2). Double and multiple deletion mutants were then obtained by standard genetic methods and verified by colony PCR analysis.
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| FY1679 | MATa/MATα ura3-52/ura3-52 TRP1/trp1Δ63LEU2/leu2 Δ1 HIS3/his3 Δ200 | Winston et al. (1995) |
| YRB1 (wt) | MATa his3 Δ200 leu2 Δ1 trp1 Δ63 ura3-52 | This work |
| YRB2 | As YRB1 except psd1 Δ∷KanMX4 | This work |
| YRB3 | As YRB1 except psd2 Δ∷KanMX4 | This work |
| YRB4 | As YRB1 except MATα cho1 Δ∷KanMX4 TRP1 | This work |
| YRB5 | As YRB1 except psd1 Δ∷KanMX4 psd2 Δ∷KanMX4 TRP1 | This work |
| MSS204 | MATa dpl1 Δ∷LEU2 TPS2-LacZ leu2-3,112 ura3-52 trp1 his4 rme1 | R.C. Dickson, University of Kentucky |
| YRB6 | MATα psd1 Δ∷KanMX4 psd2 Δ∷KanMX4 dpl1 Δ∷LEU2his3 leu2 ura3-52 | This work |
| YRB7 | psd1 Δ∷KanMX4 dpl1 Δ∷LEU2 his3/4 leu2 ura3-52 | This work |
| YRB8 | psd2 Δ∷KanMX4 dpl1 Δ∷LEU2 his3/4 leu2 ura3-52 | This work |
| KS106 | MATα eki1 Δ∷TRP1 cki1 Δ∷HIS3 leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 | Kim et al. (1999) |
| YRB9 | eki1 Δ∷TRP1 cki1 Δ∷HIS3 dpl1 Δ∷LEU2 his3 leu2 trp1-1 ura3 | This work |
| YRB10 | MATa/MATα psd1 Δ∷KanMX4/PSD1 psd2 Δ∷KanMX4/PSD2 dpl1 Δ∷LEU2/DPL1 eki1 Δ∷TRP1/EKI1 cki1 Δ∷HIS3/CKI1 his3/his3 leu2/leu2 trp1-1/TRP1 ura3/ura3 | This work |
| YRB11 | MATa/MATα psd1 Δ∷KanMX4/PSD1psd2 Δ∷KanMX4/PSD2 cki1 Δ∷HIS3/CKI1 his3/his3leu2/leu2 trp1/TRP1 ura3/ura3 | This work |
| YRB12 | psd1 Δ∷KanMX4 cki1 Δ∷HIS3 his3 leu2 ura3 | This work |
| YRB13 | psd2 Δ∷KanMX4 cki1 Δ∷HIS3 his3 ura3 | This work |
| YRB14 | psd1 Δ∷KanMX4 eki1 Δ∷TRP1 his3 leu2 ura3 | This work |
| YRB15 | psd2 Δ∷KanMX4 eki1 Δ∷TRP1 his3 leu2 ura3 | This work |
| YRB16 | psd1 Δ∷KanMX4 psd2 Δ∷KanMX4 eki1 Δ∷TRP1 his3 leu2 ura3 | This work |
| YRB17 | psd1 Δ∷KanMX4 eki1 Δ∷TRP1 cki1 Δ∷HIS3 his3 leu2 trp1-1 ura3 | This work |
| YRB18 | psd2 Δ∷KanMX4 eki1 Δ∷TRP1 cki1 Δ∷HIS3 his3 leu2 ura3 | This work |
| YRB19 | psd1 Δ∷KanMX4 cki1 Δ∷HIS3 dpl1 Δ∷LEU2 his3 leu2 trp1-1 ura3 | This work |
| YRB20 | psd2 Δ∷KanMX4 cki1 Δ∷HIS3 dpl1 Δ∷LEU2 his3 leu2 ura3 | This work |
| YRB21 | psd1 Δ∷KanMX4 eki1 Δ∷TRP1 dpl1 Δ∷LEU2 his3 leu2 trp1-1 ura3 | This work |
| YRB22 | psd2 Δ∷KanMX4 eki1 Δ∷TRP1 dpl1 Δ∷LEU2 his3 leu2 ura3 | This work |
| YRB23 | psd1 Δ∷KanMX4 psd2 Δ∷KanMX4 eki1 Δ∷TRP1 dpl1 Δ∷LEU2 his3 leu2 ura3 | This work |
| YRB24 | psd1 Δ∷KanMX4 eki1 Δ∷TRP1 cki1 Δ∷HIS3 dpl1 Δ∷LEU2 his3 leu2 trp1-1 ura3 | This work |
| YRB25 | psd2 Δ∷KanMX4 eki1 Δ∷TRP1 cki1 Δ∷HIS3 dpl1 Δ∷LEU2 his3 leu2 ura3 | This work |
| p24-3 | 3.2 kb PvuII-HindIII fragment with DPL1 cloned into YEp351 cut with SmaI and HindIII | R.C. Dickson, University of Kentucky |
| pRB1 | PSD1 ORF + about 3.6 kb 5′ and 2.2 kb 3′ flanking regions (about 7 kb insert) in YCp50 | This work |
| pRB2 | PSD2 ORF + about 8.1 kb 5′ and 7.3 kb 3′ flanking regions (about 19 kb insert) in YEp24 | This work |
| pRB3 | 2.6 kb SalI-NsiI fragment with PSD1 cloned into YEp352 cut with SalI and PstI | This work |
Table 2.
Polymerase chain reaction primers used for synthesis and checking the KanMX4 deletion cassettes
| Mutant | Primers (5′ to 3′) |
|---|---|
| psd1 Δ | S1:TGTTAGCAGATCGCTCAAATCCTTCTTGGTCGTTATTTTTTGAAG |
| AAGAAGGAAAAGCAAAGCCAGCATGcgtacgctgcaggtcgac | |
| S2:TTGATTGAACCAACATTTGTTGCACCAACAGGAGTCATGCTAAA | |
| AAATCCGTACTTCCAACTACCCAACAatcgatgaattcgagctcg | |
| P1:CGCTTGTCGCAGATAACACG | |
| P2:GGAGACCTGTTTTCTTCCGC | |
| psd2 Δ | P5′:AGAAGGAGAAAGACAAAAAAG |
| P5′L:aagctaaacagatctggcgcgccttaTTCGTCGTTGGATGCTCCTG | |
| P3′:TCGTATTTTCTGCAAGAGGAC | |
| P3′L:gtcgaaaacgagctcgaattcatcgaAAATTGGCTGTAGTTAGTGG | |
| P1:CGAAAAATACAAAGGTTATGTTAG | |
| P2:TTTGATTCCAGTGTGTATTTG | |
| cho1 Δ | S1:GTGATTGTCATTTTTAGTTGTCTATTTGATTCAATCAAAAAACAA |
| AAATAAAACTATATATTAAAAAATGcgtacgctgcaggtcgac | |
| S2:CATATAAAAGTAGAATAAAAAGTTATATGTACAAATTTTTTTTG | |
| ACGCCAGGCATGAACAAAAACTACTAatcgatgaattcgagctcg | |
| P1:TTCACACGGCACCCTCAC | |
| P2:CAGTAGCATTAGGGGGGG | |
| KanMX4 reverse:ggttgtttatgttcggatgtg |
Capitals are homologous to the respective ORF. Small letters are homologous to KanMX4.
Plasmids pRB1 and pRB2 were isolated from YCp50 (Rose et al., 1987) and YEp24 (Carlson and Botstein, 1982) yeast genomic libraries by their ability to suppress the Etn-requirement of a psd1Δ psd2Δ double deletion strain. Plasmid pRB1 carries PSD1 and pRB2 contains PSD2 as verified by PCR and restriction mapping. Standard techniques of E. coli molecular biology were used throughout the work (Ausubel et al., 1996). Plasmids were introduced into yeast cells by lithium acetate transformation (Gietz et al., 1992). To confirm synthetic lethality of the psd1Δ psd2Δ cki1Δ triple mutant, the strain was tested for loss of plasmid pRB2 by cultivation on solid synthetic medium containing 1 mg/ml 5′-fluoroorotic acid (PCR, Gainesville, FL) and 5 mM Etn, Cho, or Ser.
Yeast strains were grown under aerobic conditions at 30°C on YP medium (1% yeast extract, 2% bacto peptone) containing 2% glucose (YPD) or lactate (YPLac) as a carbon source. It has to be noted that YP media contain low amounts of Etn and Cho. Growth tests were performed on solid synthetic minimal medium (Sherman et al., 1986) containing 2% glucose, ethanol, or lactate and 2% Bactoagar (Difco, Detroit, MI). Supplemented media contained 5 mM Etn, Cho, or Ser unless otherwise stated. To study growth in liquid YP media, precultures grown to the stationary phase were diluted 1:500 (vol/vol) in fresh medium, and optical density at 600 nm was measured at the time points indicated. Respiration-deficient cells (petites) in YP medium were detected by serial dilution and plating an equal number of cells on YPD and YPLac medium.
Cell Fractionation
Total homogenates and mitochondria were prepared from spheroplasts by published procedures (Daum et al., 1982; Zinser et al., 1991). Relative enrichment of markers and cross-contamination of subcellular fractions were assessed as described by Zinser and Daum (1995).
Analytical Procedures
Lipids were extracted by the procedure of Folch et al. (1957). Individual phospholipids were separated by two-dimensional thin-layer chromatography on Silica gel 60 plates (Merck, Darmstadt, Germany) by using chloroform/methanol/25% NH3 (65:35:5; per volume) as first and chloroform/acetone/methanol/acetic acid/water (50:20:10:10:5; per volume) as second developing solvent. Phospholipids were visualized on thin-layer chromatography plates by staining with iodine vapor, scraped off the plate, and quantified by the method of Broekhuyse (1968). Protein was quantified by the method of Lowry et al. (1951) using bovine serum albumin as a standard.
Spectrophotometric quantification of mitochondrial cytochromes was carried out by the method of Watson et al. (1975) by using a Hitachi U2310 double beam spectrophotometer. Enzymatic activity of cytochrome c oxidase was measured as described by Mason et al. (1973) and that of cardiolipin synthase and phosphatidylglycerol phosphate synthase as described by Tuller et al. (1998). PtdSer decarboxylase activity was measured as reported by Kuchler et al. (1986) with minor modifications: 100 nmol of [3H]PtdSer with a specific radioactivity of 1.8 μCi/nmol was used as a substrate, and the assay was performed in 0.1 M Tris-HCl, pH 7.2, containing 10 mM EDTA.
Protein Secretion
Analysis of Gas1p maturation was performed with homogenates of cells grown to the late logarithmic phase in minimal medium in the presence of supplements as indicated. Homogenates were prepared by disintegrating cells with glass beads in a Merkenschlager homogenizer under CO2 cooling in the presence of 10 mM Tris-HCl, pH 7.2, 1 mM phenylmethylsulfonyl fluoride (Calbiochem, La Jolla, CA). Western blot analysis by using a primary rabbit antibody against Gas1p was performed as described by Haid and Suissa (1983). Immunoreactive bands were visualized by enzyme-linked immunosorbent assay with a peroxidase-linked secondary antibody (Sigma, St. Louis, MO) following the manufacturer's instructions.
Carboxypeptidase Y maturation was monitored by pulse-chase labeling and immunoprecipitation essentially as described by Munn et al. (1999). As minor modifications, cells were grown in synthetic minimal medium supplemented with 2% glucose and 5 mM Cho, and samples were taken during the chase period. Invertase secretion was assayed according to Munn et al. (1999) with the modification that cells were grown in synthetic minimal medium supplemented with 5% glucose and 5 mM Cho and induced by resuspension in synthetic minimal medium containing 0.05% glucose, 2% sucrose, and 5 mM Cho.
RESULTS
Synthesis of PtdEtn Is Essential in Yeast
To study the requirement for PtdEtn and the relative contribution of the different pathways to PtdEtn synthesis a series of haploid single and multiple deletion strains with defects in the different biosynthetic routes of PtdEtn synthesis were constructed (Table 1) and tested for their growth phenotype on defined media containing different carbon sources (Table 3). As recognized previously (Atkinson et al., 1980; Trotter et al., 1995) single deletions of PSD1 and PSD2 did not affect growth on glucose medium, but strains deleted in both PtdSer decarboxylases (psd1Δ psd2Δ) or PtdSer synthase (cho1Δ) were auxotrophic for either Etn or Cho. To study the effect of the Dpl1p-dependent salvage pathway on PtdEtn production, and to investigate whether this pathway is required for psd1Δ psd2Δ or cho1Δ mutants to grow on Cho-supplemented media, we analyzed the growth phenotype of a psd1Δ psd2Δ dpl1Δ triple deletion strain. The triple mutant was strictly auxotrophic for Etn and could not be grown by Cho supplementation alone (Table 3). Overexpression of Dpl1p from plasmid p24-3 (Table 1) in psd1Δ psd2Δ and cho1Δ strains relieved their requirement for Etn or Cho (Table 3). We conclude from these findings that a minimum of PtdEtn is required for growth on glucose, and that in psd1Δ psd2Δ or cho1Δ strains this pool of essential PtdEtn can be provided via Dpl1p-dependent sphingolipid catabolism.
Table 3.
Carbon source dependent auxotrophies of strains with defects in PtdEtn biosynthesis grown on defined solid media
| Strain | Auxotrophy on glucose | Auxotrophy on lactate/ethanol |
|---|---|---|
| Wild type | None | None |
| psd1Δ | None | Etn or Cho or Ser |
| psd2Δ | None | None |
| psd1Δpsd2Δ | Etn or Cho | Etn |
| cho1Δ | Etn or Cho | Etn |
| psd1Δ psd2Δ dpl1Δ | Etn | Etn |
| psd1Δ psd2Δ+ 2μ DPL1 | None | None |
| cho1Δ+ 2μ DPL1 | None | None |
| psd1Δ dpl1Δ | None | Etn or Cho or Ser |
| psd2Δ dpl1Δ | None | None |
| psd1Δ eki1Δ | None | Etn or Cho or Ser |
| psd1Δ cki1Δ | None | Etn |
| psd1Δ cki1Δ eki1Δ | None | Lethala |
| psd2Δ eki1Δ | None | None |
| psd2Δ cki1Δ | None | None |
| psd2Δ cki1Δ eki1Δ | None | None |
| psd1Δ cki1Δ dpl1Δ | None | Etn |
| psd1Δ eki1Δ dpl1Δ | None | Etn or Cho or Ser |
| psd2Δ cki1Δ dpl1Δ | None | None |
| psd2Δ eki1Δ dpl1Δ | None | None |
| psd1Δ psd2Δ cki1Δ | Lethal | Lethal |
| psd1Δ psd2Δ cki1Δ eki1Δ | Lethal | Lethal |
| psd1Δ psd2Δ cki1Δ dpl1Δ | Lethal | Lethal |
| psd1Δ cki1Δ eki1Δ dpl1Δ | None | Lethala |
| psd2Δ cki1Δ eki1Δ dpl1Δ | None | None |
| psd1Δ psd2Δ cki1Δ eki1Δ dpl1Δ | Lethal | Lethal |
| psd1Δ psd2Δ eki1Δ | Etn or Cho | Etn |
| psd1Δ psd2Δ eki1Δ dpl1Δ | Etn | Etn |
| cki1Δ eki1Δ dpl1Δ | None | None |
poor growth on full medium
Requirement for PtdEtn Is More Stringent on Nonfermentable Than on Fermentable Carbon Sources
In contrast to cultivation on glucose-containing media, a psd1Δ mutant strain failed to grow on media with lactate or ethanol as the carbon source unless it was supplemented with Etn, Cho, or Ser (Table 3). This defect was fully rescued by expression of Psd1p derived from the centromeric plasmid pRB1 (Table 1) in the psd1Δ background. Addition of Etn, Cho, or Ser to the medium improved growth of the psd1Δ mutant on nonfermentable carbon sources to some extent but not to the wild-type level. Whereas Etn and Cho supplementation should directly enhance PtdEtn and PtdCho formation through the Kennedy pathway, supplementation with Ser resulted in enhanced PtdSer synthesis as indicated by the rise of PtdSer from ∼10 mol% of total phospholipids in psd1Δ psd2Δ grown on Etn- or Cho-supplemented medium to 17 mol% in cells grown on Ser-supplemented medium (our unpublished results). This elevated level of PtdSer might increase the substrate level for Psd2p. The possibility that enhanced serine palmitoyl transferase-dependent synthesis of sphingoid bases (Nagiec et al., 1994) and increased hydrolysis of phosphorylated sphingoid bases by Dpl1p accounted for the Ser auxotrophy was ruled out because deletion of DPL1 in the psd1Δ mutant background did not affect growth on lactate or ethanol in the presence of Ser (Table 3). Taken together, these results indicate that enhanced synthesis of PtdSer or increased formation of Ptd-Etn or PtdCho through the Kennedy pathway can rescue growth of psd1Δ mutant cells on nonfermentable carbon sources. In contrast, psd1Δ psd2Δ or cho1Δ mutants are strictly auxotrophic for Etn on nonfermentable carbon sources (Table 3), suggesting that their capacity to synthesize PtdEtn through Dpl1p-dependent turnover of sphingoid bases was not sufficient to fulfill the elevated requirement of PtdEtn under these conditions.
Single deletions of EKI1 or CKI1, which encode the initial enzymes of the Kennedy pathway, did not affect growth of mutants deleted for PSD1 or PSD2 on glucose medium (Table 3). On lactate or ethanol containing medium, however, a psd1Δ eki1Δ double mutant was auxotrophic for Etn, Cho, or Ser, and a psd1Δ cki1Δ mutant strain was a strict Etn auxotroph (Table 3), indicating that Etn is more specifically used as a substrate by Eki1p than by Cki1p. Moreover, the psd1Δ cki1Δ dpl1Δ mutant grew on Etn-, and the psd1Δ eki1Δ dpl1Δ mutant grew on Etn, Cho, or Ser-supplementation (Table 3). As expected, a psd1Δ cki1Δ eki1Δ triple mutant failed to grow on Etn-, Cho-, or Ser-supplemented synthetic lactate or ethanol medium, because Psd2p and Dpl1p did not provide enough PtdEtn for cells grown on nonfermentable carbon sources. Thus, Psd2p in combination with either Eki1p or Cki1p was sufficient for maintaining the required pool of PtdEtn for cells grown on lactate or ethanol. In the absence of both Psd1p and Psd2p, Etn-utilization was more efficient by Cki1p than by Eki1p. A psd1Δ psd2Δ cki1Δ triple deletion was lethal (Table 3), whereas a psd1Δ psd2Δ eki1Δ triple deletion strain grew like a psd1Δ psd2Δ double mutant on lactate (except for a slightly higher requirement for Etn) and a psd1Δ psd2Δ eki1Δ dpl1Δ quadruple deletion mutant grew on fermentable and nonfermentable carbon sources supplemented with Etn. Thus, the function of Eki1p and Dpl1p in a psd1Δ psd2Δ cki1Δ triple mutant background was not sufficient for growth, whereas psd1Δ psd2Δ with intact Cki1p could grow if given appropriate supplementation.
PtdEtn Level in Mitochondria Is Growth Limiting on Nonfermentable Carbon Sources
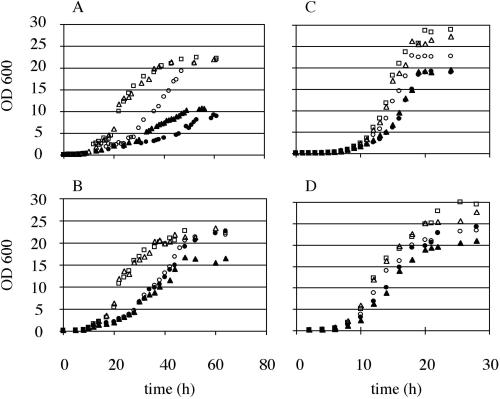
To quantify the effects of mitochondrial and extramitochondrial de novo synthesis of PtdEtn, we followed growth of mutants with defects in PtdEtn synthesis in liquid YP media (Figure 2). Consistent with the observations made on defined solid media, more Etn was required on nonfermentable than on fermentable carbon sources for strains deficient in biosynthesis of PtdEtn. Etn supplementation of YPLac enhanced growth of psd1Δ psd2Δ and cho1Δ mutants to a level comparable to that of psd1Δ but not further. Growth of psd1Δ on YPLac was not further enhanced by Etn supplementation because YPLac contains low amounts of Etn and Cho. These data suggest that, in the absence of mitochondrial production of PtdEtn, namely, in psd1Δ or cho1Δ strains, transport of PtdEtn to mitochondria becomes growth limiting. Under these conditions, the increased requirement of PtdEtn cannot be satisfied by supply with extramitochondrially synthesized PtdEtn. This notion was confirmed by determining the phospholipid composition of isolated mitochondria of the various mutant strains grown on lactate with or without supplementation of 5 mM Etn (Table 4). This analysis revealed that the growth rate of the different strains on lactate correlated with the mitochondrial PtdEtn content. In a psd1Δ strain, mitochondrial PtdEtn was reduced to ∼25% of the corresponding wild-type level; in psd1Δ psd2Δ and cho1Δ strains, PtdEtn was reduced even more dramatically to an apparently minimal level of 1–2%. This decrease of PtdEtn is compensated for by elevated levels of PtdCho, PtdSer, and phosphatidylinositol (PtdIns). Etn supplementation did not increase the PtdEtn content of mitochondria significantly, indicating that the low amount of PtdEtn available in the extramitochondrial space is not preferentially imported into mitochondria, but rather used for production of PtdCho through the methylation pathway. The predominant role of Psd1p in supplying mitochondria, relative to the total cell homogenate, with PtdEtn is apparent from the significant enrichment of PtdEtn in mitochondria in strains with intact PSD1 (Table 4). In contrast, mitochondria from psd1Δ, psd1Δ psd2Δ, or cho1Δ mutants have a PtdEtn level more comparable to that found in nonmitochondrial subcellular membranes.
Figure 2.
Growth of strains with defects in PtdEtn biosynthesis. Wild-type (□), psd1Δ (○), psd2Δ (▵), psd1Δ psd2Δ (●), and cho1Δ (▴) strains were grown on YPLac (A), YPLac supplemented with 5 mM Etn (B), YPD (C), and YPD supplemented with 5 mM Etn (D).
Table 4.
Phospholipids of strains with defects in PtdEtn biosynthesis
| Strain | Growth medium | Phospholipids in
mitochondria and cell homogenate (mol%)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA
|
PtdSer
|
PtdIns
|
PtdCho
|
PtdEtn
|
CL
|
Others
|
|||||||||
| M | H | M | H | M | H | M | H | M | H | M | H | M | H | ||
| Wild type | YPLac | 1 | 2 | 1 | 6 | 13 | 17 | 51 | 53 | 25 | 18 | 7 | 3 | 2 | 1 |
| psd1Δ | YPLac | 1 | 2 | 6 | 7 | 13 | 21 | 67 | 58 | 6 | 8 | 5 | 3 | 2 | 1 |
| YPLac +Etn | 1 | 1 | 6 | 6 | 13 | 10 | 63 | 69 | 9 | 9 | 8 | 4 | 1 | 1 | |
| psd2Δ | YPLac | 1 | 1 | 4 | 7 | 12 | 15 | 45 | 55 | 27 | 17 | 9 | 3 | 1 | 2 |
| psd1Δ psd2Δ | YPLac | 1 | 2 | 5 | 7 | 24 | 27 | 64 | 60 | 1 | 1 | 4 | 3 | 1 | 0 |
| YPLac +Etn | 1 | 2 | 9 | 10 | 1 | 11 | 67 | 69 | 3 | 3 | 9 | 4 | 1 | 1 | |
| cho1Δ | YPLac | 1 | 2 | ND | ND | 25 | 28 | 64 | 61 | 2 | 5 | 6 | 2 | 2 | 2 |
| YPLac +Etn | 1 | 2 | ND | ND | 9 | 17 | 77 | 69 | 6 | 7 | 5 | 3 | 2 | 2 | |
M, mitochondria; H, homogenate; ND, not detected.
Mean values of two independent measurements are shown. Mean deviations are less than 5%.
In mitochondria of psd1Δ and psd1Δ psd2Δ mutants, the CL content is also reduced to ∼50% of the Etn-supplemented control (Table 4). To investigate whether the low PtdEtn content in mitochondria directly affects the activity of enzymes involved in CL biosynthesis, we performed in vitro enzyme assays for CL synthase and phosphatidylglycerophosphate (PtdGro-P) synthase. After growth on YPLac without Etn supplementation specific activity of PtdGro-P synthase was twice as high in mitochondria of psd1Δ psd2Δ (0.81 ± 0.04 nmol/min × mg protein) compared with the wild-type (0.44 ± 0.03 nmol/min × mg protein), whereas the specific activity of CL synthase was similar in mitochondria of both strains (0.081 ± 0.011 nmol/min × mg protein). Thus, the observed reduction of CL is not due to reduced activities of the enzymes involved in the biosynthesis of this phospholipid.
PtdEtn Depletion Does Not Cause Obvious Damage to Mitochondrial Membranes
To examine whether enzymes of the respiratory chain depend on the mitochondrial content of PtdEtn we measured the specific activity of cytochrome c oxidase in mitochondria of psd1Δ, psd1Δ psd2Δ, cho1Δ, and psd2Δ deletion strains grown on YPLac with or without Etn supplementation. In all mutants tested this enzyme activity was similar to wild-type (0.19 ± 0.01 μmol/min × mg). Furthermore, spectroscopic analysis displayed a similar content of cytochromes aa3, b, and c in all of these strains (our unpublished results). Finally, the mitochondrial protein pattern and morphology were not affected by PtdEtn depletion either. The latter observation was made by using the green fluorescent protein (GFP)-tagged mitochondrial matrix protein CoxIVp (kindly provided by R.E. Jensen, John Hopkins University of Medicine, Baltimore, MD), and the membrane potential-dependent fluorescent dyes MitoTracker CMXRos (Molecular Probes, Eugene, OR) and 4-(4-dimethylaminostyryl)-N-methyl-pyridinium iodide (Molecular Probes) as probes. Similarly, electron microscopic analysis did not show any obvious structural alterations of mitochondria or other organelles in the psd1Δ strain (our unpublished results). These results demonstrate that the mitochondrial defect of PtdEtn-depleted strains is not due to gross changes of their mitochondrial membranes.
Reduced Level of PtdEtn Induces Formation of Respiration-deficient Cells (Petites)
An increased rate of respiration-deficient cell (petite) formation is a common phenotype of various mutants defective in lipid biosynthesis. This phenotype was described for cho1Δ (Atkinson et al., 1980) and psd1Δ mutant strains (Trotter et al., 1993), suggesting that mitochondria are more sensitive to alterations of their phospholipid composition than other organelles. Our results presented here confirm and extend these observations. Cultures of cells lacking the capacity to form mitochondrial PtdEtn (psd1Δ, psd1Δ psd2Δ, and cho1Δ) contained ∼50% petites after 2 d of growth in YPD medium, irrespective of Etn supplementation. The observation that spontaneously produced petite cells are growth arrested on nonfermentable carbon sources but remain fully viable for up to 4 d, allowed us to determine the rate of spontaneous petite formation by counting the number of petite cells at two different time points. In YPLac medium ∼10% of the cells lacking mitochondrial PtdEtn synthesis formed petites independent of growth phase and Etn supplementation (Table 5). Thus, during each cell division ∼10% of psd1Δ , psd1Δ psd2Δ, and cho1Δ cells become respiratory deficient. After three to four rounds of subculturing these strains on glucose-containing medium (YPD), petites accumulated and the entire culture became respiratory deficient.
Table 5.
Respiration deficiency (petite formation) after cultivation of cells to the midlogarithmic (midlog.) and stationary growth phase (stat.) on lactate
| Strain | Growth medium | Petites (%) midlog. | Petites (%) stat. |
|---|---|---|---|
| Wild type | YPLac | 1 | 0 |
| psd1Δ | YPLac | 13 | 9 |
| YPLac +Etn | 13 | 10 | |
| psd2Δ | YPLac | 1 | 0 |
| psd1Δ psd2Δ | YPLac | 15 | 8 |
| YPLac +Etn | 14 | 9 | |
| cho1Δ | YPLac | 14 | 9 |
| YPLac +Etn | 13 | 8 |
Mean values of two independent dilutions are shown. Mean deviations are <10%.
As a control, we examined whether the poor growth of strains with defects in PtdEtn biosynthesis on nonfermentable carbon sources was due to a reduction of their viability. For this purpose psd1Δ, psd1Δ psd2Δ, cho1Δ, psd2Δ mutant strains and wild-type were grown to mid-logarithmic growth phase on lactate (YPLac) with or without Etn supplementation and stained with acridine orange (Molecular Probes). Viability was close to 100% in all cultures, indicating that deficiency of PtdEtn directly or indirectly leads to cessation of growth on nonfermentable carbon sources but not to cell death.
Overproduction of Psd1p in Wild-Type Does Not Affect the Mitochondrial Phospholipid Composition
Because the amount of PtdEtn in mitochondria of the psd1Δ mutant seems to be growth limiting, we examined whether 1) this parameter is also growth limiting in wild-type, and 2) whether the amount of mitochondrial PtdEtn can be increased over the wild-type level by overexpression of Psd1p. The haploid wild-type strain YBR1 transformed with the 2 μ plasmid pRB3 carrying PSD1 (Table 1) and the control strain bearing the empty vector grew with almost identical rate on YPLac. Surprisingly, the phospholipid composition of mitochondria and cell homogenate was not affected by overexpression of Psd1p (our unpublished results), even though the specific activity of PtdSer decarboxylase in vitro was 13-fold increased in the cell homogenate (3.7 nmol/min × mg protein) and 17-fold in mitochondria (27.6 nmol/min × mg protein) over wild-type. These observations indicate that mitochondrial production of PtdEtn is most likely limited by the rate of import of PtdSer into mitochondria but not by the activity of Psd1p.
PtdEtn Is Required for Glycosylphosphatidylinositol (GPI)-Anchor Synthesis, but not for Secretion of carboxypeptidase Y (CPY) and Invertase
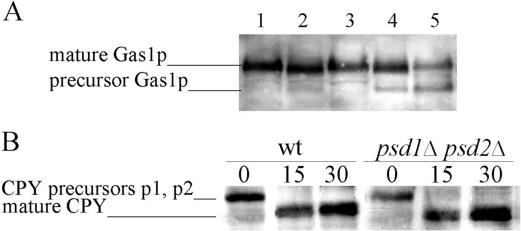
PtdEtn is the precursor for the Etn-P bridge that links the GPI-anchor to the carboxyl-terminal amino acid of proteins (Menon and Stevens, 1992). Thus, depletion of PtdEtn was expected to affect the formation of GPI-anchored proteins. To test this prediction, we studied maturation of the GPI-anchored Gas1p in strains deleted of one or both PtdSer decarboxylases on synthetic glucose medium supplemented with Etn or Cho. Western blot analysis (Figure 3A) revealed that the 100-kDa Gas1p-precursor of the endoplasmic reticulum accumulated in the psd1Δ psd2Δ double mutant. The precursor band appeared strongest, and the amount of the mature 125-kDa Golgi form of Gas1p was reduced concomitantly, when the strain was grown with Cho supplementation, i.e., when the level of PtdEtn was decreased to the minimum of less than 1 mol% of total phospholipids compared with 20 mol% of wild-type. The Gas1p precursor accumulated, although in a less pronounced way, in the Etn-supplemented psd1Δ psd2Δ mutant, supporting the view that Etn is the more efficient precursor for maintaining the PtdEtn pool in this strain.
Figure 3.
Processing of Gas1p and CPY in PtdEtn depleted cells. (A) Western blot analysis of Gas1p from wild-type (lane 1), psd1Δ (lane 2), psd2Δ (lane 3), and psd1Δ psd2Δ (lanes 4 and 5) cells grown on synthetic medium. Growth of psd1Δ psd2Δ was supplemented with Etn (lane 4) or Cho (lane 5). (B) CPY was immunoprecipitated at 0, 15, and 30 min from wild-type and psd1Δ psd2Δ cells during a pulse-chase experiment with cells grown on synthetic medium supplemented with Cho.
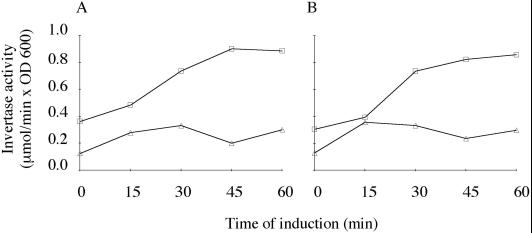
Because GPI-anchor–linked and –unlinked Gas1p precursor of the endoplasmic reticulum is not separated by gel electrophoresis, the analysis described above did not discriminate between a defect in GPI-anchor biosynthesis or a defect in sorting or transport of GPI-anchored proteins from the endoplasmic reticulum to the Golgi. For this reason, we investigated whether PtdEtn deficiency affects the secretory pathway in general by measuring the kinetics of CPY maturation and the rate of invertase secretion in psd1Δ psd2Δ grown on synthetic glucose medium supplemented with Cho. Mature CPY was formed in psd1Δ psd2Δ and wild type with similar kinetics (Figure 3B). External and internal invertase activities were also similar in both strains (Figure 4). These results demonstrate that secretion, in general, is not affected by PtdEtn deficiency. Thus, PtdEtn appears to be required for GPI-anchor biosynthesis, although we cannot exclude that the sorting of GPI-anchored proteins into secretory vesicles is specifically affected by the altered lipid environment of compartments involved in this process.
Figure 4.
Secretion of invertase in PtdEtn-depleted cells. External (□) and internal (▵) invertase activities of wild-type (A) and psd1Δ psd2Δ (B) strains grown on Cho-supplemented synthetic medium were determined after induction (see MATERIALS AND METHODS) at the time points indicated.
DISCUSSION
The spatial separation of the PtdEtn biosynthetic pathways and their different efficiencies explain why the various biosynthetic routes are not equally well suited to meet the requirements for PtdEtn in yeast on fermentable and nonfermentable carbon sources. A minimum level of PtdEtn, as formed through sphingolipid turnover, is sufficient for growth on glucose provided that PtdCho is synthesized from Cho via the CDP-Cho (Kennedy) pathway. On nonfermentable carbon sources, a higher level of cellular PtdEtn is required, and mitochondrial synthesis of PtdEtn by Psd1p becomes paramount. Under these conditions, psd1Δ psd2Δ and cho1Δ deletion mutant strains are strictly auxotrophic for Etn because the formation of endogenous Etn-P by Dpl1p is not sufficient to synthesize the amount of PtdEtn that is required. Similarly, the Psd2p pathway alone does not provide enough PtdEtn for growth of a psd1Δ strain on nonfermentable carbon sources, and increased PtdSer biosynthesis (with exogenous Ser) or the Kennedy pathway (with exogenous Etn or Cho) becomes essential. We conclude from these results 1) that PtdEtn production through the Kennedy pathway is more efficient than decarboxylation of PtdSer by Psd2p, and/or 2) that transport of PtdEtn into mitochondria is growth rate-limiting and less efficient from the site of Psd2p action than from the site where PtdEtn is synthesized through the Kennedy pathway. It has to be noted, however, that neither of these enzymatic steps has yet been unambiguously localized.
On glucose media, the Kennedy pathway is only essential when cells lack Cho1p or both Psd1p and Psd2p. In a psd1Δ psd2Δ background, deletion of CKI1 is lethal, whereas deletion of EKI1 causes only a higher requirement for Etn, especially on nonfermentable carbon sources. A psd1Δ cki1Δ double mutant does not efficiently use Cho as a source for PtdCho biosynthesis because Eki1p appears to be specific for Etn and has a rather poor Cho kinase activity. Furthermore, deletion of CKI1 (Kim et al., 1999), in contrast to deletion of EKI1, reduces the activity of the Cho and Etn transporter Ctr1p and may thus contribute to the synthetic lethality of a psd1Δ psd2Δ cki1Δ triple mutant. In contrast to psd1Δ and psd1Δ eki1Δ, the psd1Δ cki1Δ mutant does not grow on Ser-supplemented media containing nonfermentable carbon sources. In psd1Δ cki1Δ cells, the enhancement of PtdSer synthesis is probably insufficient for growth because of the limited capacity of this strain to recycle Cho formed by phospholipase D. These results also support the view that Eki1p has low Cho kinase activity, resulting in a decreased level of PtdCho in psd1Δ cki1Δ cells that cannot be compensated by increased formation of other lipids. Without taking into account specific effects of EKI1 and CKI1 deletion on transport of Etn and Cho, and a possible regulatory interaction of pathways of PtdEtn biosynthesis, we can define a ranking for the efficiency of the different PtdEtn biosynthetic routes in yeast as follows: Psd1p > Cki1p > Psd2p > Eki1p > Dpl1p.
The second part of this study was focused on possible roles of PtdEtn in yeast, especially in mitochondria. Growth analysis of the various mutant strains revealed a requirement of PtdEtn for mitochondrial function. Consistent with such a function of PtdEtn, we found that the growth defects of psd1Δ , psd1Δ psd2Δ, and cho1Δ strains on lactate medium correlated with the PtdEtn content in the mitochondria of these strains. Because Etn supplementation and thus stimulation of the Kennedy pathway did not have a pronounced effect on the PtdEtn content of mitochondria from psd1Δ, psd1Δ psd2Δ, and cho1Δ mutant strains, we conclude that in addition to its limited formation, PtdEtn is also insufficiently supplied to mitochondria, and thus becomes growth limiting, when mitochondrial synthesis of PtdEtn is absent. This observation is in line with previous results from our laboratory obtained by pulse-chase labeling, which showed that PtdEtn produced by Psd2p or via the Kennedy pathway can be imported into mitochondria, although only at a low rate (Bürgermeister et al., 2000). However, extramitochondrial PtdEtn formed in strains lacking Psd1p does not accumulate in the extramitochondrial space even under Etn supplementation, because it is more efficiently methylated to PtdCho than imported into mitochondria (our unpublished results).
Phospholipid biosynthesis is coordinately regulated in response to inositol availability. Some key enzymes of lipid biosynthesis are repressed by exogenous inositol and derepressed when inositol becomes limiting (reviewed by Henry and Patton-Vogt, 1998). Deletion of CHO1 or PSD1 results in a derepression of phospholipid biosynthetic genes and in overproduction and excretion of inositol (Opi-phenotype). Etn supplementation of cho1Δ or psd1Δ relieves this Opi-phenotype and restores regulation in response to inositol (Griac, 1997). This regulatory circuit appears to account for the increased level of PtdIns in psd1Δ psd2Δ and cho1Δ strains through derepression of IN01, encoding for inositol-1-phosphate synthase, and may also be responsible for the decreased formation of CL in psd1Δ and psd1Δ psd2Δ mutants on YPLac (Table 5). Like Ino1p, but unlike CL synthase, PtdGro-P synthase has been shown to be regulated in response to inositol (Greenberg et al., 1988; Shen and Dowhan, 1998). This regulation explains the increased in vitro activity of PtdGro-P synthase, but the unchanged activity of CL synthase, in mitochondria of psd1Δ and psd1Δ psd2Δ strains (see RESULTS). The discrepancy between in vitro activities of enzymes involved in CL formation and the decreased content of CL may be explained as a result of direct competition of PtdIns synthase, PtdSer synthase, and PtdGro-P synthase for the common substrate CDP-DAG or by noncompetitive inhibition of PtdSer synthase by inositol (Kelley et al., 1988). This hypothesis is in line with the observation that lack of PtdSer biosynthesis in cho1Δ cells results in the formation of wild-type levels of CL, while at the same time PtdIns biosynthesis is induced. Furthermore, the PtdSer content of a psd1Δ psd2Δ strain grown in the presence of Etn supplementation is significantly higher than that in cells grown without Etn supplementation, i. e., when CDP-DAG is preferentially used for PtdIns biosynthesis (Table 4). Thus, there appears to be no direct link between the growth phenotype on lactate of mutants with defects in PtdEtn biosynthesis and the CL content of mitochondria (Figure 2; Table 4), but rather an indirect link through regulatory phenomena. It is noteworthy, however, that the sum of CL and PtdEtn might be important for maintaining yeast mitochondrial function, as has been shown previously for E. coli membranes (Rietveld et al., 1993). This hypothesis is supported by the observation that a yeast strain bearing a mutation of the CL synthase gene CRD1 exhibits an elevated level of PtdEtn (Tuller et al., 1998), and the fact that deletions of CHO1 and PGS1 (PtdGro-P synthase) are synthetically lethal (Janitor et al., 1996).
No obvious defects of mitochondrial morphology, membrane potential, or respiratory enzymes were detected as a consequence of a low PtdEtn content. The growth defects of psd1Δ, psd1Δ psd2Δ, and cho1Δ strains on nonfermentable carbon sources are at least partly due to the spontaneous formation of respiration-deficient cells. We do not know at present whether the mitochondrial genome is completely lost (ρ0) or only partly deleted (ρ−) in these strains. We can also only speculate as to whether this defect is due to a reduced efficiency of anchoring the nucleoid to the inner mitochondrial membrane or to more direct effects on the replication/partitioning apparatus itself. The viability of PtdEtn biosynthesis mutants is not reduced on nonfermentable carbon sources, indicating that the severe growth defect of psd1Δ psd2Δ and cho1Δ strains is due to a dramatically decreased growth rate, but not to cell death. These results support data reported by Griac et al. (1996), who observed a similar effect with a cho1 mutant grown in the absence of Etn or Cho. The cessation of growth may result from an indirect effect of PtdEtn depletion, e.g., reduced ATP synthesis, or may be the consequence of a more direct effect, such as a membrane status-dependent control mechanism for growth.
Studies using an ept1Δ cpt1Δ double deletion mutant (Menon and Stevens, 1992) revealed that PtdEtn but not Etn, Etn-P, or CDP-Etn is the precursor for GPI-anchor biosynthesis. Our data support this result in that depletion of PtdEtn leads to a maturation defect of Gas1p. GPI-anchor biosynthesis is essential in yeast (Orlean et al., 1994), which may be one reason for the requirement of a minimum level of PtdEtn. Most remarkably, however, the general secretory pathway is not affected by PtdEtn deficiency (Figures 3B and 4). This finding is surprising because the unique biophysical property of PtdEtn to form nonbilayer structures has been thought to affect vesicle-mediated protein trafficking (de Kruijff, 1997). It seems unlikely that the residual amount of less than 1 mol% PtdEtn of total phospholipids in the psd1Δ psd2Δ strain can fulfill this biophysical requirement. Instead, it appears that in yeast PtdEtn is largely dispensable as a structural component of membranes, and that compensatory effects that are presently unknown may maintain the biophysical properties of the membranes required for cell viability.
ACKNOWLEDGMENTS
We thank G. Zellnig from the Institut für Pflanzenphysiologie, Karl-Franzens Universität Graz, Austria, for electron microscopic analysis of the psd1Δ mutant strain. The kind supply of the MSS204 strain and the p24–3 plasmid by R.C. Dickson (University of Kentucky, Lexington, KY), and of the Gas1p- and CPY-specific antibodies by H. Riezman (Biocenter Basel, Switzerland) is appreciated. This work was financially supported by the Fonds zur Förderung der Forschung Österreich (projects 12076 and 14468 to G.D., and project 13767 to R.S).
Abbreviations used:
- CDP-Cho
cytidyldiphosphate choline
- CDP-DAG
cytidyldiphosphate diacylglycerol
- CDP-Etn
cytidyldiphosphate ethanolamine
- Cho
choline
- Cho-P
cholinephosphate
- CL
cardiolipin
- CPY
carboxypeptidase Y
- CTP
cytidyltriphosphate
- Etn
ethanolamine
- Etn-P
ethanolaminephosphate
- GFP
green fluorescent protein
- GPI
glycosylphosphatidylinositol
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- PtdGro-P
phosphatidylglycerophosphate
- PtdIns
phosphatidylinositol
- PtdSer
phosphatidylserine
- Ser
serine
REFERENCES
- Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. Yeast mutants auxotrophic for choline or ethanolamine. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1996. [Google Scholar]
- Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- Broekhuyse RM. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim Biophys Acta. 1968;152:307–315. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- Bürgermeister M, Birner R, Hrastnik C, Daum G. Phosphatidylserine decarboxylation and CDP-ethanolamine pathway contribute to the supply of phosphatidylethanolamine to mitochondria of yeast. In: Op den Kamp JAF, editor. NATO-ASI Series: Protein, Lipid and Membrane Traffic. Vol. 322. Amsterdam: IOS Press; 2000. pp. 19–25. [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- de Kruijff B. Lipid polymorphism and biomembrane function. Curr Opin Chem Biol. 1997;1:564–569. doi: 10.1016/s1367-5931(97)80053-1. [DOI] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ML, Hubbell S, Lam C. Inositol regulates phosphatidylglycerolphosphate synthase expression in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4773–4779. doi: 10.1128/mcb.8.11.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P. Regulation of yeast phospholipid biosynthetic genes in phosphatidylserine decarboxylase mutants. J Bacteriol. 1997;179:5843–5848. doi: 10.1128/jb.179.18.5843-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P, Swede MJ, Henry SA. The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J Biol Chem. 1996;271:25692–25698. doi: 10.1074/jbc.271.41.25692. [DOI] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Hamamatsu S, Shibuya I, Takagi M, Ohta A. Loss of phosphatidylserine synthesis results in aberrant solute sequestration and vacuolar morphology in Saccharomyces cerevisiae. FEBS Lett. 1994;348:33–36. doi: 10.1016/0014-5793(94)00576-1. [DOI] [PubMed] [Google Scholar]
- Henry SA, Patton-Vogt JL. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- Hikiji T, Miura K, Kiyono K, Shibuya I, Ohta A. Disruption of the CHO1 gene encoding phosphatidylserine synthase in Saccharomyces cerevisiae. J Biochem. 1988;104:894–900. doi: 10.1093/oxfordjournals.jbchem.a122579. [DOI] [PubMed] [Google Scholar]
- Janitor M, Obernauerova M, Kohlwein SD, Subik J. The pel1 mutant of Saccharomyces cerevisiae is deficient in cardiolipin and does not survive the disruption of the CHO1 gene encoding phosphatidylserine synthase. FEMS Microbiol Lett. 1996;140:43–47. doi: 10.1111/j.1574-6968.1996.tb08312.x. [DOI] [PubMed] [Google Scholar]
- Kelley MJ, Bailis AM, Henry SA, Carman GM. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- Kim K, Kim KH, Storey MK, Voelker DR, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J Biol Chem. 1999;274:14857–14866. doi: 10.1074/jbc.274.21.14857. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur J Biochem. 1989;185:243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci USA. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason TL, Poyton RO, Wharton DC, Schatz G. Cytochrome c oxidase from bakers'yeast. I. Isolation and properties. J Biol Chem. 1973;248:1346–1354. [PubMed] [Google Scholar]
- McGee TP, Skinner HB, Bankaitis VA. Functional redundancy of CDP-ethanolamine and CDP-choline pathway enzymes in phospholipid biosynthesis: ethanolamine-dependent effects on steady-state membrane phospholipid composition in Saccharomyces cerevisiae. J Bacteriol. 1994;176:6861–6868. doi: 10.1128/jb.176.22.6861-6868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AK, Stevens VL. Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J Biol Chem. 1992;267:15277–15280. [PubMed] [Google Scholar]
- Mileykovskaya E, Sun Q, Margolin W, Dowhan W. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J Bacteriol. 1998;180:4252–4257. doi: 10.1128/jb.180.16.4252-4257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morein S, Andersson A, Rilfors L, Lindblom G. Wild type Escherichia coli cells regulate the membrane lipid composition in a “window” between gel and non-lamellar structures J. Biol Chem. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- Munn AL, Heese-Peck A, Stevenson BJ, Pichler H, Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol Biol Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci USA. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Miura K, Fukuda Y, Shibuya I, Ohta A, Takagi M. Phosphatidylserine synthesis required for the maximal tryptophan transport activity in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2000;64:167–172. doi: 10.1271/bbb.64.167. [DOI] [PubMed] [Google Scholar]
- Orlean P, Leidich SD, Drapp DA, Colussi P. Isolation of temperature-sensitive yeast GPI-anchoring mutants. Braz J Med Biol Res. 1994;27:145–150. [PubMed] [Google Scholar]
- Rietveld AG, Killian JA, Dowhan W, de Kruijff B. Polymorphic regulation of membrane phospholipid composition in Escherichia coli. J Biol Chem. 1993;268:12427–12433. [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- Shen H, Dowhan W. Regulation of phosphatidylglycerophosphate synthase levels in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11638–11642. doi: 10.1074/jbc.273.19.11638. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Yates R, Voelker DR. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- Tuller G, Nemec T, Hrastnik C, Daum G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild type strain grown on different carbon sources. Yeast. 1999;15:1555–1564. doi: 10.1002/(SICI)1097-0061(199910)15:14<1555::AID-YEA479>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- van der Does C, Swaving J, van Klompenburg W, Driessen AJ. Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J Biol Chem. 2000;275:2472–2478. doi: 10.1074/jbc.275.4.2472. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Watson K, Houghton RL, Bertoli E, Griffiths DE. Membrane-lipid unsaturation and mitochondrial function in Saccharomyces cerevisiae. Biochem J. 1975;146:409–416. doi: 10.1042/bj1460409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricuperohovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast. Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]