EGFR and K-ras mutation status was determined from tumor DNA extracted from cytological samples from non-small cell lung cancer patients, with results comparable with those from biopsy samples. Clinical outcomes of patients harboring mutations and their response to tyrosine kinase inhibitor therapy are also discussed.
Keywords: Carcinoma, Non-small cell lung, Cytology, Papanicolau stained smears, Mutations, Epidermal growth factor receptor–neu receptor, Genes, Ras, Erlotinib, Gefitinib
Abstract
Objective.
Epidermal growth factor receptor (EGFR) and K-ras mutations guide treatment selection in non-small cell lung cancer (NSCLC) patients. Although mutation status is routinely assessed in biopsies, cytological specimens are frequently the only samples available. We determined EGFR and K-ras mutations in cytological samples.
Methods.
DNA was extracted from 150 consecutive samples, including 120 Papanicolau smears (80%), 10 cell blocks (7%), nine fresh samples (6%), six ThinPrep® tests (4%), and five body cavity fluids (3.3%). Papanicolau smears were analyzed when they had >50% malignant cells. Polymerase chain reaction and direct sequencing of exons 18–21 of EGFR and exon 2 of K-ras were performed. EGFR mutations were simultaneously determined in biopsies and cytological samples from 20 patients. Activity of EGFR tyrosine kinase inhibitors (TKIs) was assessed.
Results.
The cytological diagnosis was adenocarcinoma in 110 samples (73%) and nonadenocarcinoma in 40 (27%) samples. EGFR mutations were identified in 26 samples (17%) and K-ras mutations were identified in 18 (12%) samples. EGFR and K-ras mutations were mutually exclusive. In EGFR-mutated cases, DNA was obtained from stained smears in 24 cases (92%), pleural fluid in one case (4%), and cell block in one case (4%). The response rate to EGFR TKIs in patients harboring mutations was 75%. The mutation status was identical in patients who had both biopsies and cytological samples analyzed.
Conclusion.
Assessment of EGFR and K-ras mutations in cytological samples is feasible and comparable with biopsy results, making individualized treatment selection possible for NSCLC patients from whom tumor biopsies are not available.
Introduction
Mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) gene in non-small cell lung cancer (NSCLC) patients are associated with clinical response to the TK inhibitors (TKIs) gefitinib and erlotinib [1–3]. These mutations are somatic and are more common in patients with clinical features known to be associated with sensitivity to TKIs, such as female gender, adenocarcinoma, Asian ethnicity. and lack of smoking history. The discovery of these mutations and their clinical significance has become a major step forward in the development of targeted therapies with EGFR TKIs such as gefitinib and erlotinib [4–6].
Activating mutations within the TK domain of EGFR are found in approximately 10%–20% of NSCLC patients and are associated with response to EGFR TKIs [5, 6]. Gefitinib showed markedly better efficacy than standard chemotherapy in this subset of patients [7, 8]. Deletions in exon 19 and the single L858R point mutation in exon 21 account for 90% of all EGFR mutations [1–3, 6]. These mutations mediate oncogenic effects by altering downstream signaling and antiapoptotic mechanisms [9] and are associated with clinical response and survival following TKI therapy. Other genetic alterations described in NSCLC, such as the T790M point mutation or insertion mutations in exon 20 of EGFR, amplification of MET, and mutations in K-ras, are associated with a lack of activity of TKIs [10–12]. As a result, assessment of mutation status has become part of the standard management of NSCLC patients [6, 10].
Unfortunately, molecular testing is often hampered by the inability to obtain appropriate tumor samples. Currently, advanced lung cancer is frequently diagnosed by fine-needle aspiration (FNA) performed via transbronchial needle aspiration (TBNA), endoscopic ultrasound (EUS), or percutaneously, guided by computerized tomography (CT) or echography [13–15]. The samples obtained are often considered inadequate to yield sufficient material to perform a complete molecular analysis [16, 17]. This is particularly relevant in patients with advanced NSCLC, in whom cytological specimens, often stained smears, are frequently the only diagnostic material available, raising the question of whether or not more invasive procedures should be performed [16]. We report our institution's experience assessing EGFR and K-ras mutations in tumor DNA extracted from cytological samples, in particular from Papanicolau-stained slides. Clinical outcomes of patients harboring mutations and their response to TKI therapy are also discussed.
Materials and Methods
Patients
Cytological samples from patients with suspected lung cancer were obtained consecutively at our institution by TBNA, EUS, CT, ultrasound-guided FNA, or blind percutaneous FNA. Rapid onsite evaluation was performed by a pathologist for all FNA procedures in order to guarantee that samples were adequate. Stained smears received from other hospitals for consultation and cytological samples obtained from body cavity fluids were also analyzed. When paraffin-embedded tumor biopsies were available, molecular analysis was performed and results were compared with those obtained from cytological samples. The following categories were used to define smoking status: smoker, >100 cigarettes per lifetime; nonsmoker, <100 cigarettes per lifetime. An institutional review board–approved protocol allows biopsy specimens to be used for research purposes. All patients signed informed consent before the procedure.
DNA Extraction
A pathologist reviewed the Papanicolau-stained slides in order to select the best slides for molecular analysis. The criterion to select adequate slides was that they showed ≥50% malignant cells. Only one slide was used for DNA extraction in each case. Prior to DNA extraction, Papanicolau-stained smears were first rinsed in alcohol and scraped into Eppendorf tubes. Slides were not destained prior to DNA extraction. DNA was extracted using Nucleospin® Tissue (catalogue no., 740952.50; Macherey-Nagel GmbH & Co. KG, Düren, Germany). DNA concentration was measured using a NanoDrop-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE).
EGFR and K-ras Mutation Analysis
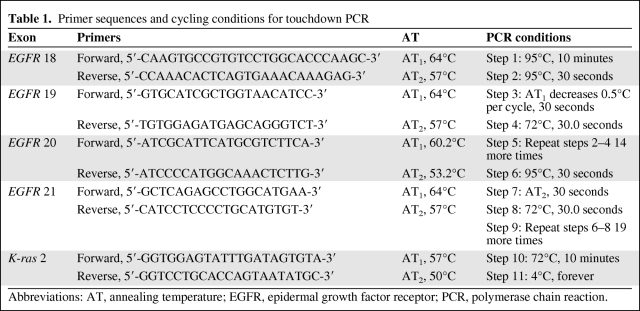
Polymerase chain reaction (PCR) and direct sequencing of exons 18–21 of EGFR and exon 2 of K-ras were performed using an ABI Prism™ 310XL DNA sequencer (Applied Biosystems, Madrid, Spain). The primer sequences used, cycling conditions, and annealing temperatures of touchdown PCR for EGFR analysis are shown in Table 1. The presence of an appropriate PCR product was confirmed by resolving the PCR products on a 2% agarose gel. PCR products were purified using the GFX™ PCR DNA and Gel Band Purification kit (GE Healthcare Bio-Sciences AB, Björkgatan, Sweden) following the manufacturer's instructions. Fragments were sequenced and analyzed in both the sense and antisense directions. DNA templates were processed for the DNA sequencing reaction using ABI Prism™ BigDye Terminator, version 3.1 (Applied Biosystems).
Table 1.
Primer sequences and cycling conditions for touchdown PCR
Abbreviations: AT, annealing temperature; EGFR, epidermal growth factor receptor; PCR, polymerase chain reaction.
Following sequencing reactions, DNA was purified using Performa® DTR Gel Filtration Cartridges (EdgeBio, Gaithersburg, MD). Sequence data were generated with the ABI Prism 310 DNA analyzer (Applied Biosystems). Mutations were identified by visual analysis of the sequenced chromatograms using SeqScape (Applied Biosystems). Sequence data were analyzed by two investigators and compared with the archived human sequence of EGFR (GenBank accession no., NG_007726.1; K-ras GenBank accession no., NG_007524.1).
Clinical Assessment and Statistical Analysis
The response rate of patients treated with TKIs was assessed using Response Evaluation Criteria in Solid Tumors [18]. The progression-free survival (PFS) interval was calculated from the date when treatment with the TKI was started to the date of progression or death from any cause. Descriptive statistics are used to present patient characteristics and results of the molecular analysis. Statistical comparisons were performed using Fisher's exact test. Survival data were analyzed using the Kaplan–Meier method [19]. Statistical analyses were performed using SPSS software (SPSS Inc., Chicago, IL). Agreement between the mutation status assessed in cytological samples and in biopsies was assessed using Cohen's κ coefficient [20]. The relationship between the κ coefficient and strength of between-method agreement may be interpreted in the following manner: 0.81–1.00, almost perfect; 0.61–0.80, substantial; 0.41–0.60, moderate; 0.21–0.40, fair; 0–0.20, slight; and <0, poor [21].
Results
Patients and Tumor Samples
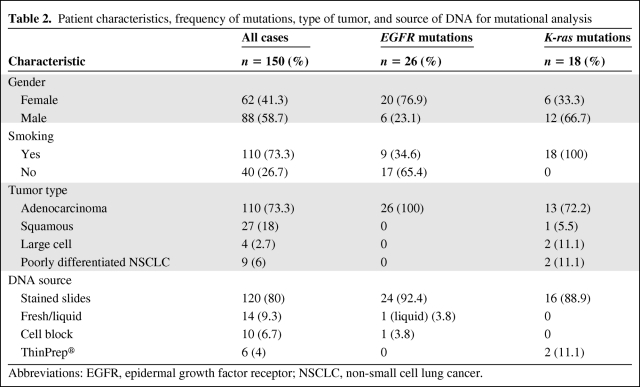
Between January 2007 and December 2009, 150 cytological samples from NSCLC patients were consecutively obtained and analyzed. Patient characteristics, tumor histology, and DNA source are shown in Table 2. Samples were obtained from FNA of primary lung tumors in 68 cases (45.3%), metastatic lymph nodes in 70 cases (46.6%), bone metastases in three cases (2%), a left adrenal metastasis in one case (0.7%), pleural fluid in six cases (4%), pericardial fluid in one case (0.7%), and bronchoalveolar lavage in one case (0.7%). TBNA was the diagnostic procedure in 104 patients, whereas EUS-FNA was used in five patients, CT-guided percutaneous FNA was used in 10 patients, and ultrasound-guided percutaneous FNA was used in three patients. In 11 patients, the pathologist performed an FNA directly on superficial lymph nodes. In eight patients, the samples were stained smears received from other hospitals for consultation, obtained by FNA from lymph nodes (six mediastinal, through TBNA, and two superficial, by direct FNA). Six samples originated from body cavity fluids, and three resulted from bronchial biopsy touch preps. DNA was extracted from 120 Papanicolau-stained smears (80%), 10 cell blocks (6.7%), nine fresh samples (6%), six ThinPrep® tests (4%), and five body cavity fluids (3.3%). DNA was successfully extracted from all samples studied. We obtained an average of 24.6 ng/μl (standard deviation, 15.3) of DNA.
Table 2.
Patient characteristics, frequency of mutations, type of tumor, and source of DNA for mutational analysis
Abbreviations: EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer.
The majority of the lesions were diagnosed as adenocarcinoma (n = 110; 73.3%) Twenty-seven patients (18%) had squamous cell carcinoma, four (2.7%) had large cell carcinoma, and nine (6%) had poorly differentiated NSCLC.
EGFR Mutation Status
We identified 26 EGFR mutations in the 150 samples analyzed. The frequency of EGFR mutations, patient characteristics, tumor type, and source of DNA for mutational analysis are shown in Table 2. All 26 mutations were described in adenocarcinomas. From the 26 mutations detected, 24 were identified in DNA from Papanicolau-stained smears. Mutations were found in 20 of 62 women (32.3%) and in six of 88 men (6.8%). EGFR mutations were identified in 17 of 40 nonsmokers (42.5%), but in only nine of 110 smokers (8.2%) (p < .001) (Table 2). A representative case is presented in Figure 1.
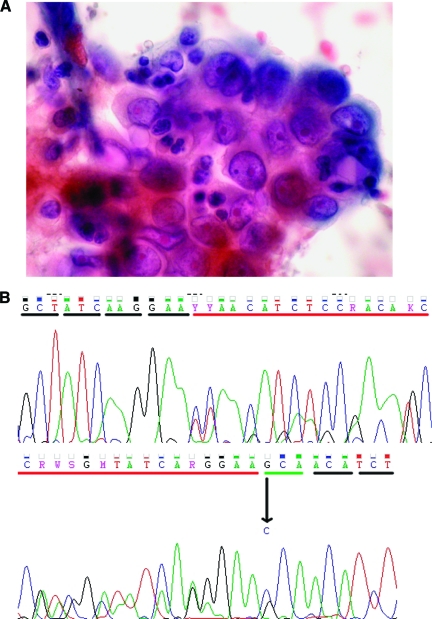
Figure 1.
Representative example of a case included in our series. (A): Papanicolau-stained smear showing lung adenocarcinoma cells obtained by fine-needle aspiration from a supraclavicular lymph node. (B): Direct sequencing of exon 19 showing a deletion and a point mutation (del L747–E749, A750P).
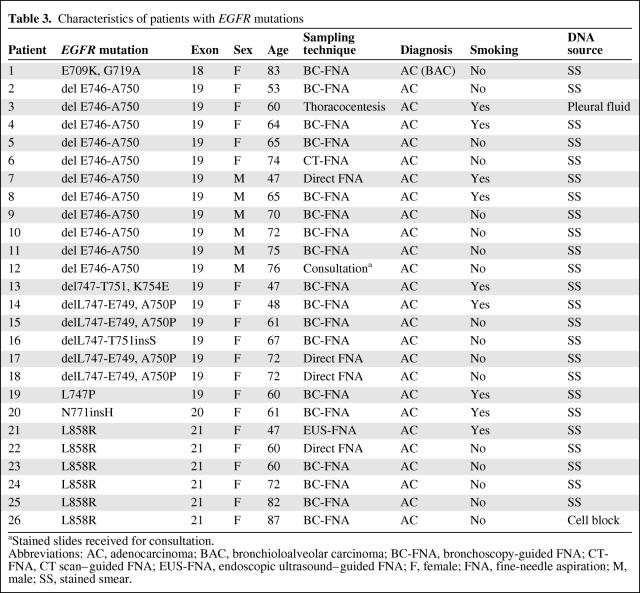
Table 3 summarizes all EGFR mutations detected as well as the characteristics of the patients and the sampling technique used. We found 18 mutations (69.2%) in exon 19, six mutations (23%) in exon 21, one mutation (3.8%) in exon 20, and one mutation (3.8%) in exon 18. In exon 19 we found 11 deletions (42.3%), one missense mutation (3.8%), five deletion plus missense mutations (19.2%), and one deletion plus insertion (3.8%). Exon 20 had an insertion, and in exon 21, all six alterations were missense mutations.
Table 3.
Characteristics of patients with EGFR mutations
aStained slides received for consultation.
Abbreviations: AC, adenocarcinoma; BAC, bronchioloalveolar carcinoma; BC-FNA, bronchoscopy-guided FNA; CT-FNA, CT scan–guided FNA; EUS-FNA, endoscopic ultrasound–guided FNA; F, female; FNA, fine-needle aspiration; M, male; SS, stained smear.
Paraffin-embedded tissue was available from 20 patients. Analysis of EGFR mutation status in those samples revealed agreement with the results obtained in cytological samples in every case (1.0 or perfect agreement, as assessed by the κ coefficient).
K-ras Mutation Status
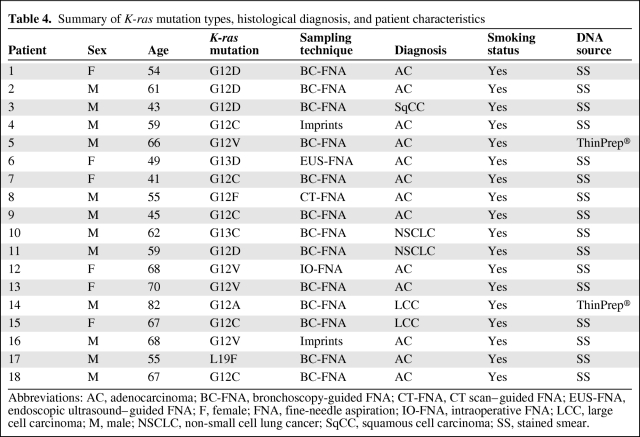
K-ras mutation status was assessed in 111 cases. Table 4 shows K-ras mutation types, diagnosis, and patient characteristics. We found K-ras mutations in 18 patients (12%). K-ras and EGFR mutations were mutually exclusive. Mutations in K-ras were observed in DNA extracted from Papanicolau-stained smears in 16 cases (89%), and from ThinPrep® tests in two cases (11%).
Table 4.
Summary of K-ras mutation types, histological diagnosis, and patient characteristics
Abbreviations: AC, adenocarcinoma; BC-FNA, bronchoscopy-guided FNA; CT-FNA, CT scan–guided FNA; EUS-FNA, endoscopic ultrasound–guided FNA; F, female; FNA, fine-needle aspiration; IO-FNA, intraoperative FNA; LCC, large cell carcinoma; M, male; NSCLC, non-small cell lung cancer; SqCC, squamous cell carcinoma; SS, stained smear.
Clinical Activity of TKIs
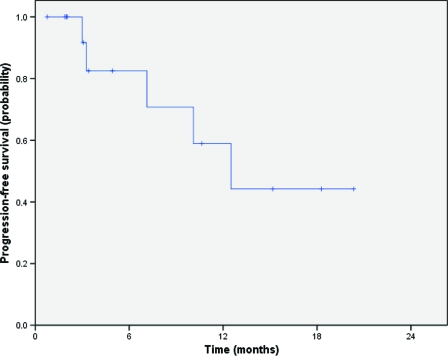
Clinical follow-up was available for 16 of the 26 patients with EGFR mutations who presented with stage IV disease. Based on the identification of such mutations, these patients received subsequent therapy with a TKI, including erlotinib [14] and gefitinib [2]. EGFR TKIs were administered as first-line therapy in eight patients (50%), as second-line therapy in seven patients (44%), and as fifth-line therapy in one patient (6%). We observed one complete response and 11 partial responses (overall response rate, 75%). In addition, one patient showed a minor response and two had ongoing stable disease at 15 and 20 months. One patient presented rapid progression at the first evaluation of disease. The median PFS duration of patients with EGFR mutations who were treated with erlotinib or gefitinib was 12.3 months (Fig. 2).
Figure 2.
Progression-free survival of patients presenting with epidermal growth factor receptor gene mutations treated with erlotinib and gefitinib.
Discussion
Molecular profiling is now considered the standard of care for patients with NSCLC [5, 6, 22]. The development of targeted therapies against the EGFR TK domain has led to the discovery of mutations and other genomic alterations that predict the efficacy of these agents. Paraffin-embedded and frozen tissue have been considered optimal samples for molecular studies. Consequently, most reports published on EGFR analyses were performed using biopsies. However, biopsies are not always available, because the diagnosis of lung cancer sometimes relies on cytological samples obtained by FNA through minimally invasive procedures in order to avoid more invasive techniques [13–15]. Therefore, the possibility of performing molecular analyses on samples obtained using minimally invasive procedures is appealing [23]. To our knowledge, our study is the first large series to show that Papanicolau-stained cytological samples are adequate for EGFR and K-ras mutation analysis. We believe this technique could be introduced into clinical practice.
The prior literature on EGFR mutation analysis relies primarily on larger tumor biopsies, but our results are comparable with regard to the frequency and distribution of EGFR mutations according to sex, histology, and prior smoking history, as well EGFR TKI activity [1–3, 6]. The 75% response rate and the median PFS interval of 12.3 months compare favorably with the 70.6% response rate and 14-month PFS interval reported in one of the most relevant series published to date [6]. Moreover, mutation status was determined in cytology as well as in biopsy samples in a subgroup of patients, and the agreement between the two methods was perfect. EGFR mutations were never observed in patients who had K-ras mutations, as has previously been reported [22].
Currently, there is ongoing debate in the oncologic community regarding the adequacy of samples obtained for molecular analysis from NSCLC patients. Although initial data suggested that cytological samples were not adequate for molecular analysis [16], recent findings suggest that minimally invasive techniques might suffice [10]. Some studies have evaluated the feasibility of assessing EGFR mutation status in paraffin-embedded cell blocks and histological cores from NSCLC patients obtained by TBNA and FNA [24–26]. Smouse et al. [27] recently showed that cytological cell blocks provide equivalent, if not higher, sensitivity when compared with surgical specimens, suggesting that the suitability of a sample should be based on tumor cell proportion and abundance, rather than on the technique used to obtain the sample. We agree with this concept, and we believe that the quality of the sample is a critical factor in order to obtain adequate results. In our study, we assessed mutations only from slides that contained ≥50% malignant cells.
To our knowledge, this is the largest series analyzing EGFR mutations in exons 18–21 as well as K-ras mutations in cytological samples. Molecular analysis results were validated with clinical outcomes as well as simultaneous assessment using traditional biopsy samples. A few small series have assessed mutations in exons 19 and 21 of EGFR using cytological samples, some of them including Papanicolau-stained smears. Boldrini et al. [28] reported their retrospective results from a small series of 23 cytological samples. Nomoto et al. [29] and Smith et al. [30] analyzed the feasibility of a high-resolution melting analysis to perform EGFR mutational analysis in small specimens, including cytological samples. Fukui et al. [31] reported their results with small diagnostic samples and biopsies from 92 patients. They reported better results from PCR analysis using cytological slides rather than biopsy specimens, regardless of the amount of tumor cells analyzed. They explained this fact by differences in the method of sample fixation between the two types of specimens, favoring methanol over formalin for mutational studies, thus highlighting the importance of preserving high-quality DNA. Savic et al. [32] studied the presence of EGFR mutations in NSCLC samples, including some Papanicolau-stained slides from NSCLC patients, using sequencing methods. None of the previous studies provided clinical information on the outcome of the patients treated with TKIs to confirm their predictive value. Oshita et al. [33] evaluated a novel heteroduplex method to study some of the most frequently described mutations in exons 19 and 21 of EGFR in cytology specimens obtained by transbronchial abrasion from 52 NSCLC patients. They assessed the clinical activity of gefitinib in 26 patients, reporting a 91% response rate in the 11 patients that had mutations. Our study added a determination of the concordance between mutation assessment in biopsies and in cytological samples.
The main limitation of our study is the limited sample size. Although we showed that assessment of mutations in cytological samples is feasible, further studies are necessary to determine the specificity and sensitivity of this technique. In addition, this methodology, as any other, requires a learning curve during which simultaneous assessment of mutations in paraffin samples, if available, can provide quality control.
Conclusion
Our results show that assessment of EGFR mutations in exons 18–21 and K-ras mutation status in cytological samples, including Papanicolau-stained slides, from NSCLC patients is feasible and results are comparable with those obtained from biopsies. Identification of EGFR mutations in cytological samples predicts clinical response to targeted therapies, thus extending the benefits of individualized treatment selection to NSCLC patients from whom tumor biopsies are not available. This finding may facilitate large-scale screening of patients with NSCLC for EGFR mutations with minimally invasive techniques, avoiding more invasive procedures. In addition, this strategy can be used for research purposes.
Acknowledgments
The authors thank Ruth Zarate, Ph.D., for her guidance in the mutational analysis; Nerea Gomez, Virginia Pulido, and Mariam Maset for their help in the management of cytological samples, patient data, and statistics; and Mercedes Aguirre for her assistance in the molecular pathology laboratory.
Author Contributions
Conception/Design: Anabel Del Barrio, Ruben Pio, Miguel Angel Idoate, Tania Labiano, Maria D. Lozano, Javier Zulueta, Jose I. Echeveste, Alfonso Gúrpide, Luis M. Seijo, Jose Luis Perez-Gracia, Salvador Martín Algarra
Provision of study material or patients: Anabel Del Barrio, Ruben Pio, Miguel Angel Idoate, Tania Labiano, Maria D. Lozano, Javier Zulueta, Jose I. Echeveste, Alfonso Gúrpide, Luis M. Seijo, Jose Luis Perez-Gracia, Salvador Martín Algarra
Collection and/or assembly of data: Anabel Del Barrio, Ruben Pio, Miguel Angel Idoate, Tania Labiano, Maria D. Lozano, Javier Zulueta, Jose I. Echeveste, Alfonso Gúrpide, Jose Luis Perez-Gracia, Salvador Martín Algarra
Data analysis and interpretation: Anabel Del Barrio, Miguel Angel Idoate, Tania Labiano, Maria D. Lozano, Jose I. Echeveste, Alfonso Gúrpide, Jose Luis Perez-Gracia, Salvador Martín Algarra
Manuscript writing: Anabel Del Barrio, Ruben Pio, Miguel Angel Idoate, Maria D. Lozano, Javier Zulueta, Jose I. Echeveste, Alfonso Gúrpide, Jose Luis Perez-Gracia, Salvador Martín Algarra
Final approval of manuscript: Anabel Del Barrio, Ruben Pio, Miguel Angel Idoate, Tania Labiano, Maria D. Lozano, Javier Zulueta, Jose I. Echeveste, Alfonso Gúrpide, Luis M. Seijo, Jose Luis Perez-Gracia, Salvador Martín Algarra
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pirozynski M. 100 years of lung cancer. Respir Med. 2006;100:2073–2084. doi: 10.1016/j.rmed.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 7.Inoue A, Kobayashi K, Usui K, et al. First-line gefitinib for patients with advanced non-small-cell lung cancer harboring epidermal growth factor receptor mutations without indication for chemotherapy. J Clin Oncol. 2009;27:1394–1400. doi: 10.1200/JCO.2008.18.7658. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 10.Sequist LV, Engelman JA, Lynch TJ. Toward noninvasive genomic screening of lung cancer patients. J Clin Oncol. 2009;27:2589–2591. doi: 10.1200/JCO.2008.20.4875. [DOI] [PubMed] [Google Scholar]
- 11.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosell R, Moràn T, Carcereny E, et al. Non-small-cell lung cancer harbouring mutations in the EGFR kinase domain. Clin Transl Oncol. 2010;12:75–80. doi: 10.1007/S12094-010-0473-0. [DOI] [PubMed] [Google Scholar]
- 13.Stigt JA, Oostdijk AH, Timmer PR, et al. Comparison of EUS-guided fine needle aspiration and integrated PET-CT in restaging after treatment for locally advanced non-small cell lung cancer. Lung Cancer. 2009;66:198–204. doi: 10.1016/j.lungcan.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Gu P, Zhao YZ, Jiang LY, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for staging of lung cancer: A systematic review and meta-analysis. Eur J Cancer. 2009;45:1389–1396. doi: 10.1016/j.ejca.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 15.Vilmann P, Puri R. The complete “medical” mediastinoscopy (EUS-FNA + EBUS-TBNA) Minerva Med. 2007;98:331–338. [PubMed] [Google Scholar]
- 16.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 17.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: Searching for the ideal method. Clin Cancer Res. 2007;13:4954–4955. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 18.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan EI, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457. [Google Scholar]
- 20.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 21.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 22.Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: Implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 23.Gazdar AF. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med. 2009;361:1018–1020. doi: 10.1056/NEJMe0905763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakajima T, Yasufuku K, Suzuki M, et al. Assessment of epidermal growth factor receptor mutation by endobronchial ultrasound-guided transbronchial needle aspiration. Chest. 2007;132:597–602. doi: 10.1378/chest.07-0095. [DOI] [PubMed] [Google Scholar]
- 25.Shih JY, Gow CH, Yu CJ, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer. 2006;118:963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Olivé I, Monsó E, Andreo F, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying EGFR mutations. Eur Respir J. 2010;35:391–395. doi: 10.1183/09031936.00028109. [DOI] [PubMed] [Google Scholar]
- 27.Smouse JH, Cibas ES, Jänne PA, et al. EGFR mutations are detected comparably in cytologic and surgical pathology specimens of nonsmall cell lung cancer. Cancer Cytopathol. 2009;117:67–72. doi: 10.1002/cncy.20011. [DOI] [PubMed] [Google Scholar]
- 28.Boldrini L, Gisfredi S, Ursino S, et al. Mutational analysis in cytological specimens of advanced lung adenocarcinoma: A sensitive method for molecular diagnosis. J Thorac Oncol. 2007;2:1086–1090. doi: 10.1097/JTO.0b013e31815ba1fa. [DOI] [PubMed] [Google Scholar]
- 29.Nomoto K, Tsuta K, Takano T, et al. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006;126:608–615. doi: 10.1309/N5PQNGW2QKMX09X7. [DOI] [PubMed] [Google Scholar]
- 30.Smith GD, Chadwick BE, Willmore-Payne C, et al. Detection of epidermal growth factor receptor gene mutations in cytology specimens from patients with non-small cell lung cancer utilising high-resolution melting amplicon analysis. J Clin Pathol. 2008;61:487–493. doi: 10.1136/jcp.2007.051425. [DOI] [PubMed] [Google Scholar]
- 31.Fukui T, Ohe Y, Tsuta K, et al. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res. 2008;14:4751–4757. doi: 10.1158/1078-0432.CCR-07-5207. [DOI] [PubMed] [Google Scholar]
- 32.Savic S, Tapia C, Grilli B, et al. Comprehensive epidermal growth factor receptor gene analysis from cytological specimens of non-small-cell lung cancers. Br J Cancer. 2008;98:154–160. doi: 10.1038/sj.bjc.6604142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshita F, Matsukuma S, Yoshihara M, et al. Novel heteroduplex method using small cytology specimens with a remarkably high success rate for analysing EGFR gene mutations with a significant correlation to gefitinib efficacy in non-small-cell lung cancer. Br J Cancer. 2006;95:1070–1075. doi: 10.1038/sj.bjc.6603396. [DOI] [PMC free article] [PubMed] [Google Scholar]