This first part of a four-part series on pharmacogenetics describes the functional impact of genetic polymorphism and provides a general background to and insight into possible clinical consequences of pharmacogenetic variability.
Keywords: Pharmacogenetics, Oncology, Anticancer drugs, Genotyping technologies
Learning Objectives
After completing this course, the reader will be able to:
Differentiate the candidate gene and genome-wide approaches to pharmacogenetic research and the impact of each on clinical study results.
Describe the clinical implications of pharmacogenetic variability and its potential role in individualized treatment of patients with cancer.
This article is available for continuing medical education credit at CME.TheOncologist.com
Abstract
Equivalent drug doses may lead to wide interpatient variability with regard to drug response, reflected by differences in drug activity and normal tissue toxicity. A major factor responsible for this variability is variation among patients in their genetic constitution. Genetic polymorphism may affect the activity of proteins encoded, which in turn may lead to changes in the pharmacokinetic and pharmacodynamic behavior of a drug, observed as differences in drug transport, drug metabolism, and pharmacodynamic drug effects. Recent insights into the functional effect of polymorphism in genes that are involved in the pharmacokinetics and pharmacodynamics of anticancer drugs have provided opportunities for patient-tailored therapy in oncology. Individualized pharmacotherapy based on genotype will help to increase treatment efficacy while reducing unnecessary toxicity, especially of drugs characterized by a narrow therapeutic window, such as anticancer drugs.
We provide a series of four reviews aimed at implementing pharmacogenetic-based drug and dose prescription in the daily clinical setting for the practicing oncologist. This first part in the series describes the functional impact of genetic polymorphism and provides a general background to and insight into possible clinical consequences of pharmacogenetic variability. It also discusses different methodologies for clinical pharmacogenetic studies and provides a concise overview about the different laboratory technologies for genetic mutation analysis that are currently widely applied. Subsequently, pharmacogenetic association studies in anticancer drug transport, phase I and II drug metabolism, and pharmacodynamic drug effects are discussed in the rest of the series. Opportunities for patient-tailored pharmacotherapy are highlighted.
Introduction to the Series
We provide a series of four reviews about pharmacogenetic variability in anticancer phase I and II drug metabolism, drug transport, and pharmacodynamic drug effects. In this series, opportunities for patient-tailored pharmacotherapy are provided, based on the current knowledge in the field of pharmacogenetics in oncology. This first of four reviews provides a general background on pharmacogenetics and discusses frequently applied methodologies and technologies in pharmacogenetic research.
Introduction to Interindividual Variability
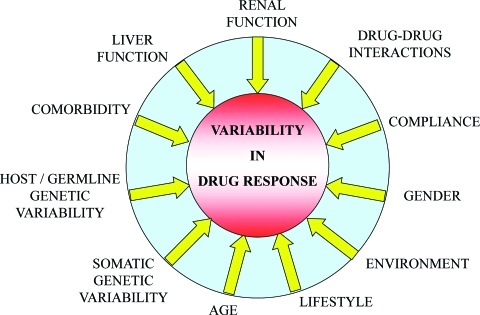
There is wide interpatient variability in the dose–effect relationship of (chemotherapeutic) drugs; some patients respond well to treatment and others do not, and the nature and severity of adverse events also shows wide variations among patients. Several host-related factors have evolved over time as determinants affecting anticancer drug treatment outcome such as age, gender, renal and liver function, concomitant medication leading to drug–drug interactions, (co-)morbidity, compliance, environment, and lifestyle (Fig. 1), of which compliance may be less relevant with (i.v. administered) chemotherapeutic drugs but may be more relevant in other areas such as, for example, with antibiotics or anti-HIV therapy. To correct for differences among subjects in drug response, dosing of selected drugs in general clinical practice is roughly divided into three age groups, that is, children (up to 16 or 18 years), adults, and the elderly (aged ≥65 years). The dose of most anticancer drugs is generally based on the individual's body weight or body surface area, but renal and liver function are also taken into account (e.g., with carboplatin) or the dose may be adapted according to plasma drug levels (e.g., with imatinib).
Figure 1.
Possible sources for interindividual variability in drug response. Besides genetic polymorphism, various additional nongenetic factors may contribute to interindividual differences in drug response.
Other sources of interpatient variability in drug response are interindividual differences in pharmacokinetics (PK), that is, drug absorption, distribution, metabolism, and elimination, and in pharmacodynamics (PD), that is, effects on drug receptors and other drug targets. Variations in the genetic constitution of genes that encode proteins involved in the PK and PD of a drug thereby significantly contribute to individual differences in drug response. Among the various biological mechanisms for genetic variability are differences in transcription factor activity, gene expression, gene silencing (epigenetics), and genetic polymorphism. Genetic polymorphisms are DNA sequence alterations consisting of single nucleotide polymorphisms (SNPs), mutations, deletions, insertions, and gene copy number variations. All types of DNA sequence alterations may lead to changes in protein structure or stability, and hence protein activity (discussed further below). Whether or not genetic variability then affects treatment outcome depends on, among other things, the functional impact of the genetic polymorphism on protein activity, and also on the relevance of a gene in the drug's pharmacological pathway and the possibility of escape pathways for drug elimination.
Types of Genetic Variability
DNA is subject to genetic polymorphism, which occurs genomewide, on average, every 1,000 bp. SNP is by far the most common genetic alteration, but small insertions, deletions, and even complete gene deletions and multiple gene copy number variants also exist [1–3]. A genetic polymorphism is defined as a minor allelic variant present in >1% of the population, otherwise it is referred to as a mutation [4]. A polymorphism is a neutral variation and can therefore be maintained in the population.
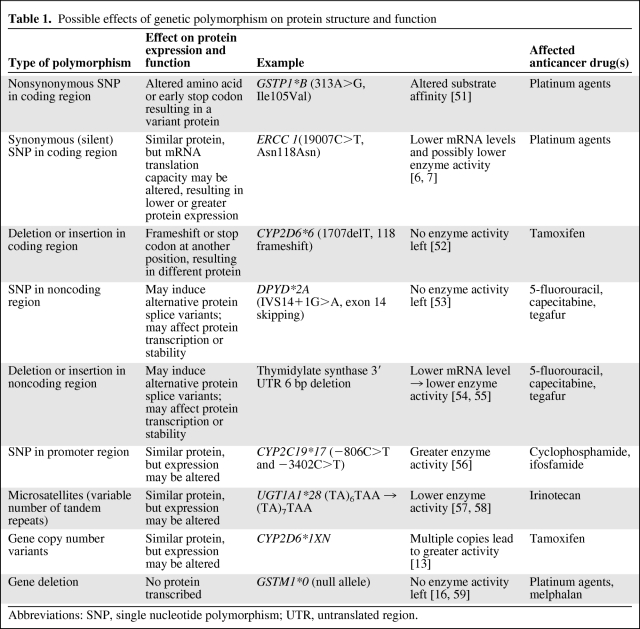
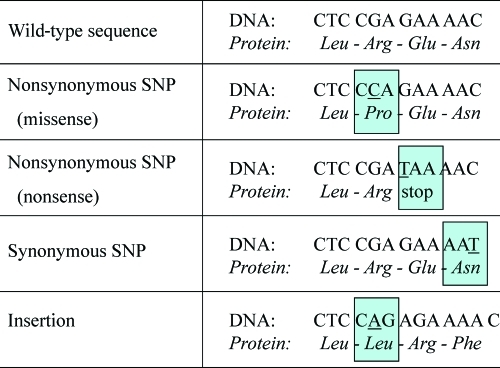
All types of genetic variability have the potential to affect protein function and activity, but how they exert their functional effect varies by type of genetic variant (Table 1). First, the functional effect of an allelic variant depends on the locus in which the genetic defect resides. SNPs, base pair deletions, and insertions may occur in coding and noncoding regions, that is, exons and introns, respectively. Exonic variants may elicit altered protein structures as a result of either substitution of an amino acid, introduction of an early stop codon, creation of an alternative splice variant, or alteration of the reading frame resulting from a frameshift (Fig. 2). Amino acid substitution is probably the most well-known effect of genetic polymorphism. Depending on the functional role of an amino acid in the protein, substitution of it may have a smaller or larger effect on the final protein activity. An example of an amino acid substitution is the isoleucine to valine substitution at position 105 (Ile105Val; 313A>G) within glutathione S-transferase P1 (GSTP1); in patients with the variant allele, GSTP1 enzyme activity is lower (Table 1). Frameshifts can be caused only by base pair deletions or insertions; for example, a deletion of 1 bp shifts the transcription of the DNA sequence. Subsequent translation produces different amino acids from that point onward (Fig. 2). The deletion of the thymine base pair at position 1707 in CYP2D6 (CYP2D6*6) is an example of a frameshift mutation that creates a protein with absent enzyme activity (Table 1).
Table 1.
Possible effects of genetic polymorphism on protein structure and function
Abbreviations: SNP, single nucleotide polymorphism; UTR, untranslated region.
Figure 2.
Effects of genetic polymorphism on the encoded protein. Dependent on its type and physical location, a genetic polymorphism may elicit changes to the primary amino acid sequence of a protein in various ways.
Abbreviation: SNP, single nucleotide polymorphism.
Although most intronic mutations have no functional effect, they can create alternative splice variants that may drastically affect protein activity, such as, for example, the IVS14+1G>A polymorphism in the gene encoding the 5-fluorouracil detoxifying enzyme dihydropyrimidine dehydrogenase. This polymorphism results in skipping of exon 14 in the translation process, and thereby in absent enzyme activity (Table 1).
A subtype and often overlooked type of polymorphism is the synonymous SNP, also termed silent polymorphism (Fig. 2). Silent polymorphisms are exonic SNPs that encode the same amino acid and therefore do not influence the primary structure of the protein. Therefore, they are often considered as irrelevant SNPs. However, at least three possible mechanisms are reported by which silent polymorphism may lead to differences in protein activity: (a) by influencing mRNA stability and structure, (b) by differences in the kinetics of translation because the codon has changed, and (c) by alternate splicing [5]. An example of a silent SNP is the 118C>T polymorphism in excision repair cross-complementing group 1, a protein involved in DNA repair. Although the wild-type (AAC) and variant (AAT) allele codons both encode the amino acid asparagine, the variant allele is associated with a 50% lower transcription level and lower mRNA levels [6, 7]. Furthermore, this polymorphism has been associated with altered clinical outcome in patients treated with platinum-based chemotherapy [8–10].
In the case of genetic polymorphism in the promoter region or in the 3′ untranslated region (3′UTR) or 5′UTR of a gene, the primary amino acid sequence of the protein also is not altered. However, protein activity may be significantly affected through altered ability or altered kinetics in protein transcription and translation. One type of polymorphism that often occurs in promoter regions is a microsatellite, also known as a variable number of tandem repeats (VNTR). These are repeats of short base pair sequences in which the number of repeats may vary among individuals. One example of a VNTR is the UGT1A1*28 polymorphism in the promoter region of UGT1A1. UGT1A1*28 shows either six or seven repeats of the nucleotide sequence TA, in which the glucuronidation activity of UGT1A1 inversely relates to the number of TA repeats.
Another type of genetic variability that contributes to various phenotypes is gene copy number variants (CNVs) [11, 12]. CNVs leave the primary amino acid sequence unchanged, but when multiple gene copies are present, the protein activity generally increases. An example of a CNV is the gene duplication of CYP2D6 (CYP2D6*1XN/CYP2D6*2XN), which subsequently result in the cytochrome P450 (CYP)2D6 ultrarapid metabolizer phenotype [13, 14]. Finally, entire gene deletions may occur. As a consequence, genes are not transcribed, thereby resulting in absent protein activity. Known frequently occurring gene deletions exist, for example, for the glutathione S-transferases GSTT1 and GSTM1 [15, 16].
Differences Between Somatic and Germline DNA
DNA analysis for pharmacogenetic purposes is usually performed with germline DNA. However, for anticancer therapy, DNA is also analyzed in tumor tissue, so-called somatic mutation analysis. The major difference between germline and somatic polymorphism is that germline polymorphism is inherited and transmits to offspring, whereas somatic polymorphism does not. The concordance rate between germline and somatic DNA may be high; this differs, however, by gene and by individual and is therefore not always extrapolatable [17]. Analysis of germline DNA in pharmacogenetics is very suitable for both PK and PD association analyses. However, in oncology, analysis of tumor tissue (somatic DNA) is especially attractive when evaluating PD effects, such as tumor response [18]. For example, somatic activating mutations in KRAS are significantly associated with the likelihood of nonresponse to monoclonal antibodies targeting the epidermal growth factor receptor (EGFR): patients who bear wild-type KRAS tumors are almost exclusively likely to respond to EGFR-targeted therapy with cetuximab or panitumumab, whereas patients with mutated KRAS tumors are significantly less likely to respond [19, 20].
Epigenetics
Another type of inherited gene transcription regulation that differs among individuals is epigenetics. Epigenetic variability does not depend on differences in the primary amino acid sequence but depends on so-called gene silencing. This is, among other things, induced by methylation of the promoter region [21, 22]. Methylation mostly occurs on so-called CpG islands, which are typically prevalent in the promoter region of genes. A CpG site is a DNA region where a cytosine nucleotide lies adjacent to a guanine, separated by a phosphate linking these two nucleosides. If CpG islands are methylated, gene expression decreases and protein activity is thereby reduced.
Adoption of Pharmacogenetics in the Clinic
Pharmacogenetic studies in clinical oncology typically analyze the relationship between genetic polymorphism and drug-related toxicity, treatment response, and survival with the (chemo)therapeutic treatment. Thereby, knowledge of the clinical impact of genetic variants may then enable patient-tailored pharmacotherapy [23]. An example of how this knowledge could be applied in clinical practice is a guideline that was developed with regard to CYP2D6 drug substrates [24]. Herein, patients are categorized as either poor, intermediate, or ultrarapid metabolizers based on their CYP2D6 genotype. Subsequently, therapeutic (dose) recommendations are provided for the individual categories for a variety of CYP2D6 substrate drugs. However, the use of pharmacogenetics in clinical practice to date, that is, genotype-based individualized drug and dose prescription, is still very limited despite the fact that thousands of pharmacogenetic association studies have been performed to date. There are only a few centers worldwide that prospectively screen, for example, for CYP2D6 variants, and make a clinical decision based on the genotype. This is partly a result of the fact that, although there may be variants that are predictive of clinical outcome, there are also genetic polymorphisms that have shown nonsignificant or even nonconsistent associations among various clinical trials. For example, contradictory results have been published for the polymorphism CYP2D6*4 in patients with breast cancer given tamoxifen [25–27]. The relationship between CYP2D6 polymorphism and tamoxifen treatment outcome is extensively reviewed in the next part of this series [28]. To demonstrate how nonconsistency in results of comparable pharmacogenetic studies may arise, it is crucial to understand the methodology of pharmacogenetic research. There are two main approaches that can be distinguished: the candidate gene approach and the genome-wide approach.
The Candidate Gene Approach
In the candidate gene approach, only a limited number of polymorphisms, which mostly reside in genes involved in the PK and PD of a drug, are associated with clinical outcome. Candidate genes are, in advance, considered to be related to the pharmacology of the drug, and these studies are therefore also termed hypothesis-driven association studies. Typical candidate genes encode, for example, drug transporters, biotransformation enzymes, or drug receptors. This is a very reasonable approach; however, thus far only a small percentage of all tested genetic variants have been identified as significant predictors of treatment outcome. A classical example of a clinically relevant candidate gene is TPMT, the gene that encodes thiopurine S-methyltransferase (TMPT). TPMT catalyzes the S-methylation of 6-mercaptopurine (6-MP) into inactive metabolites [29]. A strong genotype–phenotype relationship exists between three polymorphisms in TPMT and TPMT enzyme activity. About 80%–95% of patients with low TPMT enzyme activity are explained by the presence of TPMT*2, TPMT*3A, and TMPT*3C [30–35]. Hetero- and homozygous variant allele carriers for these SNPs present with the intermediate and poor metabolizer phenotypes, respectively. Indeed, dose reductions of mercaptopurine of up to 50% are indicated in heterozygous polymorphic carriers, and reductions up to 90% are indicated in homozygous polymorphic carriers [36–38].
Obviously however, in many cases, one single genetic trait does not sufficiently explain the wide interindividual differences in drug response. This is partially a result of the fact that the pharmacological pathway of a drug is very complex, with many PK and PD proteins involved. Differences in response to (anticancer) drugs are mostly polygenetic traits. For example, cyclophosphamide is extensively metabolized by various CYP enzymes, including CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, and CYP3A4. Subsequently, genetic deficiency of CYP2A6 will most likely not significantly influence the PK of or treatment outcome with cyclophosphamide. In addition, on the PD level, the combined activity of multiple proteins together, such as receptors and signal transduction pathways, determines the response to a drug. Moreover, in the case of chemotherapeutics, specific tumor-related proteins also are involved. Because of this complexity, the effect of a single genetic alteration is mostly not sufficiently predictive of treatment response to a drug. Namely, genetic variability in additional genes involved in the pharmacology of a drug affects treatment outcome as well, but other nongenetic factors also add to predictability [39–41].
Therefore, well-defined (prospective) clinical trials are required to determine whether genetic polymorphisms are possibly clinically relevant. Study populations need to be adequately powered to demonstrate any possible relationships, if they do exist. The population size should preferably consist of up to hundreds or even thousands of patients, but this depends, among other things, on the prevalence of the investigated polymorphism(s) and the type of parameter (e.g., toxicity or survival) that the genetic variants are associated with. To conclude, the candidate gene approach enables identification of predictive and clinically relevant polymorphisms. However, by far, not all polymorphisms have been shown to be predictive. If inconsistent associations for a genetic variant are observed, possible combination with additional polymorphisms in one or more genes might increase the predictive value for clinical outcome.
The Genomewide Approach
In contrast to the candidate gene approach, in which only a limited number of polymorphisms is tested, the genomewide approach analyzes multiple polymorphisms (mostly SNPs) across the entire human genome. Therefore, it is independent of whether or not a gene is a priori expected to be involved in the pharmacological pathway of a drug. This approach requires high-throughput genotyping technologies that are able to analyze multiple SNPs simultaneously. The number of SNPs on the array may range from a few hundred to even hundreds of thousands. These SNPs are mostly common SNPs, with a population prevalence >5%–10%, and are present throughout the whole genome. In this way, every gene is covered by several SNPs. Because up to hundreds of thousands of SNPs are analyzed, genomewide association study (GWAS) requires the use of advanced bioinformatics to handle the extensive amount of data. In addition, because of multiple comparisons within a GWAS, the data need to be corrected for multiple testing [42]. When hundreds of thousands of comparisons are performed, the p-value to reach significance may become too stringent; this is a common phenomenon in GWAS and is an important cause for replication failure.
GWAS is, however, not always genomewide. Recently, an intermediate approach in pharmacogenetic studies was introduced that uses the so-called DMET (drug metabolism enzymes and transporters) array. This array covers up to 2,000 polymorphisms within 225 genes involved in drug transport and drug metabolism. By using this reduced gene subset, compared with a larger GWAS, the need to correct for many irrelevant genes in a drug's pharmacology is eliminated, and the likelihood of obtaining false positives is thereby reduced [43].
The general methodology of GWAS is a case–control design. The case group consists of patients with a well-defined response after treatment with a specific drug. The control group is either a similar patient cohort given the same drug who did not develop that specific response or, otherwise, the control group is randomly selected from the population. The sizes of the case and control groups are mostly tens of to up to a few hundred patients. Most discriminating SNPs between cases and controls may indicate a possible relevant role for these genes in the treatment with that drug. However, because of the high number of association tests typically performed in a GWAS, positive findings always need to be confirmed in independent populations [44].
The genomewide approach differs from the candidate gene approach in that it is not hypothesis driven and it does not make use of the current knowledge about a drug's mechanism of action. Thereby, it is capable of identifying genes that were previously unknown to be of relevance. On the other hand, GWAS methodology also has a couple of disadvantages, such as high costs and the inclusion of selection bias in case and control selection. Furthermore, there is a relatively high risk (even after correction for multiple comparisons) for gaining false-positive and false-negative results because of the high number of SNPs analyzed. In addition, GWAS lacks sensitivity for rare genetic variants that are usually not covered using these types of assays [44]. A possibility for overcoming the noncoverage of rare variants is to resequence the specific genomic regions of interest to identify the causal variant allele that was associated with a certain phenotype.
The GWAS number has increased over the last years. One example of such a GWAS is a study conducted by Yang et al. [45], in which the association between >400,000 polymorphisms and treatment response in childhood acute lymphoblastic leukemia was analyzed.
GWAS is nowadays frequently applied in pharmacogenetics, but the primary area in which this methodology was used was in research in disease susceptibility. Disease susceptibility studies are focused on (genomewide) genetic differences in the prevalence of SNPs between a patient cohort with a specific disease entity and a healthy control group. The methodology for disease genetics studies uses similar genomewide screening technologies for polymorphism detection to that used in pharmacogenetic genomewide studies. Because polymorphisms can induce changes in protein activity and thereby affect human (patho)physiology, differences in the genetic constitution between a diseased and nondiseased population might identify loci that are possibly involved in the development of that disease [46–48]. Thereby, this may lead to a better understanding of the mechanism of disease and, additionally, identify new possible targets for drug development [49]. For example, a recent GWAS in disease genetics showed that several loci were associated with the risk for colorectal cancer; furthermore, these genes appeared to be especially related to mitogen-activated protein kinase signaling pathways [50].
Genotyping Technologies
A prerequisite for the routine application of pharmacogenetics in daily clinical practice is that reliable genotyping assays be available for the practicing clinician. The simplicity, sensitivity, costs, robustness, specificity, throughput (i.e., the number of reactions that can be simultaneously performed), and turnaround time of the assay are key elements for introducing pharmacogenetics successfully into the clinic. The molecular background and clinical applications of current commonly applied DNA genotyping technologies is described in detail in supplemental online data.
Conclusions and Future Perspectives
Genetic polymorphism is a frequently occurring phenomenon that is prevalent throughout the whole genome. DNA alterations may affect protein transcription, translation, and stability, which can have serious consequences for the activity of encoded proteins. As a consequence, genetic variability in genes that interact with the PK and PD of a drug may contribute to interindividual differences in drug response.
The study of pharmacogenetics is aimed at elucidating the functional and clinical impacts of genetic polymorphism. Implementation of this knowledge in clinical practice allows genotype-based drug and dose prescription for the individual patient. This enables safer and possibly more effective (chemotherapeutic) therapy. Using the candidate gene approach, a series of clinically relevant loci and allelic variants have been identified. However, results of pharmacogenetic trials have not always shown clear associations with clinical outcome. This is partially explained by differences in study design, patient selection, and treatment regimen. Most importantly, however, these observations demonstrate that variation in response to a drug does not solely rely on a few polymorphisms in genes that encode for PK- or PD-related proteins, but in fact is much more complex—a multigenetic trait. Future studies using combined predictive models including multiple genetic variants plus nongenetic factors will probably lead to clearer and additional associations with drug response. Furthermore, GWAS is becoming more common, which has the power to identify loci that were previously unknown to affect drug treatment outcome.
Besides knowledge of the functional impact of genetic polymorphism, implementation of clinical pharmacogenetics will also be boosted by the availability of rapid, robust, high-throughput, sensitive, and specific genotyping technologies. One of the various existing genotyping technologies can be chosen depending on the intended clinical application. For example, retrospective genotyping studies may suffice, with a sensitivity of a little <100% (but preferably >90%), whereas assays used for prospective pharmacogenetic testing in personalized medicine should be up to 100% specific and sensitive. This will enable the clinician to use pharmacogenetics as a tool for patient-tailored pharmacotherapy.
Furthermore, cost-effectiveness is an important factor that may determine whether genotype-based pharmacotherapy can become a standard of care in drug treatment. For treatment with highly expensive (chemotherapeutic) drugs, in particular, such as monoclonal antibodies, genotype-based selection of patients for whom the drug is most likely to be effective could prevent unnecessary toxicity and high costs. Indeed, for chemotherapeutic treatment, it has been shown that genotype-based drug and patient selection is possible and individualized pharmacotherapy is possible, as discussed in the following three parts of this series. This leads to less severe side effects and greater treatment benefits in subgroups of patients who can be selected using pharmacogenetic approaches.
Supplementary Material
Author Contributions
Conception/Design: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Collection and/or assembly of data: Maarten J. Deenen
Data analysis and interpretation: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Manuscript writing: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Final approval of manuscript: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
References
- 1.Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 2.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 3.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 4.Nebert DW. Suggestions for the nomenclature of human alleles: Relevance to ecogenetics, pharmacogenetics and molecular epidemiology. Pharmacogenetics. 2000;10:279–290. doi: 10.1097/00008571-200006000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, et al. Silent polymorphisms speak: How they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 6.Yu JJ, Lee KB, Mu C, et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 7.Yu JJ, Mu C, Lee KB, et al. A nucleotide polymorphism in ERCC1 in human ovarian cancer cell lines and tumor tissues. Mutat Res. 1997;382:13–20. doi: 10.1016/s1383-5726(97)00004-6. [DOI] [PubMed] [Google Scholar]
- 8.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 9.Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol. 2007;25:1247–1254. doi: 10.1200/JCO.2006.08.1844. [DOI] [PubMed] [Google Scholar]
- 10.Ryu JS, Hong YC, Han HS, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl ML, Johansson I, Bertilsson L, et al. Ultrarapid hydroxylation of debrisoquine in a Swedish population. Analysis of the molecular genetic basis. J Pharmacol Exp Ther. 1995;274:516–520. [PubMed] [Google Scholar]
- 14.Johansson I, Lundqvist E, Bertilsson L, et al. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci U S A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidegård J, Vorachek WR, Pero RW, et al. Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci U S A. 1988;85:7293–7297. doi: 10.1073/pnas.85.19.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh S, Mallon MA, Goodfellow P, et al. Concordance of pharmacogenetic markers in germline and colorectal tumor DNA. Pharmacogenomics. 2005;6:873–877. doi: 10.2217/14622416.6.8.873. [DOI] [PubMed] [Google Scholar]
- 18.Marsh S. Pharmacogenomics. Ann Oncol. 2007;18(suppl 9):ix24–ix28. doi: 10.1093/annonc/mdm289. [DOI] [PubMed] [Google Scholar]
- 19.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 20.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 22.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 23.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 24.Swen JJ, Wilting I, de Goede AL, et al. Pharmacogenetics: From bench to byte. Clin Pharmacol Ther. 2008;83:781–787. doi: 10.1038/sj.clpt.6100507. [DOI] [PubMed] [Google Scholar]
- 25.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 26.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 27.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deenen MJ, Cats A, Beijnen JH, et al. Pharmacogenetic variability in drug transport and phase I anticancer drug metabolism. The Oncologist. 2011;16:820–834. doi: 10.1634/theoncologist.2010-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennard L. The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol. 1992;43:329–339. doi: 10.1007/BF02220605. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner SJ, Begg EJ, Barclay ML, et al. Genetic polymorphism and outcomes with azathioprine and 6-mercaptopurine. Adverse Drug React Toxicol Rev. 2000;19:293–312. [PubMed] [Google Scholar]
- 31.Otterness D, Szumlanski C, Lennard L, et al. Human thiopurine methyltransferase pharmacogenetics: Gene sequence polymorphisms. Clin Pharmacol Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 32.Spire-Vayron de la Moureyre, Debuysère H, Sabbagh N, et al. Detection of known and new mutations in the thiopurine S-methyltransferase gene by single-strand conformation polymorphism analysis. Hum Mutat. 1998;12:177–185. doi: 10.1002/(SICI)1098-1004(1998)12:3<177::AID-HUMU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 33.Tai HL, Krynetski EY, Yates CR, et al. Thiopurine S-methyltransferase deficiency: Two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 34.Tai HL, Krynetski EY, Schuetz EG, et al. Enhanced proteolysis of thiopurine S-methyltransferase (TPMT) encoded by mutant alleles in humans (TPMT*3A, TPMT*2): Mechanisms for the genetic polymorphism of TPMT activity. Proc Natl Acad Sci U S A. 1997;94:6444–6449. doi: 10.1073/pnas.94.12.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates CR, Krynetski EY, Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: Genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 36.McLeod HL, Coulthard S, Thomas AE, et al. Analysis of thiopurine methyltransferase variant alleles in childhood acute lymphoblastic leukaemia. Br J Haematol. 1999;105:696–700. doi: 10.1046/j.1365-2141.1999.01416.x. [DOI] [PubMed] [Google Scholar]
- 37.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 38.Stanulla M, Schaeffeler E, Flohr T, et al. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. JAMA. 2005;293:1485–1489. doi: 10.1001/jama.293.12.1485. [DOI] [PubMed] [Google Scholar]
- 39.Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: Polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–920. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- 40.Efferth T, Volm M. Pharmacogenetics for individualized cancer chemotherapy. Pharmacol Ther. 2005;107:155–176. doi: 10.1016/j.pharmthera.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Hoehe MR, Timmermann B, Lehrach H. Human inter-individual DNA sequence variation in candidate genes, drug targets, the importance of haplotypes and pharmacogenomics. Curr Pharm Biotechnol. 2003;4:351–378. doi: 10.2174/1389201033377300. [DOI] [PubMed] [Google Scholar]
- 42.Johnson RC, Nelson GW, Troyer JL, et al. Accounting for multiple comparisons in a genome-wide association study (GWAS) BMC Genomics. 2010;11:724. doi: 10.1186/1471-2164-11-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burmester JK, Sedova M, Shapero MH, et al. DMET microarray technology for pharmacogenetic-based personalized medicine. Methods Mol Biol. 2010;632:99–124. doi: 10.1007/978-1-60761-663-4_7. [DOI] [PubMed] [Google Scholar]
- 44.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 45.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukaemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemer M. Genetic insights into normal and abnormal heart development. Cardiovasc Pathol. 2008;17:48–54. doi: 10.1016/j.carpath.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Rosner S, Giladi N, Orr-Urtreger A. Advances in the genetics of Parkinson's disease. Acta Pharmacol Sin. 2008;29:21–34. doi: 10.1111/j.1745-7254.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 48.El Omar EM, Ng MT, Hold GL. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene. 2008;27:244–252. doi: 10.1038/sj.onc.1210912. [DOI] [PubMed] [Google Scholar]
- 49.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 50.Lascorz J, Försti A, Chen B, et al. Genome-wide association study for colorectal cancer identifies risk polymorphisms in German familial cases and implicates MAPK signalling pathways in disease susceptibility. Carcinogenesis. 2010;31:1612–1619. doi: 10.1093/carcin/bgq146. [DOI] [PubMed] [Google Scholar]
- 51.Watson MA, Stewart RK, Smith GB, et al. Human glutathione S-transferase P1 polymorphisms: Relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 52.Saxena R, Shaw GL, Relling MV, et al. Identification of a new variant CYP2D6 allele with a single base deletion in exon 3 and its association with the poor metabolizer phenotype. Hum Mol Genet. 1994;3:923–926. doi: 10.1093/hmg/3.6.923. [DOI] [PubMed] [Google Scholar]
- 53.Meinsma R, Fernandez-Salguero P, Van Kuilenburg AB, et al. Human polymorphism in drug metabolism: Mutation in the dihydropyrimidine dehy-drogenase gene results in exon skipping and thymine uracilurea. DNA Cell Biol. 1995;14:1–6. doi: 10.1089/dna.1995.14.1. [DOI] [PubMed] [Google Scholar]
- 54.Mandola MV, Stoehlmacher J, Zhang W, et al. A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics. 2004;14:319–327. doi: 10.1097/00008571-200405000-00007. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich CM, Bigler J, Velicer CM, et al. Searching expressed sequence tag databases: Discovery and confirmation of a common polymorphism in the thymidylate synthase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:1381–1385. [PubMed] [Google Scholar]
- 56.Sim SC, Risinger C, Dahl ML, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: A balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995;333:1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 59.Abdel-Rahman SZ, el Zein RA, Anwar WA, et al. A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies. Cancer Lett. 1996;107:229–233. doi: 10.1016/0304-3835(96)04832-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.