Abstract
Objective
To study the effect of the innate cytokine, MIF, on the susceptibility and the severity of SLE in a multinational population of Caucasian and African-American patients.
Methods
We studied the association between two functional polymorphisms in the MIF gene: a −794 CATT5-8 microsatellite repeat (rs5844572) and a −173 G/C SNP (rs755622), with SLE in 3195 patients and controls. We also measured MIF plasma levels in relation to genotypes, clinical phenotypes, and TLR 7-stimulated MIF production in vitro.
Results
Both Caucasians and African-Americans with the high expression, −794 CATT7/173*C haplotype had lower SLE incidence (OR 0.63 [0.53, 0.89], p=0.001 in Caucasians, and OR 0.46 [0.23, 0.95], p=0.012 in African-Americans). By contrast, among patients with established SLE, those with nephritis, serositis, and CNS involvement had reduced frequencies of low expression MIF genotypes (−794 CATT5) when compared to patients without end-organ involvement (p=0.005 for serositis, p=0.023 for nephritis, and p=0.04 for CNS involvement). Plasma MIF levels and TLR7 stimulated MIF production in vitro reflected the underlying MIF genotype of the studied groups.
Conclusion
These data suggest that MIF, which has both pro-inflammatory properties and macrophage and B cell survival functions, exerts a dual influence on the immunopathogenesis of SLE. High expression MIF genotypes are associated with a reduced susceptibility to SLE and may contribute to an enhanced clearance of infectious pathogens. Once SLE develops however, low expression MIF genotypes may protect from ensuing, inflammatory end-organ damage.
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune inflammatory disease with a multifactorial etiology (1). Several observations support the important role of both environmental factors and susceptibility genes in disease pathogenesis. The contribution of a genetic etiology to SLE is suggested by a concordance rate of approximately 30% in monozygotic twins (2). In addition, there is a strong association between SLE incidence and particular HLA loci (3) and several non-HLA genes (4). Studies to date have not fully accounted for the total genetic contribution to SLE however, and most studies have been limited to Caucasian patients.
The cytokine, Macrophage Migration Inhibitory Factor (MIF), is appreciated to play a central regulatory role in innate immunity and in the differentiation of the adaptive response. MIF counter-regulates the immunosuppressive actions of glucocorticoids and it promotes TNFα production, leading to further MIF release and a re-entrant activation response that supports the maximal expression of pro-inflammatory mediators, matrix-degrading enzymes, and cyclooxygenases (5;6). There also is evidence for a protective effect of genetic deletion or neutralization of MIF in spontaneous mouse models of lupus (7;8).
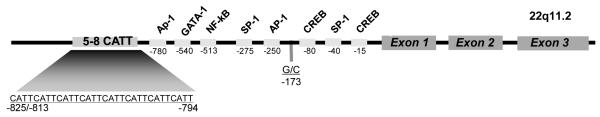
Both the circulating level and the tissue expression of MIF are elevated during inflammation, and recent data indicate that functional polymorphisms in the MIF gene influence the clinical expression of autoimmune or infectious diseases (9;10). Two polymorphisms have been identified in the MIF gene promoter: the first is a microsatellite repeat at position −794 base-pairs upstream of the MIF gene (−794 CATT5-8) where repeat length correlates with gene expression, and the second is a single nucleotide polymorphism (SNP) at position −173 base-pairs upstream of the MIF gene, where the −173*C allele is associated with higher circulating MIF levels (Figure 1). These two polymorphisms have been associated in different populations with increased clinical severity or disease susceptibility in rheumatoid and juvenile idiopathic arthritis (11;12), sarcoidosis (13), scleroderma (14), asthma (15), and inflammatory bowel disease (16).
Figure 1.
Diagram of the human MIF gene showing its three exons, predicted transcription factor binding sites, and the −173 G/C SNP (rs 755622) and −794 CATT5-8 (rs 5844572) polymorphisms. The numerical prefixes refer to nucleotide distance upstream from the transcription start site. The −173C allele, −794 CATT7 and −794 CATT 8 repeats, and −794 CATT7/−173C haplotype are associated with higher expression of MIF.
To determine the potential relationship between MIF gene polymorphisms and the systemic autoimmune disease, SLE, we conducted a multicenter observational study of 1369 SLE patients. We analyzed MIF genotype in relation to disease incidence and clinical phenotype. We examined circulating MIF and TNFα proteins in a subset of patients for their correlation with MIF genotype, and we also analyzed MIF production from peripheral blood mononuclear cells of known genotype in response to TLR7 agonism.
MATERIALS AND METHODS
Patients
Our study consisted of 1369 SLE patients and 1826 healthy controls. Patients were included in the study if they fulfilled 4 of the 11 American College of Rheumatology (ACR) revised criteria for SLE (17). Patient and control subjects were recruited locally from the United States, Sweden, and Argentina. Participants in the US were included from four academic centers: Oklahoma University (Oklahoma City, OK), The Hospital for Special Surgery (New York, NY), Upstate Medical University (Syracuse, NY), and Yale University (New Haven, CT). Participants in Sweden were recruited from Karolinska University Hospital (Stockholm, Sweden). In Argentina, patients were included from Sanatorio Parque (Rosario, Argentina). Cases and controls were divided into 2 different ethnic groups: Caucasians and African-Americans. Other ethnicities were excluded from analysis because of their relative small sample numbers. Ethnicity was determined by self-report, and in the University of Oklahoma cohort, verified by admixture analysis. The studied clinical manifestations included articular involvement, muco-cutaneous lesions or photosensitivity, renal disease, serositis, hematopoietic abnormalities, and CNS involvement. Patients were considered to have renal disease if they had one or more of the following: active urinary sediment, significant proteinuria, or histopathological evidence of lupus nephritis. Relevant Institutional Review Boards approved the study. All patients signed informed consents and the Health Insurance Portability and Accountability Act Notification where applicable.
Genotyping
Genomic DNA was isolated from serum using the easy-DNA kit (Invitrogen, Carlsbad, CA, USA). The two MIF promoter polymorphisms ( −794 CATT5-8 microsatellite repeat (rs5844572) and a −173 G/C SNP (rs755622)) were analyzed following the methodologies described previously by Wu et al.(14). We note that neither of these loci is represented in the high-density genotyping chips used previously for genome-wide association studies in SLE. Briefly, MIF −173 G/C genotyping was performed using a pre-developed TaqMan assay for allelic discrimination (Applied Biosystems, Foster City, CA, USA). The genotype of each sample was automatically attributed using fluorescence detection in an ABI Prism 7900HT instrument. Allelic discrimination was analyzed with SDS 2.1.1 software (Applied Biosystems). The MIF −794 CATT5-8 genotyping was carried out by PCR using a forward primer (5′-TGCAGGAACCAATACCCATAGG-3′) and a TET fluorescent reverse primer (TET lab5′-AATGGTAAACTCGGGGGAC-3′). Automated capillary electrophoresis on a DNA sequencer was performed on each sample and the CATT alleles were identified using Genotyper 3.7 software (Applied Biosystems).
Plasma MIF and TNFα levels
Plasma MIF levels were measured by sandwich enzyme-linked immunosorbant assay (ELISA) using specific antibodies (18). Samples were run in triplicates. TNFα levels were measured by the Ready-Set-Go ELISA kit (eBioscience, San Diego, CA, USA). Plasma MIF and TNFα levels were compared and culled from both Caucasian and African-American SLE patients and controls identified as having the high expression, −794 CATT7 /−173*C MIF haplotype (7C) or the low expression, −794 CATT5 /−173*G MIF haplotype (5G).
Peripheral Blood Leukocyte Studies
Human peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood using Ficoll-Hypaque gradient centrifugation. Cells were washed, re-suspended in RPMI 1640 medium supplemented with 10% FBS, and the mononuclear cells purified by adherence for 2 hrs (5 ×105 cells/well). Cells then were stimulated for 20 hrs with the TLR7 agonist polyU (Sigma-Aldrich, Saint Louis, MO, USA, 0.25 mg/ml) and the conditioned medium then assayed for MIF content by ELISA.
Statistical analysis
Given the population stratification of the MIF alleles under study (19), the study participants were subdivided by ethnic origin to allow appropriate comparison and analysis. The Pearson chi-square test was used to determine the homogeneity between the different Caucasian populations, and to analyze Hardy-Weinberg equilibrium. Allelic and genotypic frequencies were compared using chisquare test and (when appropriate) Fisher’s exact test for small numbers. Logistic regression was used to adjust for other covariates. Comparisons were first made between patients and controls, then between patients with and without end-organ involvement including nephritis, serositis, and CNS involvement. We compared high expression −794 CATT7 containing genotypes (−794 CATT5/7, −794 CATT6/7, −794 CATT7/7) and high expression −173*C containing genotypes (−173 C/C, −173 G/C) with low expression genotypes (non −794 CATT7 or −794 CATT5/5, −794 CATT5/6, −794 CATT6/6) and low expression −173 G/G genotypes among all groups mentioned above. The −794 CATT8 allele was found rarely (0.5%)(19), and was excluded from analysis. Studied haplotypes were reconstructed using the −173 G/C SNP and the −794 CATT5-8 repeat: 8 combinations are theoretically possible (5G, 6G, 7G, 8G, 5C, 6C, 7C, and 8C). Haplotype frequencies and their inferred probabilities were calculated using the HPlus 2.5 software (Quantitative Genetic Epidemiology Group, The Fred Hutchinson Cancer Research Center, Seattle, WA) (20). Both 8G and 8C haplotypes were not included because of their rare occurrence. The incidence of SLE associated with different MIF haplotypes as compared to 5G was calculated using logistic regression. Age, race, sex, geographic location, MIF polymorphisms, and ANA status were used as covariates. We also determined the relationship between these haplotypes and the incidence of different clinical manifestations including: nephritis (N=299), serositis (N=255), and CNS involvement (N=101). African-Americans were not included in the clinical manifestation analysis because of the small sample number of reported clinical phenotypes.
To address the issue of multiple testing, we used the permutation test in R (http://www.R-project.org/) (21). The permutation test was used in the instances where comparisons produced marginal p values.
To account for population stratification and its effect on genotype/haplotype differences, we used formulas established by Lee and Wang (22). The population stratification bias can be quantified by the confounding rate ratio (CRR). Through a mathematical calculation, Lee and Wang showed that CRR is always bound above by “U” and below by “1/U”. Where “U” is the highest odds ratio that can be attributed to the population stratification bias and “1/U” is the lowest odds ratio. “U” takes into account the highest and lowest reported prevalence (“G”) of a disease in a population affected by stratification and the highest and lowest frequency (“B”) of the genotype/ haplotype in question reported in the same population (22). If the observed odds ratio is above“U” or lower “1/U”, then these observed odds ratio cannot be explained simply by population stratification.
Power and Sample size calculation
In a study of inflammatory arthritis by Barton et al. , 438 patients and 343 controls were used to detect a 11.5% difference in −173C-containing genotypes (p=0.001), a 7.7% difference in CATT7-containing genotypes (p=0.001), and an 8.1% difference in 7C haplotypes (p=0.0001) (23). We have determined that 215, 365, and 149 patients were necessary to detect these differences respectively in a Caucasian population with a confidence interval of 95% and a power of 80%. Similarly, based on a study by Gao et al., we have determined that 129 African-American patients were needed to detect a difference of 11.1% in −173C containing genotypes with a confidence interval of 95% and a power of 80% (24). No prior studies have been conducted on CATT7 containing genotypes or haplotype differences in African-Americans. Statistical analyses were performed using SPSS v.16. Two-tailed p values ≤ 0.05 were considered statistically significant. Differences in circulating MIF and TNFα levels between groups were compared using the Student’s t test.
RESULTS
Patient Demographics
The demographic characteristics of the SLE patients and controls are shown in Table 1. There was no difference between the mean age of patients and that of controls (38.7 ± 12.6 years and 39.4 ± 12.4 years, p=0.447). Females constituted the majority of cases in both patient and control groups (89% and 86% respectively, p=0.108). The studied clinical manifestations included articular involvement (82%), muco-cutaneous lesions or photosensitivity (81%), renal disease (60%), serositis (48%), hematopoietic abnormalities (46%), and CNS involvement (19%).
Table 1.
Characteristics of SLE Patients and Healthy Controls.
| Patients | Controls | |
|---|---|---|
| Agea | 38.7 ± 12.6 (Mean ± SD) | 39.4 ± 12.4 (Mean ± SD) |
| Females | 1306 (89%) | 1569 (86%) |
| Caucasians | 1082 (74%) | 1439 (78%) |
| African-Americans | 180 (12%) | 180 (10%) |
| Arthralgia/Arthritisa | 598 (82%) | |
| Muco-cutaneous lesions/ Photosensitivitya | 590 (81%) | |
| Renal diseasea | 475 (60%) | |
| Serositisa | 380 (48%) | |
| Hematopoeitic abnormalitiesa | 256 (46%) | |
| CNS involvement/Neuropsychiatric disordersa | 153 (19%) | |
| ANA positive (titers ≥ 1:40)a | 658 (93%) | 7% |
| Anti-dsDNA positivea | 164 (75%) |
Missing data were not included in the calculation of the mean age, clinical manifestations, and the frequency of ANA and anti-dsDNA positivity.
The analysis reported herein focused on the Caucasian and African-American populations that comprised the majority of our cases. The genotype frequencies did not deviate from Hardy-Weinberg equilibrium for the polymorphisms studied in any of the ethnic groups. Significant differences in the −794 CATT genotypic and allelic distributions between Caucasians and African-Americans were noted, as previously reported, and their frequencies were similar to previously published ethnic-specific frequencies (19).
MIF Promoter Polymorphisms and SLE Incidence
To determine the potential association between MIF polymorphisms and the incidence of SLE, the frequencies of MIF alleles, genotypes, and haplotypes as well as the frequency of high expression −794 CATT7 or −173*C containing genotypes and high expression, 7C haplotypes were calculated and compared between SLE patients and healthy controls.
The −173 G/C SNP genotype frequencies for Caucasians and African-Americans are shown in Table 2. The frequency of high expression, −173*C containing MIF genotypes, which were shown previously to be associated with elevated circulating MIF levels (10;25), was lower in SLE patients than in controls in the African-American cohort (49.7% versus 64.2%, p=0.007) but not in the Caucasian cohort (32.7% versus 34.6%, p=0.36). The permutation p values were p=0.0154 for African-Americans and p=0.578 for Caucasians. This finding suggests that high-expression, −173*C containing MIF genotypes may be protective of SLE in African-Americans.
Table 2.
MIF −173 SNP Genotype Frequencies in SLE Patients and Controls Based on Ethnicity.
| Caucasians | African-Americans | |||||
|---|---|---|---|---|---|---|
| Patients (%) |
Controls (%) |
P value |
Patients (%) |
Controls (%) |
P value | |
| MIF Genotype Frequency | ||||||
| −173 GG | 701 (67.3) |
913 (65.4) |
90 (50.3) |
62 (35.8) |
||
| −173 GC | 300 (28.8) |
440 (31.5) |
64 (35.8) |
89 (51.4) |
||
| −173 CC | 41 (3.9) |
42 (3.0) |
25 (14.0) |
22 (12.7) |
||
| Total (n) | 1042 | 1395 | 179 | 173 | ||
| −173 C containing genotypes (−173 CC & −173 GC) |
341 (32.7) |
482 (34.6) |
P=0.36 | 89 (49.7) |
111 (64.2) |
P=0.007 |
The −794 CATT repeat genotype frequencies are displayed in Table 3. The high expression, −794 CATT7 containing MIF genotypes were under-represented in the Caucasian SLE group when compared with controls (21% versus 24%, p=0.049), but not in African-American SLE patients (14.5% versus 19.5%, p=0.256). The permutation p values, however, were 0.147 in Caucasians and 0.383 in African-Americans. These results, although suggestive, did not support the hypothesis that the high expression - 794 CATT7 containing genotypes protect against SLE development.
Table 3.
MIF −794 CATT5-8 Genotype Frequencies in SLE Patients and Controls Based on Ethnicity.
| Caucasians | African-Americans | |||||
|---|---|---|---|---|---|---|
| Patients (%) |
Controls (%) |
P value | Patients (%) |
Controls (%) |
P value | |
| − 794 CATT MIF Genotype Frequency | ||||||
| −794 CATT5/5 | 77 (7.7) |
76 (5.7) |
39 (22.3) |
30 (16.8) |
||
| −794 CATT5/6 | 298 (29.8) |
405 (30.5) |
48 (27.4) |
65 (36.3) |
||
| −794 CATT5/7 | 50 (5.0) |
108 (8.1) |
12 (6.9) |
13 (7.3) |
||
| −794 CATT5/8 | 1 (0.1) |
2 (0.2) |
0 | 0 | ||
| −794 CATT6/6 | 414 (41.4) |
519 (39.1) |
60 (34.3) |
48 (26.8) |
||
| −794 CATT6/7 | 148 (14.8) |
188 (14.2) |
9 (5.1) |
18 (10.1) |
||
| −794 CATT6/8 | 1 (0.15) |
5 (0.4) |
2 (1.1) |
1 (0.6) |
||
| −794 CATT7/7 | 12 (1.2) |
24 (1.8) |
4 (2.3) |
4 (2.2) |
||
| −794 CATT8/8 | 0 | 0 | 1 (0.6) | 0 | ||
| Total (n) | 1001 | 1327 | 175 | 179 | ||
| −794 CATT7 containing genotypes (− 794 CATT5/7, −794 CATT6/7, −794 CATT7/7) |
210 (21.0) |
320 (24.2) |
P=0.049 | 25 (14.5) |
35 (19.7) |
P=0.25 |
MIF haplotypes for both ethnic groups were reconstructed computationally and the estimated frequencies of all possible haplotypes and their relative odds ratio are shown in Table 4. The −173 G/C and −794 CATT polymorphisms were in linkage disequilibrium, with the −173*C allele being more frequently associated with CATT7 than with the CATT5 or CATT6 alleles (D’=0.74 for Caucasians and D’=0.80 for African-Americans). After adjusting for sex, age, geographic location, MIF polymorphisms, and ANA status, the frequency of the high expression, 7C haplotype was found to be lower in SLE patients than in controls and showed statistical significance in both Caucasians (OR 0.63 [0.53, 0.89], p=0.001) and in African-Americans (OR 0.46 [0.23, 0.95], p=0.012). The permutation p values were 0.0016 in Caucasians and 0.06 in African-Americans.
Table 4.
MIF Haplotype Frequencies in Caucasian and African-American SLE patients and Controls.
| Caucasians | African-Americans | |||||||
|---|---|---|---|---|---|---|---|---|
|
MIF Haplotype Frequency |
Patients 2n=1988 (%) |
Controls 2n=2582 (%) |
Odds ratio [95%C.I]a |
P | Patients 2n=348 (%) |
Controls 2n=348 (%) |
Odds ratio [95%C.I]a |
P |
| −794 CATT5/−173 G (5G) |
452 (22.8) |
618 (23.9) |
1 | 85 (24.0) |
71 (20.4) |
1 | ||
| −794 CATT6/−173 G (6G) |
1123 (56.5) |
1449 (56.1) |
1.08 [0.91, 1.28] |
0.395 | 140 (40.1) |
141 (40.2) |
0.85 [0.57, 1.27] |
0.43 |
| −794 CATT7/−173 G (7G) |
48 (2.4) |
26 (1.0) |
1.67 [0.82, 3.37] |
0.156 | 13 (3.5) |
3 (0.7) |
4.42 [0.99,19.74] |
0.052 |
| −794 CATT5/−173 C (5C) |
50 (2.5) |
30 (1.2) |
1.85 [0.81, 4.25] |
0.145 | 55 (15.3) |
65 (18.8) |
0.69 [0.40, 1.20] |
0.188 |
| −794 CATT6/−173 C (6C) |
146 (7.3) |
142 (5.5) |
1.24 [0.90, 1.70] |
0.194 | 37 (10.3) |
33 (9.5) |
0.88 [0.52, 1.51] |
0.648 |
| −794 CATT7/−173 C (7C) |
169 (8.5) |
317 (12.3) |
0.63 [0.53, 0.89] |
0.001 | 18 (4.8) |
35 (10.0) |
0.46 [0.23, 0.95] |
0.012 |
Odds ratio were calculated by logistic regression. The reported p values were adjusted for sex, age, geographic location, MIF polymorphisms, and ANA status.
To account for population stratification bias, we used formulas established by Lee and Wang (22), as described in the methods section. The prevalence of SLE in Caucasians is reported to be 40 to 90 per 100,000 in Caucasians and 100 to 300 per 100,000 in blacks (26;27). The frequency of the haplotype 7C in Caucasians is reported to be 5-12% (28;29) which are similar to our reported frequencies. The frequency of 7C in African-Americans has not been previously reported. Therefore, we adopted the frequencies reported in our study: 4-11%. Based on the prevalence of SLE and the frequency of 7C haplotype we calculated the highest odds ratio “U” and the lowest “1/U” that can be attributed to population stratification. In Caucasians, “U” equals 1.2 and “1/U” equals 0.83. In African-Americans, “U” = 1.33 and “1/U” = 0.75. Our observed ORs of 0.63 in Caucasians and 0.46 in African-Americans are below the lowest calculated ORs above (0.83 and 0.75 respectively) indicating that these observed ORs cannot be explained simply by population stratification. These results, taken together, further suggest that the high expression, 7C MIF haplotype confers protection against the development of SLE in both Caucasian and African-American subjects.
MIF Polymorphisms and SLE Clinical Manifestations
To determine the effect of MIF gene polymorphisms on disease manifestations, SLE patients were stratified according to the presence of documented clinical sequela. The frequencies of low expression, −794 CATT5 and −173*G containing MIF genotypes were measured and compared between patients with significant organ involvement (nephritis, serositis, and CNS involvement) and those with mild disease consisting of arthralgia/arthritis and mucocutaneous manifestations. Since the majority of the patient cohort was Caucasian, our investigation of the potential association between MIF polymorphisms and clinical manifestations focused primarily on this population in order to allow for higher sample numbers for statistical analysis.
SLE patients with nephritis, serositis, and male patients with CNS involvement had a lower frequency of low expression, −794 CATT5 containing MIF genotypes than those patients without these disease complications (36% vs 64% p=0.023 for nephritis, 42% vs 58% p=0.005 for serositis, 11% vs. 89% p=0.040 for CNS involvement). No differences were observed in the distribution of the low expression, −173*G containing MIF genotypes in patients with and without significant end-organ involvement. An analysis also was performed for the high expression −794 CATT7 and −173*C containing genotypes and 7C haplotypes, however no statistical difference was detected.
MIF Polymorphisms and Autoantibody Status
To determine the relationship between MIF polymorphisms and anti-nuclear antibody (ANA) status, the frequencies of ANA positivity (>1:40) in SLE and control subjects were determined and compared with respect to MIF genotypes and haplotypes. Both patients and healthy controls with high expression, −794 CATT7 or −173*C containing MIF genotypes as well high expression, 7C haplotypes were observed to be less likely to be ANA positive (25.8% vs. 17.7%, p=0.03 for −794 CATT7 containing genotypes, 43.8% vs. 31.8%, p=0.008 for −173*C containing genotypes, and 78% vs. 63%, p=0.0001 for 7C haplotypes). That high expression MIF alleles also are associated with a lower incidence of ANA positivity further supports the conclusion that these alleles may confer protection from SLE. Additional analyses for the potential association between anti-ds DNA or extractable anti-nuclear autoantibodies and MIF polymorphisms were not performed because of incomplete data for many patients.
Plasma Levels of MIF and TNFα
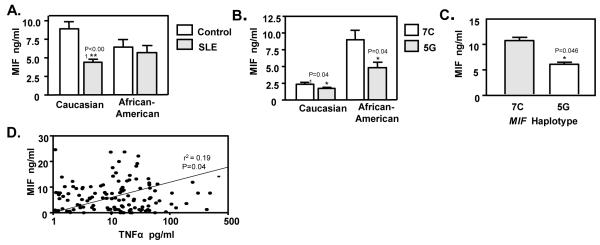
MIF levels were measured in the studied groups to determine if there was a relationship between circulating MIF concentrations and MIF haplotypes. Plasma samples were obtained at the time of study entry. The mean plasma MIF level in Caucasian SLE patients as a group was lower than in disease-free, matched controls (4.6 ± 4.6 ng/ml in patients versus 8.9 ± 7.0 ng/ml in controls [mean±SD], p<0.001, n=116 patients and 55 controls) (Figure 2A). Circulating MIF levels also were lower in the African-American patients than in controls, however this difference did not reach statistical significance (5.6 ± 6.3 ng/ml in SLE patients versus 6.4 ± 6.8 ng/ml in controls [mean±SD], p=0.580, n=44 in each group).
Figure 2.
A). MIF plasma levels in SLE patients and controls grouped by ethnicity. The columns depict the mean level of MIF in 116 Caucasian patients and 55 healthy controls, and 44 African-American patients and 44 healthy controls. Values are mean ± SEM. **p<0.001 by unpaired t test. B). MIF plasma levels in SLE patients grouped by MIF haplotype. The columns depict the mean level of MIF in 17 Caucasian patients and 41 African-American patients with either the high expression, 7C or the low expression, 5G MIF haplotype. Values are mean ± SEM. *p<0.001 by unpaired t test. C). TLR7 induced MIF release from PBMCs is regulated by MIF haplotype. The columns depict MIF released by Monocytes from Caucasian individuals with low expression, 5G (n=20) or high expression, 7C (n=10) MIF haplotype. Values are mean ± SEM. *p=0.046 by unpaired t test. D). Correlation between MIF and TNFα plasma levels in 160 Caucasian and African-American SLE patients. TNFα and MIF levels were positively correlated, r2=0.19, p=0.04 by Spearman correlation test.
We next measured plasma MIF in SLE patients identified as having a high expression, 7C MIF haplotype and compared the values with patients known to have a low expression, 5G MIF haplotype. Notably, both Caucasian and African-American patients with the high expression, 7C haplotype had higher MIF plasma levels when compared to those with the low expression, 5G haplotype (2.4 ± 1.7 ng/ml vs 1.6 ± 1.6 ng/ml [mean±SD], p=0.041 in Caucasians, and 8.9 ± 7.6 ng/ml vs. 4.8 ± 4.3 ng/ml [mean±SD], p=0.040 in African-Americans) (Figure 2B). To minimize the effect of medications and other factors on MIF plasma levels, we also measured circulating MIF concentrations in Caucasian and African-American healthy controls that we had identified as carriers of the either high expression, 7C or low expression, 5G MIF haplotypes. Of the 100 controls studied, 17 were identified as having 7C and 36 as having 5G. MIF plasma levels were higher in the high expression, 7C haplotype group (10.3 ± 8.1 ng/ml, [mean±SD]) than in the low expression, 5G group (6.4 ± 5.5 ng/ml [mean±SD], p=0.048). These data suggest that MIF haplotype influences baseline circulating MIF levels irrespective of underlying disease or autoimmunity.
MIF may be distinguished functionally from other cytokines because of its action as a regulatory mediator that sustains the activation of immune cells and the production of downstream effectors such as (30). TNFα is considered to be a major initiator of vascular endothelial cell and end-organ damage. We observed higher TNFα levels in Caucasians and African-Americans SLE patients than in controls (64.9±192.4 pg/ml in patients versus 14.3 ± 42.3 pg/ml in controls [mean±SD], p=0.05), which is in agreement with prior reports (31). Moreover, a comparative analysis of TNFα and MIF levels in SLE patients showed a weak positive correlation (r2=0.19, p=0.04) (Figure 2D). In separate analyses, we also examined circulating MIF concentrations in relation to SLE clinical manifestations. No significant associations were observed; however, this comparison may be confounded by disease activity, the effect of immunosuppressive treatment, and the influence of endogenous or administered glucocorticoids on circulating MIF levels (32)
Influence of MIF Polymorphisms on Immune Cell Activation
Activated monocytes/macrophages are an abundant source of MIF in vivo and have been shown to release MIF in response to different microbial products (6). There is evidence that viral infections and nucleic acid-containing immune complexes contribute to the immunopathology of SLE by providing recurrent or self-sustaining stimulatory signals through specific Toll-like receptors (TLRs) (33;34).
We tested the influence of MIF genotype on the ability of human peripheral blood mononuclear cells to produce MIF in response to TLR7 agonism. Monocytes were isolated from 30 healthy Caucasian controls with low expression, 5G or high expression, 7C MIF haplotypes and stimulated with the TLR7 agonist, polyU. As shown in Figure 2C, human peripheral blood monocytes with the high-expression, 7C haplotype produced almost 80% more MIF than cells with the low-expression, 5G haplotype 7C: 10.5±3.8 ng/ml vs 5G: 5.8+4.4 [mean±SD], p=0.046). These data suggest that circulating RNA, whether of viral or endogenous origin, may provide a relevant source of MIF haplotype-dependent immunologic stimulation through TLR7.
DISCUSSION
We report a significant relationship between two known promoter polymorphisms in MIF, not studied previously in genome-wide association studies, and the incidence and clinical severity of SLE (19). The presence of the high expression MIF haplotype, 7C, was associated with a lower incidence of SLE in Caucasians and to a lesser extent in African-Americans. When compared with ethnically matched controls, African-Americans with SLE had lower frequencies of high expression −173*C containing MIF genotypes. We further observed that high expression MIF genotypes are associated with a reduced incidence of ANA, which is the hallmark serologic abnormality of SLE (35). When the SLE and the control groups were stratified by MIF haplotype, a significant association between plasma MIF levels and high or low expression MIF haplotype also was evident. These results collectively suggest that a genetic predisposition to increased MIF production may confer protection against the development of an autoimmune response leading to SLE
MIF is a pro-inflammatory mediator and an upstream regulator of TNFα expression. That an increase in TNFα production may confer protection against SLE was suggested over twenty years ago by the studies of McDevitt and colleagues, who reported that monocytes with the HLA-DR3 or DR4 SLE susceptibility loci showed low levels of inducible TNFα production (36). These authors also found that TNFα administration to lupus-prone mice delayed the onset of autoimmunity (37). More recently, it has been noted that the administration of anti-TNFα therapies can lead to the development of an ANA (38), further pointing to the potentially protective role of TNFα in SLE.
Our data also indicate that patients with established SLE who have low expression, −794 CATT5 containing MIF genotypes may be protected from the development of the inflammatory clinical manifestations of serositis, nephritis, and CNS involvement. These conclusions mirror the associations that have been reported previously between high expression MIF alleles and disease severity in such inflammatory disorders as rheumatoid arthritis (10), systemic sclerosis (14), and asthma (15). Thus, once autoimmunity develops, a genetic propensity for increased MIF expression likely contributes to specific disease manifestations and end-organ damage.
What is less evident however, is how a genetic predisposition to increased MIF expression and a downstream response that includes TNFα production (6) may protect against the development of autoimmunity or clinical progression to SLE. High expression MIF alleles have been shown to be associated with improved survival from certain infections, in part by augmenting innate immune responses (28). It has been hypothesized that antecedent infections play a role in SLE by mechanisms that may involve antigenic mimicry, oligoclonal B or T cell activation, and loss of tolerance (39) Thus, a more robust, MIF-dependent anti-microbial response may promote the clearance and the timely resolution of infection, thereby protecting against the development of autoimmunity. Our in vitro data provide some support of this notion since monocytes with a high expression, 7C MIF haplotype produce more MIF upon the stimulation of TLR7. Finally, recent data support a role for excessive apoptosis or defective clearance of apoptotic cells in the immunopathogenesis of SLE. Apoptotic nuclei may overwhelm the reticulo-endothelial system, break immune tolerance, and induce autoantibody production against nuclear components (40). Higher level of MIF expression may reduce the apoptotic response during inflammation and decrease the likelihood of an autoimmune response progressing to SLE (30).
Our ELISA analyses did not reveal a significant difference in circulating MIF levels among SLE patients with different disease manifestations. Although higher MIF levels in plasma may be anticipated in patients with active end-organ disease, the measurement of MIF in the blood may be confounded by several factors. Plasma may not accurately reflect elevated levels of MIF expression in sites of tissue inflammation, and baseline MIF levels are known to vary in a circadian rhythm (41). While high-dose, exogenous glucocorticoids suppress MIF secretion, low doses may actually induce MIF release in vivo. Thus, any conclusions based on the measurement of plasma MIF must be tempered by the heterogeneity of disease activity and the different treatment protocols in patients, which include glucocorticoids. Finally, lymphopenia is a feature of SLE and may reduce circulating, immune cell sources of MIF production.
Recent genome-wide association scans have identified several gene polymorphisms that are associated with susceptibility to SLE (4). These analyses have been limited to the study of selected SNPs and have not examined microsatellite repeats such as the MIF −794 CATT site. Moreover, these studies have focused on disease susceptibility and not clinical manifestations, and they have so far been limited to Caucasian populations.
The main limitation of the present study is in its cross-sectional design. Thus, the clinical phenotype was determined up to the point in time that the study was performed. Internationally established SLE activity and damage indices also were not available for many patients; therefore, the inclusion of end-organ involvement served as our assessment of disease severity. Finally, our findings do not replicate the report of Sanchez et al. who found that, in a smaller Spanish population, there was an increased incidence of SLE in those individuals with the homozygous −173 C/C MIF genotype (29). It is possible that our conclusions differ because of sample size or unknown genetic variations between populations.
In summary, our genetic association analyses of prevalent and functional MIF polymorphisms suggest that MIF plays a dual role in SLE. In Caucasians and African-Americans, high expression MIF polymorphisms are associated with a lower incidence of SLE. In patients with established SLE however, low-expression MIF polymorphisms are associated with a lower incidence of end-organ injury. The possibility that some SLE patients demonstrate a propensity for end-organ disease based on their MIF allele may support a pharmacogenomic approach to MIF-directed that are entering clinical evaluation.
Acknowledgments
This project was supported by: The Alliance for Lupus Research, NIH AR049610, AR050498, NO1 AI50031, and The American College of Rheumatology-Research and Education Foundation. The Swedish Research Council of Medicine, the Swedish Association Against Rheumatism and the Gustaf Vth-80th Jubilee to MEAR.
Footnotes
Dr. Bucala is a co-inventor on patent applications describing the utility of MIF genotype determination. The authors have no additional financial interests.
References
- (1).Crispin JC, Liossis SN, Kis-Toth K, Lieberman LA, Kyttaris VC, Juang YT, et al. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 2010;16(2):47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35(3):311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- (3).Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, Nititham J, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5(10):e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mitchell RA, Metz CN, Peng T, Bucala R. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J Biol Chem. 1999;274(25):18100–6. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- (6).Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179(6):1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hoi AY, Hickey MJ, Hall P, Yamana J, O’Sullivan KM, Santos LL, et al. Macrophage migration inhibitory factor deficiency attenuates macrophage recruitment, glomerulonephritis, and lethality in MRL/lpr mice. J Immunol. 2006;177(8):5687–5696. doi: 10.4049/jimmunol.177.8.5687. [DOI] [PubMed] [Google Scholar]
- (8).Leng L, Chen L, Fan J, Greven D, Arjona A, Du X, et al. A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J Immunol. 2011;186(1):527–538. doi: 10.4049/jimmunol.1001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Awandare GA, Martinson JJ, Were T, Ouma C, Davenport GC, Ong’echa JM, et al. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–637. doi: 10.1086/600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Radstake TR, Sweep FC, Welsing P, Franke B, Vermeulen SH, Geurts-Moespot A, et al. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 2005;52(10):3020–3029. doi: 10.1002/art.21285. [DOI] [PubMed] [Google Scholar]
- (11).Baugh JA, Chitnis S, Donnelly SC, Monteiro J, Lin X, Plant BJ, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3(3):170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- (12).Donn R, Alourfi Z, Zeggini E, Lamb R, Jury F, Lunt M, et al. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004;50(5):1604–1610. doi: 10.1002/art.20178. [DOI] [PubMed] [Google Scholar]
- (13).Amoli MM, Donn RP, Thomson W, Hajeer AH, Garcia-Porrua C, Lueiro M, et al. Macrophage migration inhibitory factor gene polymorphism is associated with sarcoidosis in biopsy proven erythema nodosum. J Rheumatol. 2002;29(8):1671–1673. [PubMed] [Google Scholar]
- (14).Wu SP, Leng L, Feng Z, Liu N, Zhao H, McDonald C, et al. Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum. 2006;54(11):3661–3669. doi: 10.1002/art.22179. [DOI] [PubMed] [Google Scholar]
- (15).Mizue Y, Ghani S, Leng L, McDonald C, Kong P, Baugh J, et al. Role for macrophage migration inhibitory factor in asthma. Proc Natl Acad Sci U S A. 2005;102(40):14410–14415. doi: 10.1073/pnas.0507189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nohara H, Okayama N, Inoue N, Koike Y, Fujimura K, Suehiro Y, et al. Association of the −173 G/C polymorphism of the macrophage migration inhibitory factor gene with ulcerative colitis. J Gastroenterol. 2004;39(3):242–246. doi: 10.1007/s00535-003-1284-7. [DOI] [PubMed] [Google Scholar]
- (17).Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- (18).Grigorenko EL, Han SS, Yrigollen CM, Leng L, Mizue Y, Anderson GM, et al. Macrophage migration inhibitory factor and autism spectrum disorders. Pediatrics. 2008;122(2):e438–e445. doi: 10.1542/peds.2007-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhong XB, Leng L, Beitin A, Chen R, McDonald C, Hsiao B, et al. Simultaneous detection of microsatellite repeats and SNPs in the macrophage migration inhibitory factor (MIF) gene by thin-film biosensor chips and application to rural field studies. Nucleic Acids Res. 2005;33(13):e121. doi: 10.1093/nar/gni123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhao LP, Li SS, Khalid N. A method for the assessment of disease associations with single-nucleotide polymorphism haplotypes and environmental variables in case-control studies. Am J Hum Genet. 2003;72(5):1231–1250. doi: 10.1086/375140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).R: A Language and Environment for Statistical Computing. Vienna, Austria: 2011. [Google Scholar]
- (22).Lee WC, Wang LY. Simple formulas for gauging the potential impacts of population stratification bias. Am J Epidemiol. 2008;167(1):86–89. doi: 10.1093/aje/kwm257. [DOI] [PubMed] [Google Scholar]
- (23).Barton A, Lamb R, Symmons D, Silman A, Thomson W, Worthington J, et al. Macrophage migration inhibitory factor (MIF) gene polymorphism is associated with susceptibility to but not severity of inflammatory polyarthritis. Genes Immun. 2003;4(7):487–491. doi: 10.1038/sj.gene.6364014. [DOI] [PubMed] [Google Scholar]
- (24).Gao XM, Liu Y, White D, Su Y, Drew BG, Bruce CR, et al. Deletion of macrophage migration inhibitory factor protects the heart from severe ischemia-reperfusion injury: A predominant role of anti-inflammation. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2010.12.022. [DOI] [PubMed] [Google Scholar]
- (25).De Benedetti F, Meazza C, Vivarelli M, Rossi F, Pistorio A, Lamb R, et al. Functional and prognostic relevance of the −173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2003;48(5):1398–1407. doi: 10.1002/art.10882. [DOI] [PubMed] [Google Scholar]
- (26).Hopkinson ND, Doherty M, Powell RJ. Clinical features and race-specific incidence/prevalence rates of systemic lupus erythematosus in a geographically complete cohort of patients. Ann Rheum Dis. 1994;53(10):675–680. doi: 10.1136/ard.53.10.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pons-Estel GJ, Alarcon GS, Scofield L, Reinlib L, Cooper GS. Understanding the epidemiology and progression of systemic lupus erythematosus. Semin Arthritis Rheum. 2010;39(4):257–268. doi: 10.1016/j.semarthrit.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yende S, Angus DC, Kong L, Kellum JA, Weissfeld L, Ferrell R, et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. 2009;23(8):2403–2411. doi: 10.1096/fj.09-129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sanchez E, Gomez LM, Lopez-Nevot MA, Gonzalez-Gay MA, Sabio JM, Ortego-Centeno N, et al. Evidence of association of macrophage migration inhibitory factor gene polymorphisms with systemic lupus erythematosus. Genes Immun. 2006;7(5):433–436. doi: 10.1038/sj.gene.6364310. [DOI] [PubMed] [Google Scholar]
- (30).Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci U S A. 2002;99(1):345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Studnicka-Benke A, Steiner G, Petera P, Smolen JS. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. Br J Rheumatol. 1996;35(11):1067–1074. doi: 10.1093/rheumatology/35.11.1067. [DOI] [PubMed] [Google Scholar]
- (32).Foote A, Briganti EM, Kipen Y, Santos L, Leech M, Morand EF. Macrophage migration inhibitory factor in systemic lupus erythematosus. J Rheumatol. 2004;31(2):268–273. [PubMed] [Google Scholar]
- (33).Kim WU, Sreih A, Bucala R. Toll-like receptors in systemic lupus erythematosus; prospects for therapeutic intervention. Autoimmun Rev. 2009;8(3):204–208. doi: 10.1016/j.autrev.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25(3):417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- (35).Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- (36).Jacob CO, Fronek Z, Lewis GD, Koo M, Hansen JA, Mcdevitt HO. Heritable Major Histocompatibility Complex Class-Ii-Associated Differences in Production of Tumor Necrosis Factor-Alpha - Relevance to Genetic Predisposition to Systemic Lupus-Erythematosus. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(3):1233–1237. doi: 10.1073/pnas.87.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Todd JA, Acha-Orbea H, Bell JI, Chao N, Fronek Z, Jacob CO, et al. A molecular basis for MHC class II--associated autoimmunity. Science. 1988;240(4855):1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- (38).De Rycke L, Baeten D, Kruithof E, Van den BF, Veys EM, De Keyser F. The effect of TNFalpha blockade on the antinuclear antibody profile in patients with chronic arthritis: biological and clinical implications. Lupus. 2005;14(12):931–937. doi: 10.1191/0961203305lu2240rr. [DOI] [PubMed] [Google Scholar]
- (39).Rahman A, Isenberg DA. Mechanisms of disease: Systemic lupus erythematosus. New England Journal of Medicine. 2008;358(9):929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- (40).White S, Rosen A. Apoptosis in systemic lupus erythematosus. Curr Opin Rheumatol. 2003;15(5):557–562. doi: 10.1097/00002281-200309000-00006. [DOI] [PubMed] [Google Scholar]
- (41).Petrovsky N, McNair P, Harrison LC. Diurnal rhythms of pro-inflammatory cytokines: regulation by plasma cortisol and therapeutic implications. Cytokine. 1998;10:307–312. doi: 10.1006/cyto.1997.0289. [DOI] [PubMed] [Google Scholar]