Abstract
I-TASSER is an automated pipeline for protein tertiary structure prediction using multiple threading alignments and iterative structure assembly simulations. In CASP9 experiments, two new algorithms, QUARK and FG-MD, were added to the I-TASSER pipeline for improving the structural modeling accuracy. QUARK is a de novo structure prediction algorithm used for structure modeling of proteins that lack detectable template structures. For distantly homologous targets, QUARK models are found useful as a reference structure for selecting good threading alignments and guiding the I-TASSER structure assembly simulations. FG-MD is an atomic-level structural refinement program that uses structural fragments collected from the PDB structures to guide molecular dynamics simulation and improve the local structure of predicted model, including hydrogen-bonding networks, torsion angles and steric clashes. Despite considerable progress in both the template-based and template-free structure modeling, significant improvements on protein target classification, domain parsing, model selection, and ab initio folding of beta-proteins are still needed to further improve the I-TASSER pipeline.
Keywords: protein structure prediction, threading, contact prediction, ab initio folding, CASP
Introduction
The community-wide Critical Assessment of techniques for protein Structure Prediction (CASP) experiments provide a standard platform to assess the state of the art of structure modeling methods. Recent CASP experiments have witnessed considerable progress in template-based modeling (TBM),1–3 where the efforts have been mainly focused on developing better methods for template identification4–6 and on combining multiple threading templates to improve the quality of comparative models.7–11 However, an imprudent amalgamation of multiple templates can result in distorted structural models containing non-protein like secondary structures and steric clashes.12–13 Although these models may have some level of usefulness, such as fold family classification and domain boundary recognition, the local structural inaccuracies render them inapplicable for high-resolution biological applications like ligand-docking and virtual screening.14–15
Compared to TBM, not much progress has been observed in ab initio folding (or free-modeling; FM)16–18 of proteins which lack analogous templates in the Protein Data Bank (PDB)19 especially since the invention of the idea of structural fragment assembly.20–21 For I-TASSER,22–24 although the reassembly of structural fragments excised from the threading alignments often results in significantly improved models, the quality of final models is still essentially dependent on that of threading templates identified in the PDB library.
In view of these major difficulties in the field and especially the issues of the I-TASSER pipeline as reflected in the previous CASP experiments,13,25 we have been working on the development of two types of methods. First, we developed REMO26 for atomic structure construction and improvement of hydrogen-bonding network. Although REMO shows a significant ability to remove backbone clashes and to optimize the H-bonding network, it cannot improve the secondary structures and reduce side-chain atom clashes when the C-alpha traces contain severe local structure distortions. For further refinement of the I-TASSER models, we recently developed FG-MD27 which uses constrained molecular dynamic simulations to adjust the position of each atom in the protein. Second, we developed an ab initio tertiary structure prediction algorithm QUARK28–29 which assembles the protein structure models from scratch.
In CASP9, we evaluated the I-TASSER pipeline with these two new components. Analyzing the efficiency of these two new methods is the focus of this report. Although we participated in both human and server predictions, since the methods used in the two categories are essentially the same, this report will be mainly on the automated server predictions.
Materials and Methods
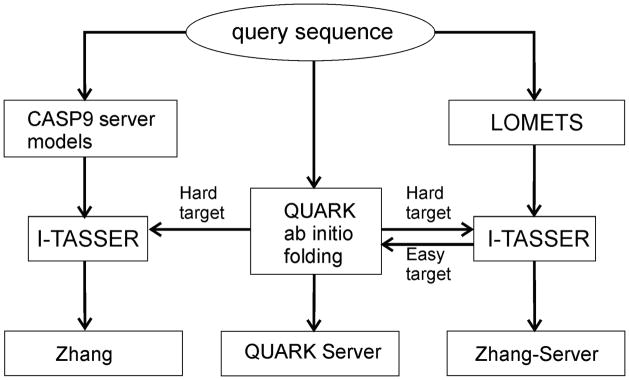
Flowchart of I-TASSER pipeline
The protein structure prediction procedure used by the human (Zhang) and the two server groups (Zhang-Server and QUARK) during CASP9 is depicted in Figure 1. The methods used by Zhang and Zhang-Server groups were based on standard I-TASSER pipeline and were essentially the same, except for that the human prediction exploited the templates in CASP9 Server Section, while Zhang-Server used threading alignments generated by LOMETS, a locally installed meta-server threading program.7
Figure 1.
Modeling flowchart by Zhang, Zhang-Server, and QUARK in CASP9.
LOMETS alignments were also used to define domain boundaries and to categorize the targets into “Easy”, “Medium”, and “Hard” category. A target was classified as “Easy”, if the Z-score of the top-scoring templates from all the threading programs was higher than the program specific Z-score cutoff (Z0); on the contrary, if none of the threading programs found a hit with a Z-score >Z0, the target was classified as “Hard”; the rest of the cases were classified as “Medium”.7 We note that there is no definite correspondence between our target categorization (Easy/Medium/Hard) and assessor’s target classifications (TBM/FM) because the prediction is blind and the scoring function of threading algorithms is imperfect.
For Easy targets, Zhang and Zhang-Server used the standard I-TASSER pipeline while for the Medium and Hard targets, they collected spatial restraints from the ab initio models by QUARK to guide the I-TASSER structure assembly simulations; the weight of the restraints was stronger for Hard targets than that for Medium targets. For Hard targets, LOMETS templates were further sorted based on their structure similarity to the QUARK model, with the purpose of fishing out better templates, and sorted templates were used as input of I-TASSER.
The QUARK server generally predicted the models by ab initio folding. But for Easy targets which have significant templates available, it uses the I-TASSER program but with a slightly different way of combination of the LOMETS threading templates (see below). All the procedures were kept fully automated.
The I-TASSER pipeline consisted of three steps: template identification, structure assembly, model selection and full-atomic model refinements, which are described below.
Template identification
The target sequences were first threaded through a non-redundant PDB structure library to identify template structures that may have a similar structure or similar structural motif as the query protein. The threading in I-TASSER is conducted by LOMETS7, which includes 8 individual threading programs: FUGUE,30 HHSEARCH,4 MUSTER,5 PROSPECT2,31 SAM-T02,32 SPARKS2,33 PPA-I23 and SP334. In Zhang-server, the I-TASSER pipeline used 6 threading programs (HHSEARCH, MUSTER, PROSPSECT2, SPARKS2, PPA-I and SP3) which on average had a better performance in our benchmarking tests. The QUARK server (for Easy targets) used all the 8 LOMETS threading programs plus two in-house threading programs PPA-II and PPA-III23 to include more diverse alignments. In the human prediction (Zhang), threading alignments generated by HHSEARCH, MUSTER, PROSPECT2, PPA-I and the models submitted by five CASP servers (Zhang-Server, QUARK, RAPTORX, Baker-ROBETTASERVER, HHpredA) were used for collecting spatial restraints and structural fragments.
For Hard targets, most of the top templates have a Z-score < Z0 and their ranking order in LOMETS is close to random. A recent study35 showed that the folds present in current PDB library are nearly complete for single-domain proteins, indicating that appropriate templates should be detectable for almost all the proteins. Considering this, we compare the top 5 QUARK ab initio models to the top 20 templates identified by each threading programs and then re-rank the templates of all LOMETS programs based on their TM-score to the closest QUARK models. We used the best TM-score rather than the average TM-score to all five QUARK models because a significant match of the threading template to any ab initio models can be considered as a meaningful hint to speculate that the template might have some right aspects since threading and QUARK are two distinct approaches. Furthermore, only 20 templates were selected because we assume that a reasonable threading program should have a meaningful alignment in its top 20 hits; including more templates will increase number of false positive templates after sorting. We note that although we use the QUARK models to re-rank the templates, only the original LOMETS alignments will be input into the I-TASSER simulations.
The purpose of the template sorting is to exploit the ab initio models to fish out correct templates from the PDB pool. Indeed, we noticed that even a partially folded QUARK model (i.e. only part of the structure is correct) can sometimes identify templates with correct full-length fold. Since I-TASSER uses top 20 (for Easy targets) or top 50 (for Medium/Hard targets) threading templates, a correctly sorted list with better threading alignments put at the top, will improve the final I-TASSER models.
Structure assembly simulations
In I-TASSER, continuous fragments are excised from threading templates and exploited for assembling full-length models,23,36 while the unaligned loop regions are built by ab initio modeling procedure37 on a lattice system. The I-TASSER force field includes four components: (1) general knowledge-based statistical terms from the PDB (Cα/side-chain correlations,37 hydrogen-bond38 and hydrophobicity39); (2) spatial restraints collected from threading templates7; (3) sequence-based Cα/Cβ/side-chain center contact predictions by SVMSEQ40–41; (4) distance map from segmental threading42. As mentioned above, for Medium and Hard target, I-TASSER also uses spatial restraints collected from QUARK models.
SVMSEQ is a Support Vector Machine (SVM) based residue-residue contact prediction algorithm that only uses sequence information.40 It was trained using local window features (position-specific scoring matrices, secondary structure types and solvent accessibility predictions) and in-between segment features (residue separations, secondary structure of the contacting residues, and state distributions of the contacting residues). Nine sets of contact predictions are generated based on the three atom types (Cα, Cβ and side-chain center), each with three different contact cutoffs (6, 7 and 8Å). All nine ab initio contact predictions are used as restraints during I-TASSER simulation, with weights proportional to their confidence.41
I-TASSER structure assembly procedure consists of two sets of iterative Monte Carlo simulations.23 The first round uses the threading templates as initial structures. In the second round, the simulations start from the cluster centroids generated by SPICKER43 which clusters all the trajectories from the first set of simulations. Spatial restraints, which are collected from the PDB structures hit by TM-align44 using the cluster centroids as query structures, are also incorporated in the I-TASSER simulations. The purpose of the second stage is to refine the local geometry as well as to improve the global topology of the SPICKER centroids.
Model selection and refinements
The structures in low-temperature replicas of I-TASSER and QUARK simulations are clustered by SPICKER.43 Cluster centroids are generated by averaging the Cα coordinates of all the clustered decoys. Next, full-atomic models are constructed by REMO26 from these cluster centroids, while optimizing the hydrogen-bonding network, where a H-bonding list is pre-constructed based on secondary structure predictions and the 3D backbone model. REMO can quickly build the initial full-atomic models from Cα traces but often the models have local structure and side-chain atom distortions. Finally, all the models generated by REMO are submitted to FG-MD (Fragment-Guided Molecular Dynamics),27 with the purpose of improving the local geometry and hydrogen-bonding, and reducing backbone and side-chain steric clashes in the model. FG-MD simulations are carried out in vacuum, as implemented in LAMMPS45 package. The force field consists of energy terms from Amber99,46 Cα repulsive potential, statistical hydrogen-bonding potential, and distance restraints collected from both the template and structural fragments searched by TM-align44 in the PDB library, using initial I-TASSER/QUARK models as the probe. The distance restraints are generated by combination of distance maps from initial model, TM-align global template and TM-align fragments at each location. FG-MD refinement simulation is the last step of structure predictions pipeline used by both I-TASSER and QUARK servers and the refined models are used as final models for submission.
QUARK ab initio structure assembly
QUARK is an ab initio structure prediction method that uses atomic-level knowledge-based force field and replica-exchange Monte Carlo simulation to generate high quality 3D structures.28–29 For a given target, QUARK first generates a set of small structural fragments of 1–20 residues by “gapless threading” of the target sequence through the PDB library and ranks them based on a composite scoring function consisting of sequence and structure profiles, predicted secondary structure and torsion angles. The range of fragment length was optimized based on a large scale benchmarking test.47 The “gapless threading” here refers to a procedure to scan the query sequence fragments along the PDB structure from N- to C-terminal without gap/insertion allowed. This is essentially different from the normal threading algorithms which include gaps/insertions in the query-template alignments. Also, the normal threading programs usually use the entire sequence of query proteins rather than the fragmental sequences except for special purpose.42
Top 200 fragments at each residue position are exploited for assembling full length 3D models using replica-exchange Monte Carlo simulations. The protein chain here is described by a reduced model, consisting of main-chain atoms and side-chain center to reduce the number of explicitly treated degrees of freedom and the intra-molecular interactions in the polypeptide chain. The structure assembly simulations are guided by a composite force field which consists of atomic-level and knowledge-based energy terms along with distance profiles collected from the fragment library. Energy terms such as H-bond potential, excluded volume and statistical potentials are calculated based on the pairwise atomic distances in the reduced model.
Two types of movements are implemented in the Monte Carlo simulations: (a) continuous movements which include bond-length, bond-angle and torsion-angle modifications, and segmental rotation, shift and perturbation; (b) discontinuous movements including helix repack, beta repair, beta-turn reform and fragment replacement by that from the template fragment library. During the fragment replacement movements, more shorter fragments are attempted to substitute the old ones as the simulation runs longer. This is for the purpose of increasing the acceptance rate, since when decoy structures become more compact it is much harder to accept new big movements.
Twenty independent Monte Carlo runs are implemented; each run has 40 replicas covering 500 simulation cycles. Decoys from 10 low temperature replicas are clustered by the SPICKER program. The final atomic models are constructed from the cluster centroids by REMO26 and FG-MD,27 as described above.
Results
Based on CASP assessor’s definitions, 116 CASP targets were split into 147 domains and assessed in the server section. Among the 147 domains, 118 are template-based modeling (TBM) targets, while 29 domains are free-modeling targets (FM, including 3 TBM/FM targets). As some of the targets had very close templates in the PDB library, only 78 domains were evaluated in the human section. Because much more targets were tested in the server section and the methods used by our server and human predictions are identical, we will mainly focus on automated server predictions.
I-TASSER draws templates closer to native
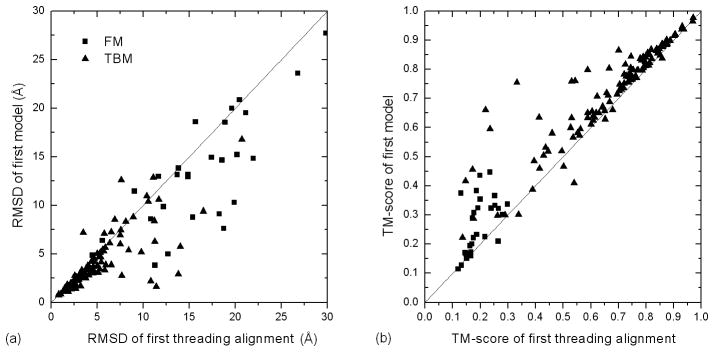
I-TASSER fragment assembly simulations are guided by spatial information collected from LOMETS threading templates. Accordingly, a comparison between the final model and initial templates is important to analyze whether the initial template structures were brought closer to the native states in the final predicted model. Figure 2 shows a head-to-head comparison of structure similarity (RMSD and TM-score) between the Zhang-Server models and LOMETS threading templates, where all the values are calculated in the same threading aligned region. As shown in Figure 2, I-TASSER simulations improved the template structures for 129 out of 147 test cases according to RMSD. The average RMSD and TM-score of the top threading template in the aligned regions are 7.3Å and 0.578, with an average alignment coverage of 91%. After the I-TASSER refinement simulations, the structure of the same aligned region had an RMSD of 6.0 Å and TM-score of 0.634, a 10% TM-score increase without considering the effect of the enlarged coverage. Here and afterwards, the percentage of the TM-score improvement is calculated based on the relative difference, i.e. the ratio of the absolute difference to the value before the improvement (10% in this case). This should not be confused with the ratio of the absolute difference versus the best possible model with a TM-score=1 (5.6% in this case).
Figure 2.
Comparison between the first Zhang-Server models and the threading templates by LOMETS. RMSD (a) and TM-score (b) values of the template and the final model are calculated in the same set of aligned residues.
The average result of comparison between the final predicted models and the initial templates is summarized in Table I. As shown in Column 4, the threading alignments of the TBM targets often have higher alignment coverage than that of FM targets. If we compare the average TM-score of the first threading templates to the first I-TASSER model (Columns 5 and 8), the improvements of the I-TASSER model over threading are 8% and 36% for TBM and FM targets on the threading aligned regions, respectively. The larger improvement observed for FM targets is mainly because template detection for FM targets is by definition more difficult and most of the top threading alignments are incorrect, thus leaving more room for improvement by I-TASSER simulations. Meanwhile, when comparing the best template with the best models, the improvements are 3% for TBM targets and 28% for FM targets. Here and afterwards, “the best template” refers to the best template identified by LOMETS threading, which should not be confused with the “best possibly templates” in the PDB library.
Table I.
Comparison of threading templates and Zhang-Server final models.
| First (best)template | First (best) model | |||||||
|---|---|---|---|---|---|---|---|---|
| Nta | Ralib | Covc | TM-scorealie | Ralib | Ralld | TM-scorealie | TM-scoreallf | |
| TBM | 118 | 4.7(4.0) | 93%(93%) | 0.671(0.721) | 3.8(3.4) | 4.3(3.9) | 0.724(0.743) | 0.755(0.773) |
| FM | 29 | 17.7(16.5) | 85%(82%) | 0.194(0.237) | 14.9(13.8) | 15.2(14.1) | 0.264(0.303) | 0.312(0.361) |
| All | 147 | 7.3(6.4) | 91%(91%) | 0.578(0.627) | 6.0(5.5) | 6.4(5.9) | 0.634(0.657) | 0.667(0.691) |
Number of targets.
RMSD to native of the threading aligned region (Å).
Coverage of the aligned region.
RMSD to native of the full-length model (Å).
TM-score to native of the threading aligned regions.
TM-score to native of the full-length model.
Although above RMSD and TM-score improvements have been mainly calculated in the threading aligned regions, the I-TASSER simulations generated improvements through the whole chain. To count this, we list the full-chain TM-score data in the last column of Table 1, where the TM-score increases over the first (best) template are 13% (7%) and 61% (52%) for the TBM and FM targets, respectively.
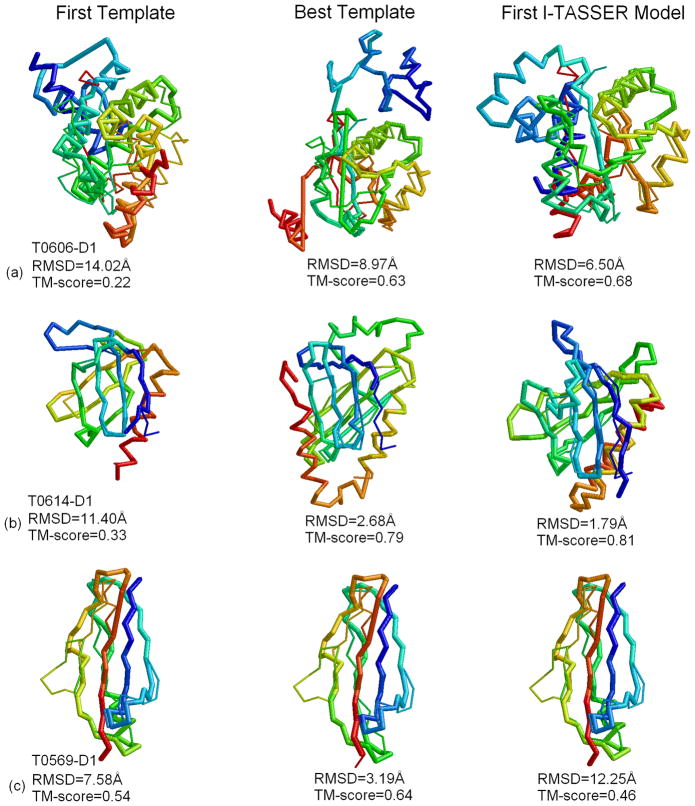
Figure 3 shows three typical examples which reflect both gain and loss in the I-TASSER simulations. For the two TBM targets, T0606-D1 and T0614-D1, both first LOMETS templates have an incorrect fold (shown in left panel of Figure 3a and b). The best templates were detected by LOMETS but with a very low rank (shown in the middle panel). The final Zhang-Server models have a higher accuracy than the best templates for both cases. The global topology of Zhang-Server model1 for T0606-D1 (Figure 3a right panel) was significantly improved (TM-score=0.68) over the best identified template (TM-score=0.63), because I-TASSER simulations correctly folded the C-terminal region (146–157) of the protein, which was incorrectly oriented in the threading template. The C-terminal region of the best threading template is isolated and disconnected with the other aligned region. I-TASSER simulation makes the two parts continuous and compact guided by the force field especially the terms of hydrophobicity and distance restraints that were collected from multiple templates. Similarly, the I-TASSER simulations improved the local beta-sheet packing for T0614-D1, resulting in Zhang-server Model1 (Figure 3b right panel) have a lower (1.79Å) RMSD and higher TM-score (0.81) than the best identified template (RMSD=2.68 Å; TM-score=0.79).
Figure 3.
Examples of the predicted models (thick backbone) superimposed on the native structures (thin backbone). (a) T0606-D1; (b) T0614-D1; (c) T0569-D1. The first template, best template in top 10, and the first Zhang-Server model are listed in the Left, Middle, and Right panel respectively. Blue to red runs from N- to C-terminal.
T0569 is a TBM target where I-TASSER simulation deteriorated the quality of the template (Figure 3c) where the first Zhang-Server model has a lower TM-score (0.46) than the initial template (0.54) in the threading aligned regions. However, if we examine the five submitted models, the fifth Zhang-Server model has a higher TM-score (0.57) than the initial template. The main reason is that most threading algorithms hit 2kvzA as their first hit (which has a TM-score of 0.4), while only 2 programs found the closest structural template 3i57A (TM-score=0.64). This example highlights the typical issue of model ranking in I-TASSER when the majority threading programs consistently hit the incorrect (usually the second best) template while the best template is hit only by the minority of the threading programs.
Human versus server predictions
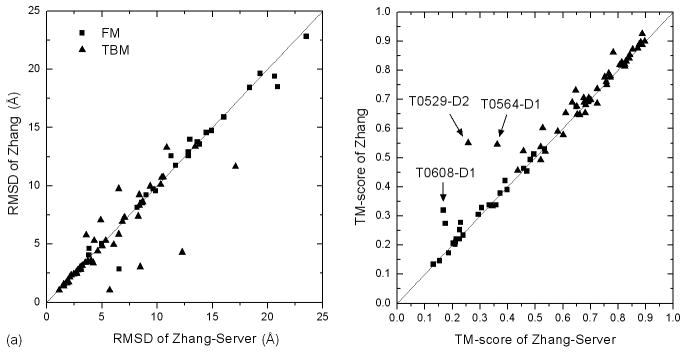
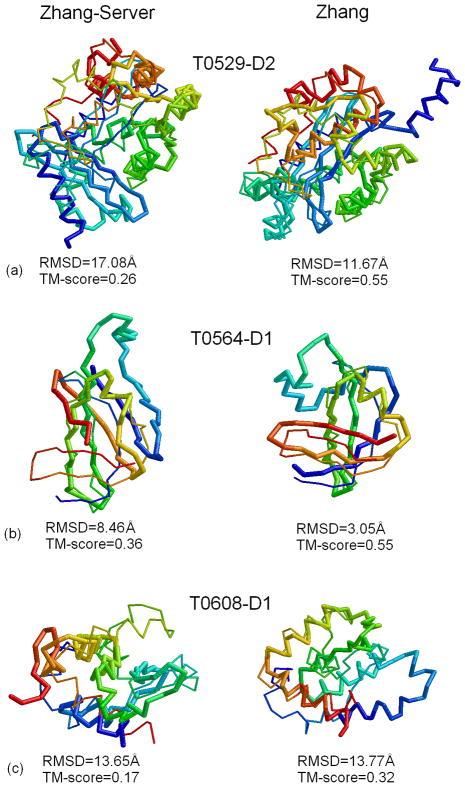
Although both Zhang and Zhang-Server predictions were based on an automated I-TASSER modeling procedure, human predictions have on average a higher TM-score mainly because the human predictions had a broad range of template selections from the CASP servers which have on average a better quality than the LOMETS threading, especially for FM targets. A head-to-head comparison between Zhang and Zhang-Server in Figure 4 shows how much the better templates influence the I-TASSER modeling results. Obviously there are more proteins, in which the human models have higher TM-score or lower RMSD than that for the sever models. The average TM-scores of the human models for the FM and TBM targets are improved by 5% and 3% respectively, over the server models. Next, we look into several cases (T0529, T0564 and T0608) in more detail which reveal the major reasons of the improvement in our human predictions.
Figure 4.
Comparison between the first predicted models by Zhang-Server and Zhang for 78 human targets.
T0529 is a large protein of 569 residues, consisting of two-domains. The N-terminal domain is a FM target, while the C-terminal domain is a TBM target. Based on whole-chain threading alignments generated by LOMETS, the query sequence was split into three-domains; however, all the domains were still classified as Hard targets by LOMETS. The final models were generated by assembling the models of the three individual domains. Since the sequence was split based on automated domain splitting procedure, the domain boundaries were defined incorrectly. Subsequently, correct templates for the second domain were not identified by LOMETS, resulting in a low accuracy final model (TM-score=0.26, left panel of Figure 5a). During the human prediction, it was noticed that multiple server models showed convergence in the second domain region and the domain boundaries were therefore correctly identified. After the correct domain splitting, correct templates like 1y97A were hit by multiple threading programs and the final model submitted by “Zhang” had a TM-score=0.55 (right panel of Figure 5a).
Figure 5.
Examples of structure modeling by Zhang-Server (Left) and Zhang (Right). (a) T0529-D2; (b) T0564-D1; (c) T0608-D1. Models (thick backbone) are superimposed on the native structure (thin backbone) with blue to red running from N- to C-terminal.
T0564 is a small single-domain beta protein (89 residues) which includes 2 beta-hairpins and 2 pairs of long-range beta sheets (Figure 5b). The first Zhang-Server model has a TM-score=0.36, while the second Zhang-Server model has a TM-score=0.56. The incorrect ranking of models is again because the best templates (1gutA and 1wjjA) were hit by only a minority of threading programs. In the human prediction, more correct templates from the CASP servers were included which helped to improve the accuracy of spatial restraints, resulting in the biggest I-TASSER cluster having the correct fold.
T0608 is a two-domain α+β protein of 279 residues. The first domain T0608-D1 is a short α-helical domain with no detectable templates, while the second domain is a TBM target with 2gu1A as the closest structure template. Zhang-Server attempted to fold this protein as a whole chain, which resulted in an incorrectly predicted topology (TM-score=0.17) for T0608-D1 (shown in the left panel of Figure 5c). During the human prediction, the target was split into two domains, because the I-TASSER ab initio routine is better suited for handling small single-domain proteins.23 Moreover, models generated by QUARK were of a better quality than the threading templates, which further improved the quality of spatial restraints, resulting in first human model with a much improved TM-score=0.32, although it was still far from satisfactory (right panel of Figure 5c).
Template re-ranking by QUARK model improves the quality of top templates
During the server modeling, threading alignments of 30 targets were re-ranked based on their structure similarity to QUARK ab initio models, in which 11 were TBM targets and 19 were FM targets. These are the targets which were judged by LOMETS as Hard targets. Nevertheless, there are indeed some targets which have good templates detected by some threading programs with low rank. In these cases, QUARK-based re-ranking may help improve the template selection. To evaluate the effect of re-ranking, in Table II we list the TM-scores of the templates with and without re-ranking. Average TM-score of the QUARK ab initio models used for re-ranking templates, which has an alignment coverage 100%, is shown in column 11.
Table II.
Comparison of threading templates with and without re-ranking.
| LOMETS templates | Templates with re-ranking | QUAd | Mode | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First | Besta | First | Besta | ||||||||
| Type | Nt | Covb | TMc | Covb | TMc | Covb | TMc | Covb | TMc | TMc | TMc |
| TBM | 11 | 85% | 0.361 | 89% | 0.507 | 97% | 0.437 | 95% | 0.522 | 0.380 | 0.542 |
| FM | 19 | 91% | 0.214 | 88% | 0.304 | 90% | 0.273 | 92% | 0.326 | 0.311 | 0.349 |
| All | 30 | 88% | 0.268 | 88% | 0.379 | 92% | 0.333 | 93% | 0.398 | 0.336 | 0.420 |
The best in top 10 templates.
Coverage of the threading alignment over the target sequence.
Abbreviation of TM-score.
The first model by QUARK ab initio folding.
The first submitted model in Zhang-Server.
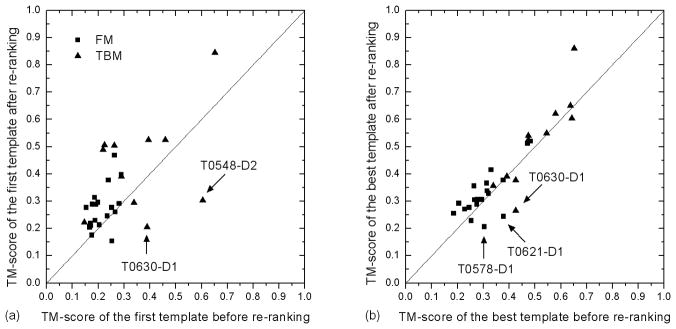
For the TBM targets, the average TM-score of the first template after re-ranking was increased from 0.361 to 0.437, showing an overall improvement of 21%. The absolute increase of TM-score is 0.076. However, the TM-score of the best in top 10 templates was improved only marginally from 0.507 to 0.522, an improvement of 3%. Figure 6 shows the scatter plot of TM-score of the QUARK re-ranked templates versus the original threading templates compared to the native structures. For the first template, only two TBM targets (T0548-D2 and T0630-D1) had an obviously worse TM-score than the original template (Figure 6a). For the best in top 10 templates, only T0630-D1 has an obviously lower TM-score (Figure 6b). If we compare the QUARK model and the re-ranked templates, the TM-score of the first template (0.437) is 15% even better than the TM-score of the full-length QUARK model (0.380) which was used as the reference to select the templates. As we observed, the QUARK ab initio models sometime matches only part of the template which are supposed to be correct; but the entire region of the selected template structure turns out to be correct, which has a higher TM-score than the QUARK models. Here, the part of the consensus structure between the ab initio model and the threading template served as a fingerprint to pick up the entire template.
Figure 6.
Comparison of the threading templates before and after re-ranking by QUARK models. (a) TM-score of the first template. (b) TM-score of the best in top 10 templates.
For the FM targets, since no threading alignments have a significant Z-score, the original ranking of templates based on Z-score is usually unreliable because the correlation of the TM-score and Z-score is very weak in this region of Z-score.7 In this situation, a structural similarity between threading template and the model built by ab initio simulations can be more meaningful. As shown in Table II, the TM-score of the templates sorted by the QUARK models is higher for both the first and the best templates than that of the original templates. Figure 6 shows that there are dominantly more points of Hard targets above the diagonal line. The overall TM-score improvements after template re-ranking for FM targets are 28% and 7% for the first and the best in top 10 templates, respectively (Table II).
There are only three targets, including two FM targets (T0578-D1 and T0621-D1) and one TBM target (T0630-D1), where the best template after re-ranking is worse than the best template in the original ranking. In all these cases, native structures contain several beta-sheets with long-range contacts. These examples highlight the inability of QUARK to fold beta-proteins of complex topology, because the current ab initio programs tend to build all beta-proteins into beta-hairpins of short-range contacts, which are often worse than those generated by threading procedure. Therefore, re-ranking procedure does not work well for the beta-proteins.
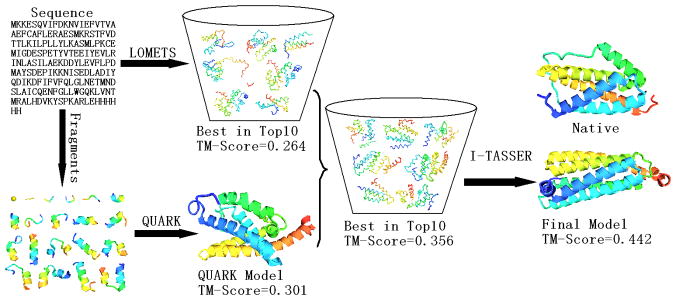
Figure 7 illustrates the procedure of I-TASSER modeling for the Hard targets by combining the LOMETS and QUARK-based template re-ranking. The shown example is from a FM target T0618, with 6 helices and 1 short beta-hairpin. The long loop (15–73) between the 2nd and 3rd helices spans around the helices 4 and 5, which constitute a helix bundle of complex topology. Before re-ranking, the top 10 templates found by LOMETS all have very low coverage and wrong topology with the best template has a TM-score=0.26. The ab initio model generated by QUARK has a TM-score=0.30 with the helices 1, 2, 6 approximately correctly packed. Subsequently, the LOMETS templates are sorted based on the TM-score to the QUARK model, resulting in the best template in top 10 having a TM-score=0.36. Furthermore, I-TASSER simulations are guided by restraints from QUARK model as well as re-ranked top templates, resulting in a much improved model with a TM-score=0.44. According to Table II, the average TM-score of the first I-TASSER model is 0.420 which is around 57% higher than the first LOMETS template for the 30 Hard targets.
Figure 7.
Flowchart of the I-TASSER simulation for Hard targets with LOMETS templates sorted by QUARK models. The example shown is from T0618-D1.
Ab initio contact prediction by SVMSEQ
The topology of protein structures can be specified by the residue-residue contact maps. For TBM targets, when most of the top threading alignments by LOMETS are correct, template-based contact maps have a high accuracy. For FM targets, the coverage and accuracy of the contact map are usually low since template alignments are often diverged and incorrect. Therefore, the sequence based contact predictions may become useful to protein structure predictions. Table III shows a comparison between the Cα and side-chain contacts by LOMETS and SVMSEQ40 for all the 147 domains. For TBM targets, the average accuracy and coverage by LOMETS are 0.632 and 0.728 for Cα, which are much higher than that of the SVMSEQ (0.348 and 0.279). For the FM targets, however, both the coverage and accuracy of the Cα and side-chain contacts by the SVMSEQ predictions are higher than that of the template-based contact predictions. Especially, there were a substantial number of new native contacts (18 Cα and 19 side-chain contacts) which were not predicted by LOMETS, which demonstrates a complement between the ab initio contact prediction and the template-based contact prediction. A combination of these two complementary contacts predictions will result in significantly improved accuracy of contact restraints which is essential for I-TASSER structure assembly.41
Table III.
Sequence-based versus template-based contact predictions.
| Cα contacts | Side-chain contacts | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOMETS | SVMSEQ | LOMETS | SVMSEQ | |||||||||
| NPa | Acc | Cov | Acc | Cov | NNb | NPa | Acc | Cov | Acc | Cov | NNb | |
| TBM | 308 | 0.632 | 0.728 | 0.348 | 0.279 | 5 | 371 | 0.626 | 0.554 | 0.284 | 0.193 | 15 |
| FM | 206 | 0.143 | 0.185 | 0.194 | 0.197 | 18 | 243 | 0.183 | 0.133 | 0.185 | 0.149 | 19 |
| All | 287 | 0.532 | 0.617 | 0.317 | 0.263 | 8 | 345 | 0.536 | 0.468 | 0.264 | 0.184 | 16 |
Number of contact pairs in native structure.
Number of new native contact pairs predicted by SVMSEQ but not by LOMETS.
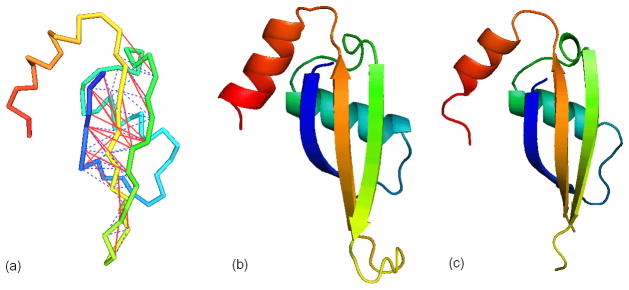
T0604-D1, the N-terminal domain of VP0956 protein from Vibrio parahaemolyticus, is a typical example showing the help of the sequence-based contact prediction on protein structure modeling. This domain is a FM target and most ab initio folding programs failed to fold this protein, mainly because of long-range paired beta-sheets. However, Zhang-Server built a high-resolution model with a RMSD 2.66Å to native. The whole chain threading of T0604 by LOMETS has no reliable alignment and only 9 out of 92 Cα contact predictions was correct, where the total number of contacting pairs (Cα distance <8Å) in the native structure is 121. Even running LOMETS on the T0604-D1 sequence, the accuracy of template-based Cα contacts is only 15.2%. SVMSEQ predicts 97 contacting pairs, out of which 63 are correct, and 55 are new contacts. The correct and wrong contacts are shown in red and blue respectively in Figure 8a. The correct contacts mainly occur along the beta pairs and some within the loop regions. By comparing the Zhang-Server model and the native structure in Figures 8b and 8c, it is observed that the three beta strands are almost perfectly packed and the two alpha helices in the model have exactly the same orientations as in the native structure.
Figure 8.
Structure modeling result for the FM target T0604-D1. (a) Contact predictions by SVMSEQ with the true positive contacts in solid red lines and the false positive contacts in dash blue lines. (b) The first Zhang-Server model. (c) The experimental structure.
Model quality improved by FG-MD
The models generated after I-TASSER simulation are reduced protein models, where each residue is represented by its Cα and side-chain center of mass. Construction of full atomic models from the Cα traces, while retaining the global topology, is a non-trivial problem, especially, considering that the SPICKER cluster centroids are average structures and contain severely distorted local structures.
There are a number of available tools which can be used for atomic structure construction and refinement.26,48–50 In Table IV, we show a comparison between the quality of the final models constructed from SPICKER cluster centroids by three different procedures, i.e. PULCHRA,50 REMO,26 and FG-MD,27 where FG-MD started the atomic-level refinement from the REMO models. Since all the targets were submitted by Zhang-Server as full-length models, for simplicity, the evaluation is done on 116 full-length targets without splitting them into domains.
Table IV.
Model quality by PULCHRA, REMO and FG-MD on 116 targets.
| RMSD(Å) | TM-score | #HB | HB-score | Clash-score | MPscore | |
|---|---|---|---|---|---|---|
| PULCHRA | 7.658 | 0.663 | 128 | 0.349 | 331.2 | 4.653 |
| REMO | 7.656 | 0.664 | 137 | 0.371 | 250.6 | 4.247 |
| FG-MD+REMO | 7.364 | 0.673 | 193 | 0.478 | 9.0 | 2.942 |
Although the PULCHRA and REMO models on average have similar RMSD and TM-score, REMO models include 7% more hydrogen bonds than PULCHRA models, where H-bonds are calculated by HBPLUS.51 If we define HB-score as the ratio of the number of the native H-bonds in the model to the total number of H-bonds in the native structure, the HB-score of the REMO models is 6.3% higher than that of the PULCHRA models. Compared to REMO, the models generated after FG-MD refinement show a small improvement in RMSD (by 0.29Å) and TM-score (by 1%), while the HB-score of the models increased on average by 28.8%; this is mainly attributed to the additional H-bonding energy terms introduced in the MD simulations.27
Another important ability of the atomic structure construction is to remove the steric clashes. To evaluate this ability, we define a clash-score which counts the total number of clashes between every pair of atoms including hydrogen atoms. Here, we use HAAD52 to add hydrogen atoms in all three models by PULCHRA, REMO and FG-MD. Since none of these models are specifically optimized for clashes based on HAAD H-atoms, this comparison should be more objective than by considering the clashes of heavy atoms only. As a result, PULCHRA has the weakest ability in excluding the steric clashes, which has on average 331.2 clashed pairs in the model. The REMO models have 32% less clashes (250.6) than the PULCHRA models, while FG-MD further reduces the number of clashes by 28 times compared to REMO. Most of the clashed pairs in FG-MD were from the H-atoms added by the HAAD program, which are not optimized by MD simulations. The average number of clashes of our submitted models is 9.0, which is comparable to 10.3, the number of clashes in CASP9 experimental structures whose hydrogen atoms are also added by HAAD.
Finally, we exploit the standard MolProbity program53 to evaluate the overall quality of our atomic models. MolProbity provides a MPscore for each structural model, which is a log-weighted combination of the number of various structural outliers, including side-chain rotamer outliers, Ramachandran outliers, and steric clashes. A structure with a numerically lower MPscore indicates better quality.53 As shown in Table IV, the average MPscore of the FG-MD models is 2.942 which is 44% lower than that of the REMO models where the MPscore of the REMO model is 10% lower than that of the models by PULCHRA.
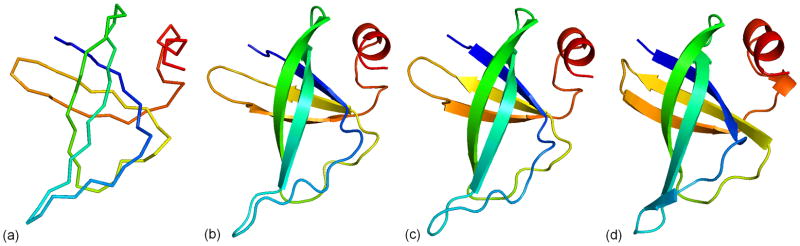
In Figure 9, we show an example from Target T0530 to demonstrate the procedure of atomic structure construction and refinement. The protein has a coiled beta-hairpin structure (Figure 9d). Figure 9a shows the starting Cα trace model generated after SPICKER clustering. This model has a high TM-score of 0.811 to native, but contains 44 Cα clashes. Starting from this initial structure, full-atomic model is generated by REMO (Figure 9b), which has a similar TM-score of 0.809 to native and 33 hydrogen bonds (HB-score=0.44). Although REMO removed all the Cα clashes in the model, it introduced 82 atomic clashes between other heavy atoms. Figure 9c shows the final model generated after FG-MD refinement simulation, starting from the REMO model. The FG-MD simulations not only removed all the atomic clashes, but also improved the main-chain topology (TM-score=0.818). The number of H-bonds in the FG-MD model is also higher with an improved HB-score of 0.49.
Figure 9.
Modeling result of the target T0530. (a) Cα trace generated by the SPICKER cluster centroid. (b) Full atomic model by REMO from the Cα trace. (c) Final model refined by FG-MD. (d) The experimental structure.
Despite the ability of FG-MD in improving the local structures, the average improvement on the global backbone topology by FG-MD is small (with a TM-score increasing ~0.2% compared to the I-TASSER models). The major driven force for the observed template improvements is attributed to the use of multiple threading templates.3
Discussions
The I-TASSER CASP9 pipeline, without human intervention, showed encouraging results on protein tertiary structure modeling, which is required for the genome-scale applications. Compared to the last CASP experiments, progress was observed in ab initio folding and high-resolution refinement with the development of QUARK and FG-MD. For distantly homologous proteins, QUARK ab initio models provided a reasonable structural framework for generating spatial restraints and for re-ranking of threading templates; while sequence-based ab initio contact predictions by SVMSEQ further helped guide the I-TASSER fragment assembly simulations. Finally, FG-MD improved the local quality and sometimes main-chain topology of the predicted models, thus improving the biological usability of the predicted structures.
Generally, threading-based structural assembly method works well when appropriate templates are detected, while QUARK-based ab initio folding generates better models for free-modeling targets. However, correct determination of target type (TBM or FM) is critical for choosing the appropriate methodology for structure modeling. The challenge is at the weakly homologous modeling region, where reasonable templates can be available even when the threading alignment scores are low. Correspondingly, these templates are often ranked low. We found that a combination of the threading alignment score (Z-score) and the structural similarity (i.e. average pairwise TM-score) between the templates identified by different threading programs provides a more accurate classification of the targets.15 For FM targets that contain beta-sheets with long-range contacts and complex topology, we observed that the template-based modeling generates better results than ab initio folding. This on one hand advises us the way to choose the appropriate method for FM target modeling; on the other hand, it highlights the inability of ab initio folding method to model complicated beta-sheet topology, which is the major problem we want to solve in the next step of QUARK development. One possible way to address the problem is to use the predicted residue contacts (e.g. by SVMSEQ) or beta-sheets (e.g. by BETApro54 or ASTRO-FOLD55–56) to guide the fold assembly simulation. In the first principles method ASTRO-FOLD, which performs ab initio folding without using database templates, hydrophobic contacts are maximized for the prediction of beta-sheet topology through solving a combinatorial optimization problem. The predicted contacts have demonstrated the ability of improving the tertiary structure ensemble of beta and alpha+beta proteins.
Model selection is a classic issue in the CASP experiment. I-TASSER has the advantage in refining the models, which are on average significantly better than the initial threading templates; mainly attributed to the use of consensus spatial restraints collected from multiple templates. However, I-TASSER sometimes fails to select the best model as the first model, when the best template is detected only by minority of threading programs and the majority of the threading alignments consistently hit an incorrect template. Here, the second condition is essential for I-TASSER’s failure in selecting the best template, while it was observed that I-TASSER can often pick up the best template if the templates by the majority of the threading programs are diverged. When the majority of the threading programs hit a common (incorrect or second best) template, the consensus restraints can be too strong and distract the template selection of the I-TASSER modeling.
Incorrect domain splitting is another long-standing issue in protein structure prediction. Threading-based domain splitting, i.e. based on template structure and unaligned regions in threading alignment, has been shown as a powerful method for domain detection. However, for the extremely difficult targets, iterative threading might become necessary. T0529 is one such example, where the I-TASSER server infers incorrect domain boundary because most of the LOMETS programs generate weak alignments for the entire query sequence. In the human prediction, the region of 378–569 emerges as an independent domain with the alignments of a higher confident score in the second round of LOMETS when threading was run on the roughly split sequence based on the first round; this eventually results in the correct detection of the C-terminal domain, a template-based domain target.
Several protein targets in CASP9 were solved in their quaternary structure form. For example, T0629-D2 is a long tail fiber protein from bacteriophage T4, with three identical protein chains intertwined together to form an elongated six-stranded antiparallel beta-strand structure. The core of this structure is stabilized by the alternate hydrophilic and hydrophobic regions, where the hydrophilic residues also form coordination site for seven iron ions. All the structure modeling methods failed to generate a reasonable structure for this domain, highlighting the need to extend the current tertiary structure modeling method for quaternary structure modeling to model these complex protein structures.
Acknowledgments
The project is supported in part by the NSF Career Award (DBI 0746198), and the National Institute of General Medical Sciences (GM083107, GM084222).
Abbreviations
- RMSD
root mean squared deviation
- MD
molecular dynamics
References
- 1.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(S8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 2.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77 (Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 5.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72(2):547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng J, Xu J. Boosting Protein Threading Accuracy. Lect Notes Comput Sci. 2009;5541:31. doi: 10.1007/978-3-642-02008-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19(8):1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struct Biol. 2008;8:18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Wang Q, Barz B, He Z, Kosztin I, Shang Y, Xu D. MUFOLD: A new solution for protein 3D structure prediction. Proteins. 2010;78(5):1137–1152. doi: 10.1002/prot.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floudas CA. Computational methods in protein structure prediction. Biotechnol Bioeng. 2007;97(2):207–213. doi: 10.1002/bit.21411. [DOI] [PubMed] [Google Scholar]
- 12.Fischer D. 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51(3):434–441. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(S8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 14.Arakaki AK, Zhang Y, Skolnick J. Large scale assesment of the utility of low resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y. Protein structure prediction: when is it useful? Curr Opin Struct Biol. 2009;19(2):145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69(S8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 17.Ben-David M, Noivirt-Brik O, Paz A, Prilusky J, Sussman JL, Levy Y. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77 (Suppl 9):50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- 18.Floudas CA, Fung HK, McAllister SR, Monnigmann M, Rajgaria R. Advances in Protein Structure Prediction and De Novo Protein Design: A Review. Chemical Engineering Science. 2006;61:966–988. [Google Scholar]
- 19.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowie JU, Eisenberg D. An evolutionary approach to folding small alpha-helical proteins that uses sequence information and an empirical guiding fitness function. Proc Natl Acad Sci U S A. 1994;91(10):4436–4440. doi: 10.1073/pnas.91.10.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268(1):209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 22.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77(S9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009;76(3):665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Zhang Y. High-resolution protein structure refinement using fragment guided molecular dynamics. 2010. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu D, Zhang Y. QUARK ab initio protein structure prediction I: Method developments. 2010. Submitted. [Google Scholar]
- 29.Xu D, Zhang Y. QUARK ab initio protein structure prediction II: results of benchmark and blind tests. 2010. Submitted. [Google Scholar]
- 30.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Xu D. Protein threading using PROSPECT: design and evaluation. Proteins. 2000;40(3):343–354. [PubMed] [Google Scholar]
- 32.Karplus K, Karchin R, Draper J, Casper J, Mandel-Gutfreund Y, Diekhans M, Hughey R. Combining local-structure, fold-recognition, and new fold methods for protein structure prediction. Proteins. 2003;53 (Suppl 6):491–496. doi: 10.1002/prot.10540. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Zhou Y. Single-body residue-level knowledge-based energy score combined with sequence-profile and secondary structure information for fold recognition. Proteins. 2004;55(4):1005–1013. doi: 10.1002/prot.20007. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58(2):321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci USA. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: a new approach to ab initio protein structure prediction. Biophys J. 2003;85(2):1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proc Natl Acad Sci U S A. 2006;103(8):2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33(10):3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics. 2008b;24(7):924–931. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Szilagyi A, Zhang Y. Improving protein structure prediction using multiple sequence-based contact predictions. 2010. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Zhang Y. Recognizing protein substructure similarity using segmental threading. Structure. 2010;18(7):858–867. doi: 10.1016/j.str.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem. 2004;25(6):865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plimpton S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J Comput Phys. 1995;117:1–19. [Google Scholar]
- 46.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 47.Xu D, Zhang Y. What is the optimal fragment length for ab initio protein structure assembly? 2010. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J Mol Biol. 1991;218(1):183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 49.Levy-Moonshine A, Amir el AD, Keasar C. Enhancement of beta-sheet assembly by cooperative hydrogen bonds potential. Bioinformatics. 2009;25(20):2639–2645. doi: 10.1093/bioinformatics/btp449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotkiewicz P, Skolnick J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J Comput Chem. 2008;29(9):1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Roy A, Zhang Y. HAAD: A quick algorithm for accurate prediction of hydrogen atoms in protein structures. PLoS One. 2009;4(8):e6701. doi: 10.1371/journal.pone.0006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng J, Baldi P. Three-stage prediction of protein beta-sheets by neural networks, alignments and graph algorithms. Bioinformatics. 2005;21 (Suppl 1):i75–84. doi: 10.1093/bioinformatics/bti1004. [DOI] [PubMed] [Google Scholar]
- 55.Klepeis JL, Floudas CA. ASTRO-FOLD: a combinatorial and global optimization framework for Ab initio prediction of three-dimensional structures of proteins from the amino acid sequence. Biophys J. 2003;85(4):2119–2146. doi: 10.1016/s0006-3495(03)74640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajgaria R, Wei Y, Floudas CA. Contact prediction for beta and alpha-beta proteins using integer linear optimization and its impact on the first principles 3D structure prediction method ASTRO-FOLD. Proteins. 2010;78(8):1825–1846. doi: 10.1002/prot.22696. [DOI] [PMC free article] [PubMed] [Google Scholar]