Abstract
Previous studies of mRNA for classical glutathione peroxidase 1 (GPx1) demonstrated that hepatocytes of rats fed a selenium-deficient diet have less cytoplasmic GPx1 mRNA than hepatocytes of rats fed a selenium-adequate diet. This is because GPx1 mRNA is degraded by the surveillance pathway called nonsense-mediated mRNA decay (NMD) when the selenocysteine codon is recognized as nonsense. Here, we examine the mechanism by which the abundance of phospholipid hydroperoxide glutathione peroxidase (PHGPx) mRNA, another selenocysteine-encoding mRNA, fails to decrease in the hepatocytes and testicular cells of rats fed a selenium-deficient diet. We demonstrate with cultured NIH3T3 fibroblasts or H35 hepatocytes transiently transfected with PHGPx gene variants under selenium-supplemented or selenium-deficient conditions that PHGPx mRNA is, in fact, a substrate for NMD when the selenocysteine codon is recognized as nonsense. We also demonstrate that the endogenous PHGPx mRNA of untransfected H35 cells is subject to NMD. The failure of previous reports to detect the NMD of PHGPx mRNA in cultured cells is likely attributable to the expression of PHGPx cDNA rather than the PHGPx gene. We conclude that 1) the sequence of the PHGPx gene is adequate to support the NMD of product mRNA, and 2) there is a mechanism in liver and testis but not cultured fibroblasts and hepatocytes that precludes or masks the NMD of PHGPx mRNA.
INTRODUCTION
mRNAs for selenoproteins harbor one or more UGA codons for the nonstandard amino acid selenocysteine (Sec; Stadtman, 1996; Sunde, 1997). Sec incorporation requires a cis-acting mRNA stem-loop structure termed the selenocysteine insertion sequence (SECIS) element, at least one if not several of which are located in the 3′ untranslated region (UTR) of mammalian selenoprotein mRNAs (reviewed in Low and Berry, 1996; Atkins et al., 1999), as well as a number of trans-acting factors that govern generation or function of the specialized selenocysteyl-tRNASec (Lee et al., 1989; Low et al., 1995, 2000; Ding and Grabowski, 1999; Copeland et al., 2000). One important determinant of Sec codon usage as a site for Sec incorporation is the concentration of dietary selenium (Se), an essential trace element required for the synthesis of selenocysteyl-tRNASec (reviewed in Stadman, 1996). Prolonged Se deficiency reduces the abundance and activities of all selenoproteins to extents that vary with the particular protein and tissue in which the protein is expressed, consistent with the importance of Sec to selenoprotein synthesis and enzyme activity (reviewed in Sunde, 1997).
Because the Se-dependent incorporation of Sec at a UGA codon by the SECIS pathway is thought to be in competition with translation termination, it is possible that all selenoprotein mRNAs are natural substrates for the mRNA decay pathway called mRNA surveillance or nonsense-mediated decay (NMD; Maquat, 1995, 2000; Li and Wilkinson, 1998; Hentze and Kulozik, 1999). The effect of Se deficiency on selenoprotein mRNA abundance has been most thoroughly studied for classical glutathione peroxidase 1 (GPx1) mRNA, the half-life of which has been found to decrease in the liver of intact animals and in every cultured cell tested (Chada et al., 1989; Hill et al., 1992; Bermano et al., 1995, 1996a; Weiss and Sunde, 1997, 1998; Moriarty et al., 1998). Regardless of the Se concentration, GPx1 mRNA abundance decreases when the sole Sec codon is converted to a UAA nonsense codon, which almost always directs translation termination, and increases when the Sec codon is converted to a UGC cysteine codon, which almost never directs translation termination, indicating that translation termination at the Sec codon results in NMD (Moriarty et al., 1998). Notably, NMD is the only mechanism by which Se concentration detectably affects GPx1 mRNA abundance (Moriarty et al., 1998). According to a recently established rule, termination codons elicit NMD when located >50–55 nucleotides (nt) upstream of the 3′-most exon-exon junction of an mRNA (Nagy and Maquat, 1998, and references therein). Consistent with this rule, 1) the Sec codon of GPx1 mRNA resides 105 nt upstream of the sole exon-exon junction, 2) moving the intron to a position 59 nt downstream of the Sec codon still allows for NMD, but 3) moving the intron to a position 43 or 15 nt downstream or 83 nt upstream of the Sec codon eliminates NMD (Sun et al., 2000).
Despite the prediction that all selenoprotein mRNAs could be natural substrates for NMD, not all appear to be. As an example, although severe Se deficiency in rats (0.003 mg of Se/kg) decreases the abundance of GPx1 mRNA in liver and heart to barely detectable levels, the level of mRNA for phospholipid hydroperoxide glutathione peroxidase (PHGPx) in the two tissues is apparently unaffected and in the thyroid is increased by ∼50% (Bermano et al., 1995). A priori, these data indicate that PHGPx mRNA is resistant to NMD. Given that the sole Sec codon resides 105 nt upstream of the third of six exon-exon junctions, it may be an exception to the rule established for which termination codons elicit NMD.
To investigate the mechanism for the apparent immunity of PHGPx mRNA to NMD, the sole TGA Sec codon within the pig PHGPx gene was converted to either a TAA nonsense codon or a TGT cysteine codon. Results demonstrate that PHGPx mRNA is, in fact, a substrate for NMD in NIH3T3 fibroblasts or H35 hepatocytes. Furthermore, Se deficiency augments the efficiency with which Sec codon-containing mRNA is subject to NMD. NMD is not attributable to the experimental conditions under which the pig PHGPx gene was expressed because Se deficiency also augments the efficiency with which the endogenous PHGPx mRNA of H35 cells is subject to NMD. Therefore, the absence of a detectable change in PHGPx mRNA abundance in either the liver or testis of Se-deficient rats must be attributable to a mechanism, absent in mouse NIH3T3 fibroblasts and rat H35 hepatocytes, that either masks or precludes the NMD of PHGPx mRNA.
MATERIALS AND METHODS
Rats, Diets, and Insolation of Hepatocytes and Testes
Long-Evans hooded rats were fed Se-deficient or Se-supplemented diets, and hepatocytes were isolated from perfused liver as previously described (Moriarty et al., 1998). Testes were removed and frozen in liquid nitrogen.
Generation of pmCMV-PHGPx, pPHGPx, pSP-rPHGPx, and Mutagenized Derivatives
The full-length pig PHGPx gene was removed as a ∼10,000-kbp NotI-NotI fragment from a bacteriophage λ vector (kindly provided by Leopold Flohé, Braunschweig, Germany) and inserted into the NotI site of pBluescript SK+ (Stratagene, La Jolla, CA). To generate pmCMV-PHGPx, the pig PHGPx gene in pBluescript SK+ was polymerase chain reaction (PCR)-amplified as two fragments: a 1.0-kbp fragment that extends from the 5′ UTR into exon 2 by using primers 5′-CTGCGGGATCCTGGCG-3′(sense, where underlined nucleotides specify a BamHI site and italicized nucleotides are mutagenic) and 5′-GGCTGAGAATTCGTGC-3′(antisense, where underlined nucleotides specify an EcoRI site), and a 1.4-kbp fragment that extends from exon 2 into 3′-flanking DNA by using primers 5′-GCACGAATTCTCAGCC-3′ (sense, where underlined nucleotides specify an EcoRI site) and 5′-CAGGGGTGAGAAGCTTACCGGC-3′(antisense, where underlined nucleotides specify a HindIII site). The two fragments were digested with BamHI and EcoRI or EcoRI and HindIII, respectively, and then were inserted simultaneously into the BamHI and HindIII sites of pBluescript-KS(−). The construction of pmCMV-PHGPx was completed by inserting the 550-bp XbaI-BamHI fragment that includes the entire mouse cytomegalovirus (mCMV) promoter into the XbaI and BamHI sites. Codon 72 was changed from TGA to either TGT or TAA by oligonucleotide-directed mutagenesis (by using antisense primers 5′-CGTCTTGCCACATTGAG-3′ or 5′-CGTCTTGCCTTATTGAG-3′, respectively). Nonsense mutations within codon 28 or 34 were introduced using mutagenic antisense primers 5′ GGGACGCTTACTGCAAGG 3′ or 5′ CACATCGCTAGTCGTCG 3′, respectively.
To generate pPHGPx, the 1.65-kbp BamHI-EcoRI fragment that extends from 480 bp upstream of the 5′-most series of pig PHGPx transcription initiation sites into exon 2 was cloned into the corresponding sites of pBluescript-KS(−). The resulting 1.42-kbp XbaI-AvrII fragment that extends from upstream of the pig PHGPx promoter (i.e., from vector sequences) into intron 1 was used to replace into the corresponding region of pmCMV-PHGPx in the context of 72 (TGA), 72(TGT), 72(TAA), 28Ter, or 34Ter.
pSP-rPHGPx(TGA), which harbors wild-type rat PHGPx cDNA driven by the SP6 RNA promoter, was generated by inserting the Klenow-filled 697-bp BanI-XhoI fragment, which extends from 5 bp upstream of ATG(27) into the 3′ UTR, at the Klenow-filled XbaI and SacI sites of pGEM-4Z (Promega, Madison, WI).
Coupled Transcription-Translation of pSP-rPHGPx cDNAs In Vitro
One of several test pSP-rPHGPx cDNA plasmids (5 μg) and reference luciferase DNA (1 μg; Promega) were mixed with the TNT Wheat Germ Extract System (25 μl; Promega) so as to generate 35S-labeled protein after transcription by SP6 RNA polymerase. To remove 35S-labeled proteins that comigrate with PHGPx, a portion (12 μl) of each reaction was brought to pH 5 by using 1 M acetic acid (16.2 μl), incubated on ice for 30 min, and subsequently centrifuged at 12,000 × g and 4°C for 15 min. Pellets were dissolved in 60 μl of 1× SDS sample buffer (Promega), and a portion (10 μl) was denatured at 80°C and electrophoresed in a 10% SDS-polyacrylamide gel. 35S-labeled PHGPx and luciferase were quantitated using Phosphor-Imaging (Molecular Dynamics, Sunnyvale, CA) and visualized using autoradiography. The level of PHGPx was divided by the number of methionine residues per molecule to control for variations between the length of different PHGPx proteins and then normalized to the level of luciferase to control for labeling variations between reactions.
Cell Transfections
For experiments that did not vary Se concentration, mouse NIH3T3 fibroblast and rat H35 hepatoma cell lines were maintained in minimal essential medium (MEM) (Life Technologies, Gaithersburg, MD) containing 10% fetal bovine serum (FBS). Cells were transiently transfected with the designated plasmid DNAs (pmCMV-PHGPx or pPHGPx test plasmids [1.4 μg] and the pmCMV-TPI reference plasmid [0.6 μg]) by using either calcium phosphate (Moriarty et al., 1998) or LipofectAMINE PLUS Reagent (Life Technologies; Sun and Maquat, 2000). For experiments in which 1 μg of either pCI-Neo-hUPF1 Wt or pCI-Neo-hUPF1 R844C (Maquat and Li, 2001) were included, the amounts of transfecting pmCMV-PHGPx and pmCMV-Gl (Zhang et al., 1998b) were 1 and 0.2 μg, respectively. For experiments that varied Se concentration, cells were transfected as described above and transferred 12 h later to Se-deficient or Se-supplemented Dulbecco's MEM. Se-supplemented medium contained 7 ng/ml sodium selenite, 5 μg/ml insulin, and 5 μg/ml transferrin (Figure 6; Bermano et al., 1996a) or 0.7 or 3.5 ng/ml sodium selenite, and either 10% FBS or 5 μg/ml insulin and 5 μg/ml transferrin (Figure 7).
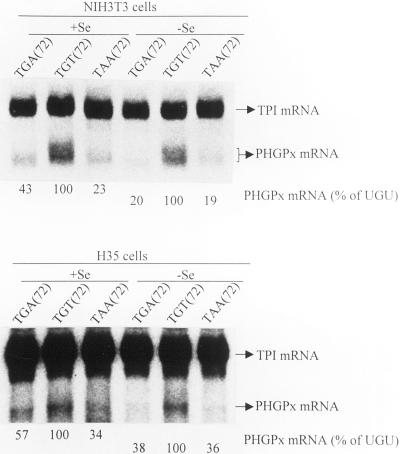
Figure 6.
Se deficiency augments the NMD of PHGPx mRNA harboring the Sec codon in NIH3T3 and H35 cells. NIH3T3 and H35 cells were treated and analyzed as described in the legend to Figure 4 except that the cells were cultured 12 h after transfection in either Se-deficient (−Se) or Se-supplemented (+Se) medium. Values did nor vary by >5–7% in two independently performed experiments.
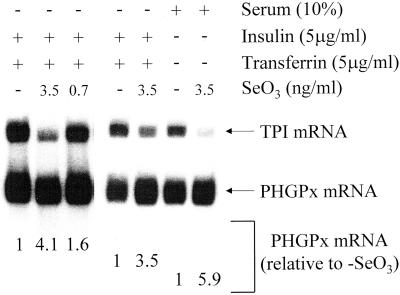
Figure 7.
Se deficiency also augments the NMD of endogenous PHGPx mRNA in untransfected H35 cells. Untransfected H35 cells were cultured as described in the legend to Figure 6 except that insulin and transferrin were replaced by 10% FBS where specified and the concentration of sodium selenite was as specified. For each lane, the level of endogenous PHGPx mRNA was normalized to the level of endogenous TPI mRNA, and normalized levels were expressed relative to the level in the absence of sodium selenite (SeO3), which was defined as one for cells cultured in insulin + transferrin as well as cells cultured in the presence serum.
RNA Isolation, Northern Blot Hybridization, and RT-PCR Analysis
Total or nuclear and cytoplasmic RNAs were isolated from hepatocytes and cultured cells as described (Moriarty et al., 1998). Total testis RNA was isolated by thawing the frozen testes and extraction in Trizol reagent (Life Technologies). Northern blot hybridizations and the coupled reverse transcription-PCR (RT-PCR) were performed as described (Moriarty et al., 1998; Zhang et al., 1998b). However, rat PHGPx RNA was detected by blot hybridization by using a uniformly labeled 217-bp NcoI-XhoI fragment that extends from exon 6 into the 3′ UTR of rat PHGPx cDNA (Figure 1) or a ∼750-bp EcoRI-EcoRI fragment of rat PHGPx cDNA (Figure 7), pig PHGPx RNA was detected by blot hybridization using a uniformly labeled 230-bp AlwNI-AlwNI fragment from the pig PHGPx gene that consists of 110-bp of exon 7 plus 3′-flanking DNA, rat PHGPx RNA was detected by RT-PCR using primers 5′-CGGCCTAAGGCCCTACAAGTGTTGG-3′ (sense) and 5′-GGGTGGACGAGACCAGACCTGGAAGGAGGC-3′ (antisense), and rat triosephosphate isomerase (TPI) RNA was detected by blot hybridization by using an 830-bp NcoI-NdeI fragment from human TPI cDNA (Maquat et al., 1985).
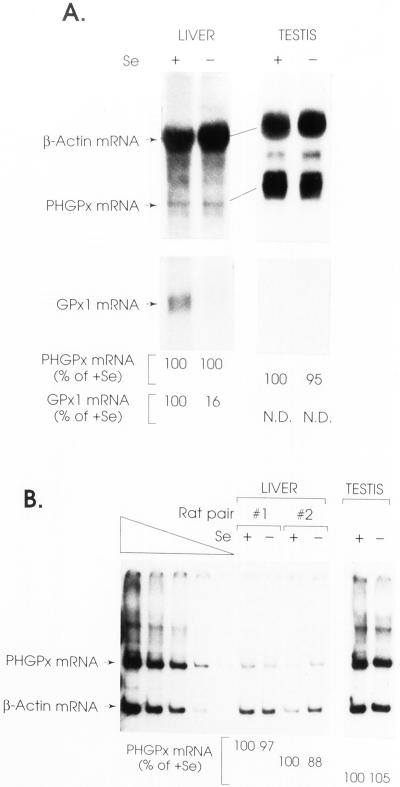
Figure 1.
Hepatocytes and testes from rats fed a Se-deficient diet for 12 wk have the same level of PHGPx mRNA as the corresponding tissues from rats fed a Se-supplemented diet. Rat pairs and hepatocyte RNAs were identical to those described in Moriarty et al. (1998). (A) Total RNA (25 μg) was electorphoresed in agarose, transferred to a nylon membrane, and hybridized to 32P-labeled cDNAs for rat PHGPx (a gift from Donna Driscoll; Pushpa-Rekha et al., 1995), β-actin (Moriarty et al., 1998), and GPx1 (Moriarty et al., 1998). The levels of PHGPx and GPx1 mRNAs were normalized to the level of β-actin mRNA and presented as a percentage of the normalized level in Se-fed animals, which was defined as 100%. N.D., not detectable. Results are shown for one pair of animals. (B) RT-PCR analysis of PHGPx and β-actin mRNAs was as described (Moriarty et al., 1997, 1998), except that PHGPx mRNA was analyzing using primers described in MATERIALS AND METHODS. The left-most four lanes assay a serial dilution of testis RNA and demonstrate that the analysis is quantitative.
RESULTS
Se Deficiency Fails to Reduce the Abundance of PHGPx mRNA in Rat Liver and Testis
Having demonstrated previously that the level of classical GPx1 mRNA is reduced in the liver of rats fed a Se-deficient diet relative to the level in the liver of rats fed a Se-supplemented diet (Moriarty et al., 1998), we analyzed the levels of PHGPx mRNA in the liver and testes of the same rats. Quantitations of β-actin mRNA, which is insensitive to Se concentration (Moriarty et al., 1998), and GPx1 mRNA, which varies with Se concentration, served as controls. The level of PHGPx mRNA, measured by Northern blot hybridization or a coupled RT-PCR, was virtually unchanged in either tissue by the change in dietary Se concentration (Figure 1), consistent with similar studies that also used rats (Bermano et al., 1995; Lei et al., 1995). As shown previously, the level of GPx1 mRNA in the liver of Se-deficient rats was reduced to 16% the level in the liver of Se-supplemented rats (Bermano et al., 1995; Lei et al., 1995; Moriarty et al., 1998), and in testis the level of PHGPx mRNA was significantly higher than the level of GPx1 mRNA (Lei et al., 1995; Figure 1). The insensitivity of PHGPx mRNA abundance to Se concentration is unexpected for three reasons. First, rat PHGPx and rat GPx1 mRNAs are 43.5% identical (Ho et al., 1988; Pushpa-Rekha et al., 1995). Second, at least for the PHGPx gene of pig, which is the only characterized PHGPx gene, the sole Sec codon resides within exon 3 and is followed by four introns, all of which reside >50–55 bp downstream of the Sec codon (Brigelius-Flohéet al., 1994). In fact, the distance between the Sec codon and the closest downstream intron for both PHGPx and GPx1 genes is 105 bp, and this intron has been shown to be critical for the NMD of GPx1 mRNA (Moriarty et al., 1997; Weiss and Sunde, 1998; Sun et al., 2000). Third, based on studies of GPx1 mRNA translation (Moriarty et al., 1998), there is no reason to think that the cotranslational insertion of Sec into PHGPx protein would not be in competition with translation termination and, therefore, NMD.
Evidence That the Apparent Resistance of PHGPx mRNA to NMD Is Not Due to Accumulation of Nascent Peptide at the Codon Preceding the Sec Codon
In theory, resistance to NMD could arise if translation fails to terminate when Sec failed to be incorporated at the UGA Sec codon. For example, release factors, which are likely to be required for NMD (Czaplinski et al., 1998), may not be involved if the nascent PHGPx peptide fails to be released from its mRNA template when the Sec codon reaches the A site of the translating ribosome. A precedent for this is provided by the 22-codon upstream open reading frame (uORF2) of the human cytomegalovirus UL4 transcript leader. The Geballe lab has shown that translation of uORF2 functions in cis to inhibit translation of the downstream ORF: the nascent uORF2 peptide accumulates linked to tRNAPro, the tRNA that decodes the final codon of uORF2, and results in the stalling of ribosomes at the end of uORF2 (Degnin et al., 1993; Cao et al., 1996a,b; 1998). For a similar situation to apply to PHGPx mRNA, incorporation of Sec at the UGA codon would have to relieve stalling and allow for synthesis of full-length protein.
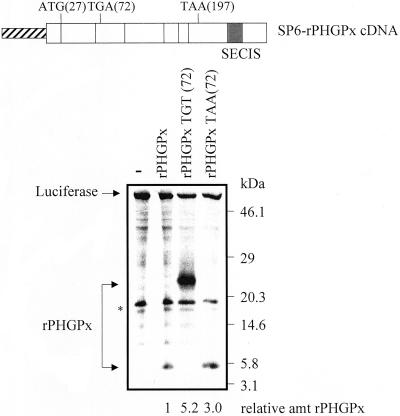
To determine whether peptide release is inhibited at the UGA Sec codon of PHGPx mRNA when the Sec codon is recognized as nonsense, PHGPx mRNA initiating translation at the second [AUG(27)] of two initiation codons [where the first is AUG(0)] and harboring the UGA(72) Sec codon or either a UGU (cysteine; Cys) or UAA (nonsense) codon in its place were synthesized and translated in vitro. In rat, the second initiation codon is the predominant site of translation initiation in somatic cells (Pushpa-Rekha et al., 1995). The first initiation codon is used in testis (Pushpa-Rekha et al., 1995) and thought to allow targeting to mitochondria, subsequent cleavage by a mitochondrial peptidase and, finally, targeting to the mitochondrial intermembrane space (Roveri et al., 1992). Notably, both somatic-cell (liver) and germ-cell (testis) PHGPx mRNA is immune to NMD (Figure 1). Experiments with a TNT reticulocyte lysate that coupled transcription and translation (Promega) generated full-length PHGPx protein from the Cys-containing construct but no detectable protein from the Sec-containing construct (our unpublished data). The absence of detectable full-length PHGPx protein from the Sec-containing construct was expected because incorporation of Sec into protein is essentially undetectable even in reticulocytes made from rabbits fed Se-supplemented diets (Moriarty and Reddy, personal communication). The absence of detectable protein terminating at UGA(72) (either free or attached to tRNA) suggests that the truncated protein was likely targeted for ubiquitin-mediated decay. When the experiments were repeated using a wheat germ extract, which is devoid of the ubiquitin-mediated decay pathway, the Cys-containing transcript generated full-length protein (Figure 2, middle arrow), and UGA(72)-containing or UAA(72)-containing transcripts generated truncated proteins of the size predicted for translation termination at either the Sec or nonsense codon, respectively (i.e., proteins not attached to tRNA; Figure 2, lower arrow). These data suggest that the apparent resistance of PHGPx mRNA to NMD is not due to accumulation of the nascent peptide at the codon preceding the Sec codon when the Sec codon is recognized as nonsense.
Figure 2.
rPHGPx nascent peptide generated by the coupled transcription-translation of rPHGPx (TGA) cDNA by using wheat germ extracts does not accumulate at the codon preceding the Sec codon when the Sec codon is recognized as nonsense. The structure of the SP6-rat(r)PHGPx cDNA and positions of important codons are shown. The diagonally stripped box specifies the bacteriophage SP6 promoter, open boxes represent PHGPx exons, and the darkened region of the last exon indicates the SECIS. ATG(27) specifies the translation initiation codon used primarily in somatic tissues (Pushpa-Rekha et al., 1995), TGA(72) specifies the sole Sec codon [which in derivative constructs was changed to either TGT(72) or TAA(72)], and TAA(197) specifies the normal termination codon. Test pSP-rPHGPx plasmids (9 μg) harboring either the usual Sec (TGA) codon, a Cys (TGT) codon, or a nonsense (TAA) codon and reference luciferase DNA (2 μg; Promega) were incubated in the presence of [35S]methionine in 25 μl of the TNT Wheat Germ Lysate System (Promega). By so doing, cDNAs were transcribed by SP6 RNA polymerase, and product RNAs were then translated. A fraction (12 μl) was precipitated by reducing the pH to 5.0 by using acetic acid and subsequently electrophoresed in acrylamide. The level of PHGPx from each plasmid was normalized to the level of luciferase, and normalized values were then calculated relative to the amount of normalized protein from the TGA-containing construct (after accounting for the number of radioactive amino acids per molecule), which was defined as 1. The asterisk denotes an unidentified protein that was produced in extracts without exogenous DNA (−).
PHGPx mRNA Is a Substrate for NMD in Cultured NIH3T3 and H35 Cells
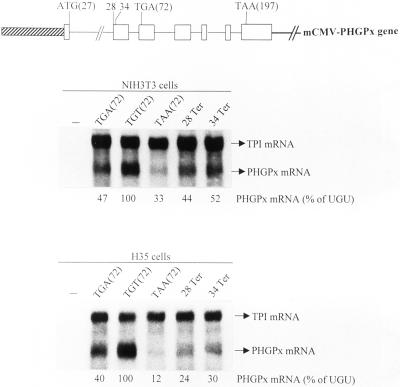
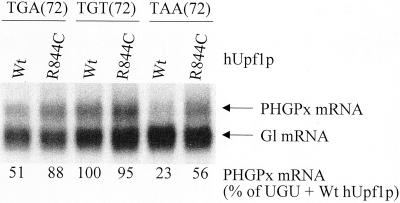
Because deletion of all introns from genes for either triosephosphate isomerase or GPx1 abrogates the NMD of product mRNA that prematurely terminates translation (Moriarty et al., 1998; Zhang et al., 1998a), and because the rat gene for PHGPx has yet to be isolated, we chose to study the pig PHGPx gene (Brigelius-Flohéet al., 1994). To begin to understand why PHGPx mRNA is apparently not subject to NMD at Se concentrations that result in the NMD of GPx1 mRNA, the transcribed portion of the pig PHGPx gene was inserted behind the mCMV promoter so that ATG(27) provided the start site for translation (Figure 3). Mouse NIH3T3 fibroblasts or rat H35 hepatocytes were then transiently transfected with a test pmCMV-PHGPx plasmid that harbored either TGA(72), TGT(72), or TAA(72), and a reference pmCMV-TPI plasmid (Zhang et al., 1998a) that produced mRNA for TPI and was used to control for variations in transfection efficiency and RNA recovery. PHGPx and TPI transcripts were detected by blot hybridization using conditions that did not detect RNA endogenous to NIH3T3 or H35 cells (Figure 3). Unexpectedly, the levels of UGA(72)-containing and UAA(72)-containing mRNAs were, respectively, only 47 and 33% the level of UGU-containing mRNA in NIH3T3 cells and only 40 and 12% the level of UGU-containing mRNA in H35 cells (Figure 3). Because the UGA codon is recognized as nonsense less efficiently than the UAA codon but more efficiently than the UGU codon, the simplest interpretation of these findings is that PHGPx mRNA is subject to NMD in either cell type. Therefore, cultured cells do not recapitulate the apparent immunity of PHGPx mRNA to NMD observed for rat liver or testis. To determine whether nonsense codons at other positions also elicit a reduction in mRNA abundance, codon 28 was changed from TGC to TAA (28Ter) or codon 34 was changed from TGG to TAG (34Ter). Each nonsense codon resulted in a reduction in mRNA abundance (Figure 3). In keeping with the idea that the reduction in abundance is a reflection of NMD, overexpression of a dominant-negative (R844C) form of human (h) Upf1 protein (Sun et al., 1998) increased the levels of PHGPx mRNA harboring either UGA(72) or UAA(72) approximately twofold but had no effect on the level of PHGPx mRNA harboring UGU(72), whereas overexpression of a wild-type (Wt) form of hUpf1 protein had no effect on the level of any of the three mRNAs (Figure 4).
Figure 3.
RNA blot hybridization indicates that pmCMV-PHGPx harboring either the TGA Sec codon or one of several nonsense codons generates mRNA that is reduced in abundance in NIH3T3 and H35 cells. Structures of the mCMV-PHGPx gene and positions mutated in the various derivative alleles are shown. The diagonally stripped box specifies the mCMV promoter, open boxes represent PHGPx exons, intervening lines represent introns, and the right-most bold line represents PHGPx 3′-flanking DNA. Codons for translation initiation, Sec, and translation termination are specified as determined for rPHGPx sequences (see legend to Figure 2). NIH3T3 or H35 cells that had been propagated in MEM plus 10% FBS were either not transfected (−) or transiently transfected with the designated pmCMV-PHGPx test plasmid and the pmCMV-TPI reference plasmid, total RNA was isolated, and PHGPx and TPI transcripts were quantitated by blot hybridization. The level of each PHGPx mRNA was normalized to the level of TPI mRNA and subsequently calculated as a percentage of the normalized level of PHGPx UGU (72) mRNA, which was defined as 100. Values did nor vary by >7–8% in three independently performed experiments.
Figure 4.
RNA blot hybridization indicates that pmCMV-PHGPx harboring either the Sec codon or a TAA codon in its place generates mRNA that is subject to NMD. Cos cells were transfected with the specified pmCMV-PHGPx test plasmid, the pmCMV-Gl reference plasmid, and either pCI-Neo-hUPF1 Wt or pCI-Neo-hUPF1 R844C, and PHGPx and Gl transcripts were quantitated as described in the legend to Figure 3. The level of each PHGPx mRNA was normalized to the level of Gl mRNA and subsequently calculated as a percentage of the normalized level of PHGPx UGU (72) mRNA in the presence of Wt hUpf1 protein (p), which was defined as 100. Values did nor vary by >7–8% in two independently performed experiments.
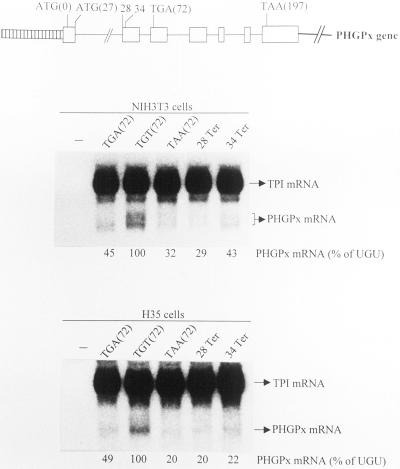
One possible explanation for the failure of the transient transfections to recapitulate the metabolism of PHGPx RNA observed for rat liver and testis could be that the transfections were performed with an incomplete PHGPx gene. To determine whether inclusion of the promoter and first 53 transcribed bp that are missing from pmCMV-PHGPx confer immunity to NMD, NIH3T3 and H35 cells were transiently transfected with test PHGPx alleles (Figure 5) and the reference mCMV-TPI allele. PHGPx alleles were identical to mCMV-PHGPx alleles except that they harbored the full-length pig PHGPx gene, including transcriptional regulatory sequences. Notably, multiple transcripts were evident for each PHGPx allele (Figure 5), most likely reflecting heterogeneity at the 5′ end known to be characteristic of rat PHGPx mRNA in somatic cells (Thimmalapura et al., 1995). Our finding essentially no difference when comparing the expression level of each of the various PHGPx alleles with the expression level of the corresponding mCMV-PHGPx allele (compare Figures 3–5) indicates that the full-length PHGPx gene also generates mRNA in cultured cells that is subject to NMD.
Figure 5.
pPHGPx harboring either the TGA Sec codon or one of several nonsense codons generates mRNA that is subject to NMD in NIH3T3 and H35 cells. NIH3T3 and H35 cells were treated and analyzed as described in the legend to Figure 3 except that pPHGPx plasmids were used in the place of pmCMV-PHGPx plasmids. Values did nor vary by >5–7% in two independently performed experiments.
Se Deficiency Augments the NMD of PHGPx mRNA in NIH3T3 or H35 Cells
In theory, the apparent immunity of PHGPx mRNA to NMD in liver and testis of rats fed a Se-deficient diet could be due to up-regulation of PHGPx gene expression under Se-deficient conditions that compensates for the down-regulation characteristic of NMD. A precedent for this is provided by the finding that Se deficiency results in a significant (but unquantified) increase in the abundance of gastrointestinal GPx1 mRNA in hepatocytes (Wingler et al., 1999). Notably, any up-regulation of PHGPx mRNA abundance by Se deficiency must be posttranscriptional because run-on assays of isolated rat liver nuclei failed to reveal any effect of Se on the PHGPx transcription rate (Bermano et al., 1995). To examine the possibility of up-regulation, NIH3T3 and H35 cells were transfected with a test pPHGPx plasmid and the reference pmCMV-TPI plasmid, transferred 12 h later to Se-deficient or Se-supplemented medium that contained insulin and transferrin in the place of serum, and harvested after an additional 24 h (Moriarty et al., 1998). Relative to Se supplementation, Se deficiency reduced the level of UGA-containing PHGPx mRNA approximately twofold but was of little consequence to the level of either UGU-containing or UAA-containing PHGPx mRNA in either cell type (Figure 6). This finding, plus the finding that NMD was evident for both UGA-containing and UAA-containing mRNAs regardless of the Se concentration (Figure 6), indicates that Se deficiency augments rather than masks or inhibits NMD in NIH3T3 and H35 cells.
To rule out the possibility that NMD was due to experimental artifact induced either by expressing a pig gene in rodent cells or by using transient expression as a means to study PHGPx gene expression, the effect of Se concentration on the level of endogenous PHGPx mRNA produced in untransfected H35 cells was examined. Results indicated that the addition of Se to cells cultured in medium containing either 10% FBS or insulin and transferrin also increased the level of endogenous PHGPx mRNA: the higher the level of Se, the higher the level of PHGPx mRNA (Figure 7). Therefore, there must be inherent differences between the metabolism of PHGPx transcripts in rat tissues and cultured cells.
DISCUSSION
We demonstrate here that the level of PHGPx mRNA in rat liver and testis is insensitive to variations in dietary Se concentration (Figure 1), consistent with previous studies (Bermano et al., 1995; Lei et al., 1995). Insensitivity, however, was not evident in transient transfections of NIH3T3 or H35 cells with PHGPx alleles that included one or both translation initiation codons (Figures 3 and 5). Experiments in which the TGA(72) Sec codon was converted to either a TAA nonsense codon or a TGT Cys codon and assayed under Se-supplemented or Se-deficient conditions indicated that only the TGA(72)-containing alleles generate mRNA that is sensitive to the concentration of Se (Figure 5). Our finding that Se deficiency also reduced the level of PHGPx mRNA produced from the genome of untransfected H35 cells (Figure 7) rules out the possibility that the transient transfections artificially induced the response to Se concentration. The decrease in PHGPx mRNA abundance as a consequence of Se deficiency can be attributed to NMD for at least three reasons. First, regardless of Se concentration, the level of UAA(72)-containing mRNA was lower than the level of UGA(72)-containing mRNA, and the level of UGA(72)-containing mRNA was lower than the level of UGU(72)-containing mRNA. This result is an indication of NMD because the efficiency with which each codon directs translation termination is UAA > UGA > UGU, regardless of Se concentration. Second, Se concentration affected only the level of UGA(72)-containing mRNA. Third, the level of UGA(72)-containing mRNA, like UAA(72)-containing mRNA but not UGU(72)-containing mRNA, was increased by the expression of a dominant-negative form of hUpf1 protein (Figure 4), which is known to abrogate NMD (Sun et al., 1998). We conclude that PHGPx mRNA that derives from a full-length gene contains all of the cis-acting sequences required to support NMD.
Our finding that the PHGPx gene can generate mRNA that is subject to NMD was unexpected and implies that rat liver and testis but not NIH3T3 and H35 cells are characterized by one or more trans-acting factors that either inhibit or mask the NMD of PHGPx mRNA. These factors do not generally affect selenoprotein transcripts because GPx1 mRNA is subject to NMD in both rat tissues, as it is in cultured cells (Chada et al., 1989; Hill et al., 1992; Bermano et al., 1995; Weiss and Sunde, 1997, 1998; Moriarty et al., 1998). Notably, Bermano et al. (1996a) reported that PHGPx mRNA is not reduced in abundance in Se-deficient H4 hepatoma cells when produced from intron-less PHGPx cDNA. Their finding is consistent with the demonstrated role of introns in NMD (Moriarty et al., 1998; Zhang et al., 1998a).
While our work was under review, Fletcher et al. (2000) reported (as data not shown) that the addition of Se to untransfected McArdle 7777 cells cultured in medium containing 10% FBS substantially increased the level of endogenous PHGPx protein as detected by Western blot analysis but had no effect on the level of endogenous PHGPx mRNA. We would not predict these results for PHGPx mRNA based on our studies of untransfected H35 cells (Figure 7). In fact, we found that the addition of Se to untransfected McArdle 7777 cells cultured in medium containing 10% FBS did increase the level of endogenous PHGPx mRNA. For example, the addition of sodium selenite to 7 ng/ml resulted in a fourfold increase in the level of PHGPx mRNA (our unpublished data). The basis for the discrepancy between our results and those of Fletcher et al. (2000) is unclear.
In theory, immunity of PHGPx mRNA to NMD in rat liver and testis could reflect the failure of PHGPx to be released from its mRNA template when the UGA Sec codon reaches the A site of the translating ribosome. The ability of translationally active wheat germ extracts to synthesize truncated proteins of the size predicted for translation termination at either UGA(72) or UAA(72) (i.e., proteins not attached to tRNA) rules out this possibility for wheat germ (Figure 2). The situation in rat tissues remains unknown and may be difficult to assess given that truncated PHGPx is undetectable in reticulocytes lysates (our unpublished data), as are many truncated proteins due to ubiquitin-dependent proteolysis (reviewed in Peters et al., 1998). Alternatively, immunity of PHGPx mRNA to NMD in rat liver and testis could be attributable to very efficient Sec incorporation at UGA(72) regardless of Se concentration, possibly due to trans-acting factors in these tissues that function together with a specialized SECIS element. For example, translation termination could be obviated by efficient “channeling” of tRNASec to UGA(72). Such a scenario was proposed after the PHGPx SECIS element was shown to direct the incorporation of 75Se modestly (1.3-fold) more efficiently than the GPx1 SECIS element: each SECIS element was used to substitute for the SECIS element of type I iodothyronine deiodinase, and the resulting hybrid cDNAs were expressed in cultured cells under Se-deficient conditions (Bermano et al., 1996b). Subsequent studies that assayed Sec incorporation as read-through into a luciferase reading frame confirmed that the PHGPx SECIS is four- and sevenfold more efficient than the GPx1 SECIS under Se-deficient and Se-replete conditions, respectively (Wingler et al., 1999). This finding was extended more recently with the demonstration that the SECIS-binding protein SBP2, which recruits a specialized elongation factor for Sec-specific tRNA, preferentially stimulates Sec incorporation directed by the PHGPx SECIS element relative to other SECIS elements (Low et al., 2000).
Nevertheless, studies in addition to those reported here offer no evidence that the PHGPx SECIS confers immunity to NMD in cultured cells: hybrid genes in which the 3′ UTR of GPx1 mRNA was substituted for that of PHGPx mRNA generated mRNA susceptible to NMD under either Se-adequate or Se-deficient conditions (Weiss and Sunde, 1998). Furthermore, it is difficult to conceive of a SECIS element so effective that it would function with comparable efficiency regardless of the Se concentration. More likely, the apparent insensitivity of PHGPx mRNA abundance to Se concentration in rat liver and testis is due to the inactivation of NMD regardless of the Se concentration or to a Se-induced pathway that counterbalances the effects of NMD. As one example, NMD could be inactivated by one or more factors in rat liver and testis that function together with a stabilizing element analogous to the element located downstream of GCN4 uORF4 of Saccharomyces cerevisiae, which precludes NMD by an unknown mechanism when translation terminates at uORF4 (Ruiz-Echevarria et al., 1998). Experiments that test the Se-dependence of PHGPx gene expression in rats harboring the TAA(72)-containing PHGPx allele or one of a series of hybrid PHGPx/GPx1 alleles in place of the endogenous PHGPx gene should offer important insight into PHGPx RNA metabolism.
ACKNOWLEDGMENTS
We thank Leopold Flohé for the pig PHGPx gene, Donna Driscoll for rat PHGPx cDNA, Heinz Baumann for the rat H35 hepatoma cell line, Paul Copeland and Donna Driscoll for helpful discussions, and Yasuhito Ishigaki for help preparing some of the figures for publication. This work was supported by Public Health Service Research Grant GM-59614 (to L.E.M.).
REFERENCES
- Atkins JF, Matsufuji S, Gesteland RF. Dynamics of the genetic code. In: Gesteland RF, Cech TC, Atkins JF, editors. The RNA World. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 637–673. [Google Scholar]
- Bermano G, Arthur JR, Hesketh JE. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996a;387:157–160. doi: 10.1016/0014-5793(96)00493-0. [DOI] [PubMed] [Google Scholar]
- Bermano G, Arthur JR, Hesketh JE. Role of the 3′ untranslated region in the regulation of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase gene expression by selenium supply. Biochem J. 1996b;320:891–895. doi: 10.1042/bj3200891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohé R, Aumann K-D, Blocker H, Gross G, Kiess M, Kloppel K-D, Maiorino M, Roveri A, Schuckelt R, Ursini F, Wingender E, Flohé L. Phospholipid-hydroperoxide glutathione peroxidase. J Biol Chem. 1994;269:7342–7348. [PubMed] [Google Scholar]
- Cao J, Geballe AP. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996a;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Geballe AP. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996b;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Geballe AP. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- Chada C, Whittney C, Newburger PE. Post-transcriptional regulation of glutathione peroxidase gene expression by selenium in the HL-60 human myeloid cell line. Blood. 1989;74:2535–2541. [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–322. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnin CR, Schleiss MR, Cao J, Geballe AP. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Grabowski PJ. Identification of a protein component of a mammalian tRNASec complex implicated in the decoding of UGA as selenocysteine. RNA. 1999;5:1561–1569. doi: 10.1017/s1355838299991598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM. Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA. 2000;6:1573–1584. doi: 10.1017/s1355838200000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Hill KE, Lyons PR, Burk RF. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem Biophys Res Commun. 1992;185:260–263. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- Ho US, Howard AJ, Crapo JD. Nucleotide sequence of a rat glutathione peroxidase cDNA. Nucleic Acids Res. 1988;16:5207. doi: 10.1093/nar/16.11.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BF, Worland PJ, Davis JN, Statman TC, Hatfield DL. Identification of selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- Lesson A, Mehta A, Singh R, Chisolm GM, Driscoll DM. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biol Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- Low SC, Grundner-Culemann E, Harney JW, Berry MJ. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 2000. pp. 849–868. [Google Scholar]
- Maquat LE, Chilcote R, Ryan PM. Human triosephosphate isomerase cDNA and protein structure: studies of triosephosphate isomerase deficiency in man. J Biol Chem. 1985;160:3748–3753. [PubMed] [Google Scholar]
- Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intron-less genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA. 1997;3:369–1373. [PMC free article] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be cytoplasmic nonsense-mediated mRNA decay. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. When nonsense affects mRNA abundance: a rule for termination codon position within intron-containing genes. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Peters J-M, King RW, Deshaies RJ. In: Ubiquitin and the Biology of the Cell. Peters J-M, Harris JR, Finley D, editors. New York: Plenum; 1998. pp. 345–387. [Google Scholar]
- Pushpa-Rekha TR, Burdsall AL, Oleksa LM, Chisolm GM, Driscoll DM. Rat phospholipid-hydroperoxidase glutathione peroxidase: cDNA cloning and identification of multiple transcription and translation start sites. J Biol Chem. 1995;45:26993–26999. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- Roveri A, Casasco A, Maiorino A, Dalan MP, Calligaro A, Ursini F. Phospholipid hydroperoxide glutathione peroxidase of rat testis. Gonadotropin dependence and immunocytochemical identification. J Biol Chem. 1992;267:6142–6246. [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, González CI, Peltz SW. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;675:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Sun X, Moriarty PM, Maquat LE. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J. 2000;19:4734–4744. doi: 10.1093/emboj/19.17.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1p protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde RA. Selenium. In: O'Dell L, Sunde RA, editors. Handbook of Nutritionally Essential Mineral Elements. New York: Marcel Dekker; 1997. pp. 493–556. [Google Scholar]
- Weiss SL, Sunde R. Selenium regulation of classical glutatione peroxidase expression requires the 3′ untranslated region in Chinese hamster ovary cells. J Nutr. 1997;127:1304–1310. doi: 10.1093/jn/127.7.1304. [DOI] [PubMed] [Google Scholar]
- Weiss SL, Sunde R. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler K, Böcher M, Flohé L, Kollmus H, Brigelius-Flohé R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur J Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998a;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-meditated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998b;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]