Abstract
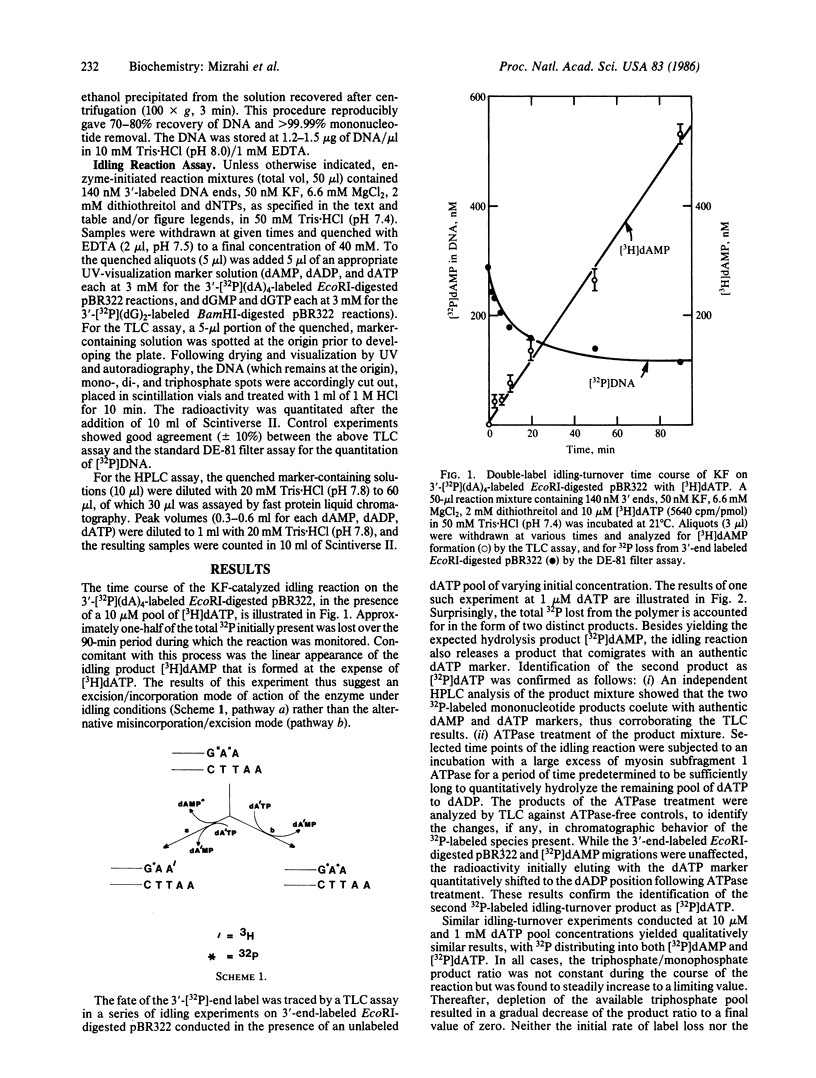
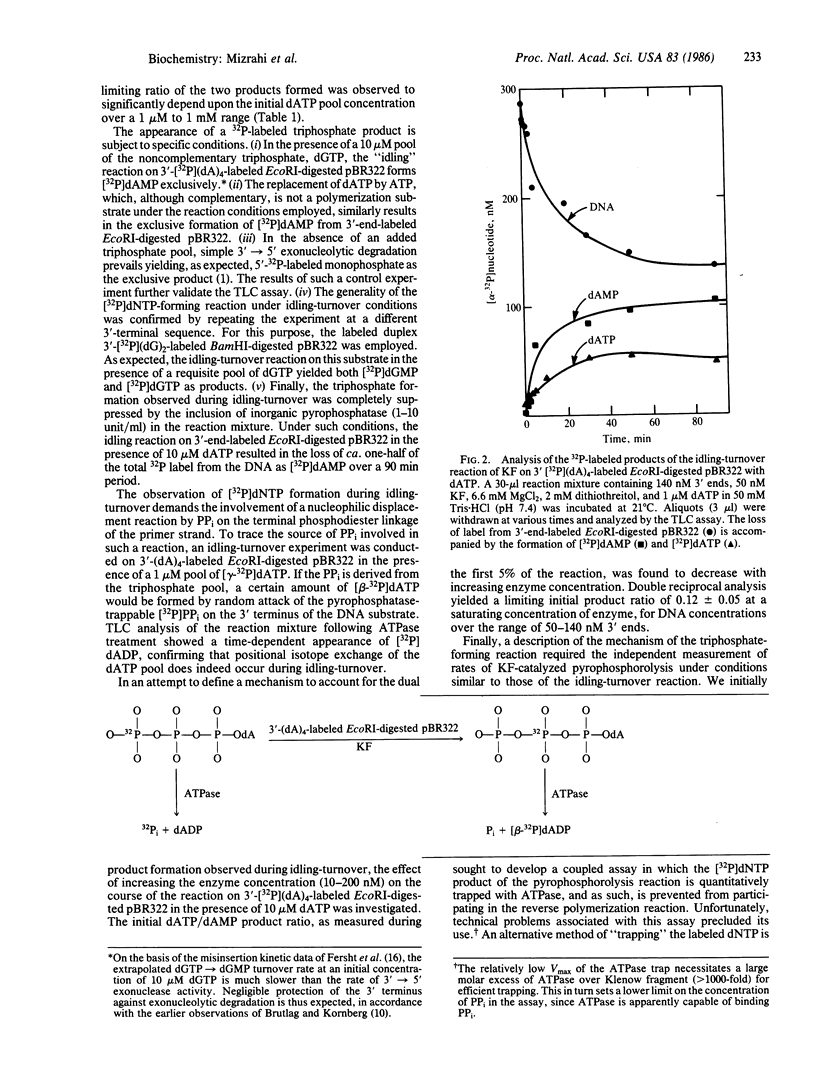
The mechanism of the idling-turnover reaction catalyzed by the large (Klenow) fragment of Escherichia coli DNA polymerase I has been investigated. The reaction cycle involved is one of excision/incorporation, in which the 3' deoxynucleotide residue of the primer DNA strand is partitioned into its 5'-mono- and 5'-triphosphate derivatives, respectively. Mechanistic studies suggest the 5'-monophosphate product is formed in the first step by simple 3'----5' exonucleolytic cleavage. Rapid polymerization follows with the concomitant release of inorganic pyrophosphate. In the second step, the 5'-triphosphate product is generated by a pyrophosphorolysis reaction, which, despite the low concentration of pyrophosphate that has accumulated, occurs at a rate that is comparable with that of the parallel 3'----5' hydrolysis reaction.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bessman M. J., Muzyczka N., Goodman M. F., Schnaar R. L. Studies on the biochemical basis of spontaneous mutation. II. The incorporation of a base and its analogue into DNA by wild-type, mutator and antimutator DNA polymerases. J Mol Biol. 1974 Sep 15;88(2):409–421. doi: 10.1016/0022-2836(74)90491-4. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Bryant F. R., Johnson K. A., Benkovic S. J. Elementary steps in the DNA polymerase I reaction pathway. Biochemistry. 1983 Jul 19;22(15):3537–3546. doi: 10.1021/bi00284a001. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Eckstein F. A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate. J Biol Chem. 1979 Aug 10;254(15):6889–6893. [PubMed] [Google Scholar]
- Clayton L. K., Goodman M. F., Branscomb E. W., Galas D. J. Error induction and correction by mutant and wild type T4 DNA polymerases. Kinetic error discrimination mechanisms. J Biol Chem. 1979 Mar 25;254(6):1902–1912. [PubMed] [Google Scholar]
- Deutscher M. P., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 28. The pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):3019–3028. [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Tsui W. C. Kinetics of base misinsertion by DNA polymerase I of Escherichia coli. J Mol Biol. 1983 Apr 25;165(4):655–667. doi: 10.1016/s0022-2836(83)80272-1. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Branscomb E. W. Enzymatic determinants of DNA polymerase accuracy. Theory of coliphage T4 polymerase mechanisms. J Mol Biol. 1978 Oct 5;124(4):653–687. doi: 10.1016/0022-2836(78)90176-6. [DOI] [PubMed] [Google Scholar]
- Gupta A. P., Benkovic P. A., Benkovic S. J. The effect of the 3',5' thiophosphoryl linkage on the exonuclease activities of T4 polymerase and the Klenow fragment. Nucleic Acids Res. 1984 Jul 25;12(14):5897–5911. doi: 10.1093/nar/12.14.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. P., Benkovic S. J. Stereochemical course of the 3'----5'-exonuclease activity of DNA polymerase I. Biochemistry. 1984 Nov 20;23(24):5874–5881. doi: 10.1021/bi00319a029. [DOI] [PubMed] [Google Scholar]
- Gupta A., DeBrosse C., Benkovic S. J. Template-prime-dependent turnover of (Sp)-dATP alpha S by T4 DNA polymerase. The stereochemistry of the associated 3' goes to 5'-exonuclease. J Biol Chem. 1982 Jul 10;257(13):7689–7692. [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., Dube D. K., Beckman R. A., Koplitz M., Gopinathan K. P. On the fidelity of DNA replication. Nucleoside monophosphate generation during polymerization. J Biol Chem. 1981 Apr 25;256(8):3978–3987. [PubMed] [Google Scholar]
- Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985 Jul 16;24(15):4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. A centrifuged-column procedure for the measurement of ligand binding by beef heart F1. Methods Enzymol. 1979;56:527–530. doi: 10.1016/0076-6879(79)56050-9. [DOI] [PubMed] [Google Scholar]


