Abstract
Understanding the underlying mechanisms by which a normal cell avoids the oncogenic potential of MUC1 signaling requires further definition of the pathways by which the MUC1 cytoplasmic tail is processed in both normal and tumor-derived cells. In the present study we describe the processing pathway initiated by TACE/ADAM17 cleavage of MUC1. Utilizing the human uterine epithelial cell line, HES, derived from normal endometrium, we show that endogenous full length MUC1 undergoes regulated intramembranous proteolysis mediated by presenillin-dependent γ-secretase. Cytokine-stimulated HES cells exposed to γ-secretase inhibitors accumulated a membrane-associated 15kDa fragment of the MUC1 C-terminal subunit (CTF15). Inhibitors of TACE/ADAM17–mediated shedding inhibited accumulation of MUC1–CTF15 and MUC1 ectodomain release to a similar extent consistent with MUC1–CTF15 being a product of TACE/ADAM17 action. Reduction of catalytically active γ-secretase complex by nicastrin siRNA treatment also resulted in CTF15 accumulation. Furthermore, mature nicastrin, the substrate receptor for γ-secretase, co-immunoprecipitated with CTF15 in the presence of γ-secretase inhibitors indicating the formation of CTF15: nicastrin complexes. MUC1–CTF15 accumulation in response to γ-secretase inhibition was demonstrated in both normal and tumor-derived cells from humans and mice indicating that this processing pathway exists in many cell contexts. We did not detect products of MUC1 cleavage by γ-secretase in the presence of various proteasomal inhibitors indicating that subsequent degradation is either non-proteasomal or extremely efficient. We suggest that this efficient pathway attenuates potential signaling mediated by cytoplasmic tail fragments.
Keywords: MUC1, γ-SECRETASE, ENDOMETRIUM, EMBRYO IMPLANTATION
MUC1 is a heavily glycosylated type I transmembrane protein expressed at the apical surface of many normal secretory epithelial cells [Gendler, 2001]. Due to its elevated expression in a variety of human malignancies and its ensuing oncogenic potential [Hanisch and Muller, 2000; von Mensdorff-Pouilly et al., 2000], considerable effort has been expended in determining MUC1’s suitability as both a prognostic indicator for disease progression and therapeutic target for disease attenuation. Inherent to the success of these efforts is an understanding of the metabolic events by which a tumor cell processes MUC1, how these events differ from those occurring in normal cells, and how these differences may contribute to the initiation, maintenance, and/or progression of the pathologic state.
Full length MUC1 (MUC1F) is composed of three structural domains: (i) an extracellular domain (ECD) containing extensively O-glycosylated tandem repeat 20 amino acid sequences; (ii) a hydrophobic transmembrane domain (TMD) and; (iii) a cytoplasmic tail domain (CTD). MUC1F initially is synthesized as a single polypeptide chain in both normal and malignant cells. Shortly after synthesis in the endoplasmic reticulum, MUC1 undergoes an autoproteolytic cleavage within the SEA module located within the ECD, 58 amino acids beyond the N-terminus of the TMD [Levitin et al., 2005; Macao et al., 2006]. The resulting two subunits form a heterodimer [Ligtenberg et al., 1992] through a stable non-covalent association (metabolic complex) that persists as MUC1 transits through the golgi apparatus, undergoing O-glycosylation, and arriving at the cell surface [Ligtenberg et al., 1992; Julian and Carson, 2002]. The ectodomain containing the tandem repeats is released from a subset of cell surface MUC1F. MUC1F ECD release is dependent on the activity of at least two sheddases, TACE/ADAM 17 [Thathiah et al., 2003] and MT1-MMP [Thathiah and Carson, 2004]. No soluble form of MUC1F ECD is released in the absence of sheddase activity in normal cells. Although the association of the two subunits of the MUC1F metabolic complex is theoretically reversible, dissociation under physiologic conditions has not been demonstrated. However, NM23-H1, released into medium only from tumor-derived cells, interacts with the ECD portion of MUC1F C-terminal subunit, disrupting the metabolic complex and displacing the N-terminal subunit [Mahanta et al., 2008].
While early studies focused on the fate of the MUC1 ECD, current interest has centered on processing and the ultimate fate of the TMD and CTD of MUC1F. The potential involvement of the MUC1 CTD in signaling events, both indirectly via activation of signaling pathways [Schroeder et al., 2001; Lillehoj et al., 2004; Thompson et al., 2006] and directly when translocated to the nucleus [Li et al., 2003; Singh et al., 2007] or mitochondria [Ren et al., 2004, 2006], necessitates elucidation of the intervening metabolic events. Translocation to both intracellular locales requires internalization followed by transport, presumably endosomal. Endosomal transport has been established only for internalization of MUC1 C-terminal subunit initiated by ligand engagement of EGF receptor [Pochampalli et al., 2007; Liu et al., 2008]. Yet translocation to any intracellular site requiring retention of the TMD will likely involve endosomal transport. While retention of the TMD is necessary for insertion of MUC1 CTD into the mitochondrial outer membrane [Ren et al., 2006], a MUC1 CTD construct lacking the TMD proved capable of nuclear entry [Leng et al., 2007]. Although not necessary for nuclear entry, the TMD does not prevent it since all forms of the C-terminal subunit derived from full length MUC1 have been detected in nuclear extracts [Leng et al., 2007]. When the MUC1 carboxy terminal (CT) form detected at the intracellular destination is the intact C-terminal subunit, the endosomal compartment responsible for transport must be non-degradative. The predominant form detected at both locales is the intact N-glycosylated C-terminal subunit. Investigators have noted the presence in tumor-derived cells of additional C-terminal forms lacking N-glycosylation [Ren et al., 2004; Ramasamy et al., 2007]. Attenuation of signaling would require both internalization and sequestration from cytoplasmic adaptors or degradation. While cleavage of the MUC1 TMD could release a soluble CTD, no reports have described a normal or tumor-derived cell context which produces a soluble MUC1 CTD from endogenous full length MUC1. Thus, cleavage of the TMD could serve as a trigger for CTD destruction, attenuating any possibility of further participation in signaling events, either positively or negatively. Alternatively, TMD cleavage could release a soluble CTD.
Previously we demonstrated that TACE/ADAM17 is one of the sheddases responsible for MUC1 cleavage and release of MUC1F ectodomain [Thathiah et al., 2003]. Some products of TACE/ ADAM17 cleavage serve as substrates for regulated intramembrane proteolysis executed by presenillin-dependent γ-secretase [Medina and Dotti, 2003]. There are few requirements for a γ-secretase substrate; however, with one exception [Meyer et al., 2003] they are monomeric type I transmembrane proteins. Cleavage and release of the majority of the ectodomain is a necessary prerequisite [Struhl and Adachi, 2000]. The resulting ectodomain “stub” recognized by nicastrin, the substrate receptor for the active γ-secretase complex, cannot exceed 50 amino acids and must have an unobstructed amino terminus [Shah et al., 2005]. The amino terminus of the membrane-anchored C-terminal subunit of the MUC1F metabolic complex is obstructed by association with the larger N-terminal subunit. Inspection of the ECD of the C-terminal membrane anchored subunit of the MUC1F metabolic complex indicates that even if the complex were to dissociate, the membrane-anchored subunit is an unlikely candidate as a γ-secretase substrate since its ectodomain (58 amino acids) is too large. In contrast, TACE/ ADAM17 cleaves MUC1 C-terminal subunit within the 58 amino acid ECD [Thathiah and Carson, 2004] resulting in a ECD “stub” of 27 amino acids, that is, a size consistent with that of other substrates recognized by nicastrin [Shah et al., 2005].
During cytokine stimulation of normal cells, which resulted in overexpression of MUC1F and increased shedding [Thathiah and Carson, 2004], we detected two additional forms of the MUC1 C-terminal subunit. These forms appeared to be similar to the non-N-glycosylated C-terminal forms produced by tumor-derived cells [Ren et al., 2004; Ramasamy et al., 2007; Mahanta et al., 2008]. In the present study we describe the metabolic processing pathway initiated by sheddase cleavage which involves these forms. The product of sheddase cleavage of endogenous MUC1F serves as a substrate for γ-secretase in both normal and tumor-derived cells. The product of γ-secretase cleavage is undetectable and appears to be rapidly degraded in both normal and tumor-derived cells.
EXPERIMENTAL PROCEDURES
MATERIALS
The γ-secretase inhibitors, L685,458 and S2188, and proteasomal inhibitors, MG101, MG132, and lactacystin, were purchased from Sigma (St. Louis, MO). Recombinant human interferon γ and recombinant human tumor necrosis factor α (TNF α) were purchased from Roche Applied Science (Indianapolis, IN). Mouse monoclonal MUC1 antibodies 214D4 and HMFG1 recognizing epitopes in the MUC1 tandem repeat sequences were provided as hybridoma medium by Dr. John Hilkens (Netherlands Cancer Institute, Amsterdam, The Netherlands) and Dr. Sandra Gendler (Mayo Clinic, Scottsdale, AZ), respectively. Peptide affinity purified rabbit polyclonal MUC1 antibody, CT1, raised against a 17 amino acid peptide at the carboxy terminus of MUC1 cytoplasmic tail, was generated and characterized as described previously [Pimental et al., 1996; Julian and Carson, 2002]. Rabbit polyclonal anti-nicastrin was purchased from Affinity Bioreagents (Golden, CO). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG was purchased from Sigma. HRP-conjugated donkey anti-rabbit IgG and HRP-conjugated sheep anti-mouse IgG were purchased from Jackson Immunologicals (West Grove, PA). ECL reagent Super Signal West Dura was purchased from Pierce (Rockford, IL).
CELL LINES, CULTURE, AND TREATMENT CONDITIONS
The human uterine epithelial cell line, HES, derived from normal human endometrium, was provided by Dr. Douglas Kniss (Ohio State University, Columbus, OH) and maintained in high glucose Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) charcoal-stripped, heat inactivated fetal bovine serum (HyClone; Logan, UT), 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen), and 1µM sodium pyruvate (Invitrogen). T47D and ZR75-1 cell lines, obtained from the American Tissue Culture Collection, were maintained in RPMI 1640 (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Primary cultures of isolated mouse uterine epithelium were generated from random cycling 8-week-old virgin females (ICR strain) as previously described [Pimental et al., 1996], and cultured on Matrigel®-coated Anocell 10 culture dish inserts (Whatman, Clifton, NJ) in Dulbecco’s modified Eagle’s medium/Ham’s F12 (1:1) (Invitrogen) supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin. For experimental treatments, cell lines were grown in 24-well plates in the media described above. When the cells reached confluence, 25ng/ml human recombinant TNF α and 200 U/ml human recombinant interferon gamma (IFNγ) were added with either vehicle (0.5% [v/v] dimethyl sulfoxide) or the indicated concentrations of L685,458, S2188, MG101, MG132, or Lactacystin. After the indicated period of culture, medium was removed and the cell layer was lysed. When shedding was to be examined, cells were rinsed twice in serum-free medium and cultured for the time indicated in serum-free medium before treatments were initiated in serum-free medium. The TACE/ADAM17 inhibitor, TAPI, obtained from Dr. John Doedens and Dr. Roy Black (Immunex, Seattle, WA), was added at a final concentration of 50µM as described previously [Thathiah et al., 2003].
SAMPLE PREPARATION
To examine shed/secreted proteins, medium from treated cells was collected, and centrifuged for 15 min at 10,000g at 4°C. After the addition of fetal bovine serum protein (50 µg) as carrier to the clarified media, the samples were brought to 10% (w/v) trichloracetic acid (TCA) and precipitated overnight at 4°C. The precipitates were rinsed in 100% acetone, air dried, and redissolved in equal volumes of sample extraction buffer (SEB; 0.05M Tris, pH 7.0, 8 M urea, 1.0% [w/v] sodium dodecyl sulfate, 1% [w/v] β-mercaptoethanol, and 0.01% [w/v] phenylmethylsulfonyl fluoride) and Laemmli sample buffer [Laemmli, 1970] containing 10µl/ml protease inhibitor cocktail (Sigma, P-8340). To examine cell-associated proteins, the cells were subjected to a “hot lysis”: addition of 250 µl/well of boiling 0.5% (v/v) Nonidet P-40 in phosphate buffered saline (PBS) minus calcium and magnesium and containing 1mM ethylenediaminetetraacetic acid and 10 µl/ml protease inhibitor cocktail (Sigma, P-8340). After 5 min, the cellular material was scraped into the lysis buffer. Alternately, for co-immunoprecipitation experiments, lysis was performed on ice for 1 h. Insoluble material was removed by centrifugation for 10 min at 10,000g at 4°C. The clarified lysate was used for immunoprecipitation or precipitated by 10% (w/v) TCA and redissolved as described above. When immunoprecipitation was not to be performed, cell-associated proteins were solubilized in SEB, precipitated by 10% (w/v) TCA and redissolved as described above (total cell protein).
IMMUNOPRECIPITATION
One hundred microliters fresh lysate (representing 40% of lysate from one confluent well of a 24-well plate) was incubated with 40 µl (∼0.22 µg IgG) 214D4 overnight at 4°C and antibody complexes were removed by incubation with preblocked protein G agarose (Kirkegaard and Perry Laboratories, Gaithersburg, MD). The resin was pelleted by centrifugation and the resulting supernate was analyzed or subjected to a second immunoprecipitation with the indicated antibody (HMFG1 or CT1) followed by removal of antibody complexes with preblocked protein G agarose. The resin pellets containing antigen/antibody complexes were rinsed twice with0.5% (v/v) NP-40inPBS – Ca2+-Mg2+andonceinPBS – Ca2+-Mg2+ Antibody complexes were extracted with equal volumes SEB and Laemmli sample buffer containing 10 µl/ml (v/v) protease inhibitor cocktail (Sigma, P-8340). Post-immunoprecipitation supernates were subjected to TCA precipitation as described above for lysates.
N-DEGLYCOSYLATION
Lysate (200 µl) was incubated with or without 7 U of PNGase F (Sigma) for 36 h at room temperature. Samples were subsequently precipitated with 10% (w/v) TCA at 4°C and processed for gel analysis as described above.
MEMBRANE ISOLATION
Confluent cultures of HES cells treated for 48 h with cytokines with or without 1 µM L685,458 were scraped into hypotonic buffer (5mM Tris pH 7.5, 1mM EDTA, 0.25M sucrose, 10 µl/ml protease inhibitor cocktail, with or without 1 µM L685,458) and incubated on ice for 1 h. The swollen cells were homogenized with 30 strokes of a Dounce homogenizer (pestle A), until >95% of cells were disrupted (assessed by visual microscopic examination). The homogenate was centrifuged sequentially at 4°C for 10 min at 1,000g, 20 min at 10,000g, and 2 h at 100,000g. The final supernatant was subjected to TCA precipitation as described above for the lysates. Pellets were solubilized in SEB and Laemmli sample buffer (v/v, 1:1) containing 10ml/ml protease inhibitor cocktail (Sigma, P-8340). Inclusion of L685,458 in the homogenization buffer was necessary to preserve CTF15 during preparation.
GEL ELECTROPHORESIS AND WESTERN BLOTTING
Lysates or immunoprecipitates were applied to 15% (w/v) acrylamide SDS–PAGE for detection of CT fragments or 10% (w/v) acrylamide for detection of MUC1-ECD. Samples were transferred to nitrocellulose and probed as previously described [Julian and Carson, 2002]. For densitometric analyses, images were generated from appropriately exposed films using an Alpha Imager 2200 (Alpha Innotech Corp.) and analyzed using Scion Image (Scion Corporation, Frederick, MD). Individual bands were quantified by multiplying the mean density (after subtracting background for the same area) by area (defined in square inches containing 5,625 pixels per square inch). When multiple bands were recognized by an antibody, the total was defined as the sum of all individual for the sample. Statistical analyses were performed using one-way ANOVA and the Tukey-Kramer multiple comparisons test via the GraphPad Instat program (GraphPad Software, Inc., San Diego, CA).
REDUCTION OF NICASTRIN EXPRESSION BY TRANSIENT siRNA TRANSFECTION
HES cells, grown in the absence of antibiotics to 40–60% confluence, were transfected with nicastrin siRNA duplex or GAPDH siRNA duplex (Dharmacon, Lafayette, CO) using RNAi MAX (Invitrogen) in OPTI-MEM according to the manufacturer’s instructions. siRNA duplex Nct-1045 was directed to the target sequence 5′-AAGGGCAAGTTTC-CCGTGCAGTT-3′ of nicastrin. Consecutive transfections were necessary to sustain the reduced level of nicastrin through the treatment. Three days after the initial transfection, a second transfection was performed (cells were not replated). Cytokine treatment was initiated with addition of recovery media. The γ-secretase inhibitor, L-685,458, was included at a final concentration of 1mM in the treatment of a parallel set of samples as a positive control to determine the maximal amount of CTF15 fragment that could be accumulated due to catalytic inhibition. Additional controls, including mock-transfected cells and cells transfected with siRNA duplex to GAPDH (5′-UGGUUUACAU-GUUCCAAUA-3′), received the same treatments.
RESULTS
Identification of the MUC1 substrate for γ-secretase cleavage
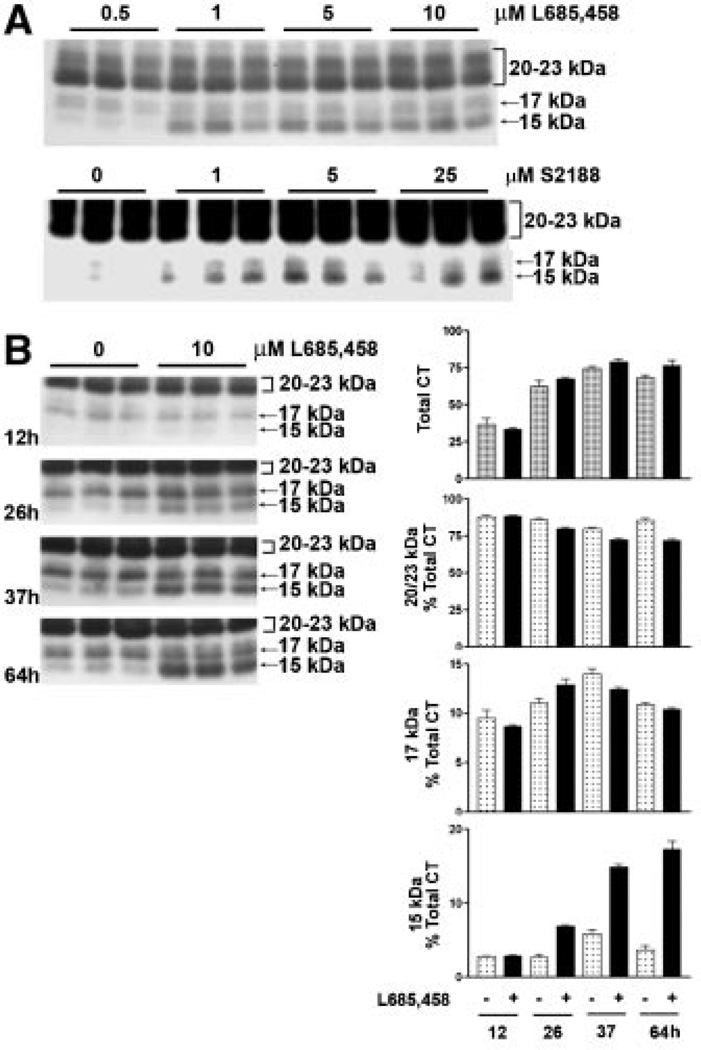
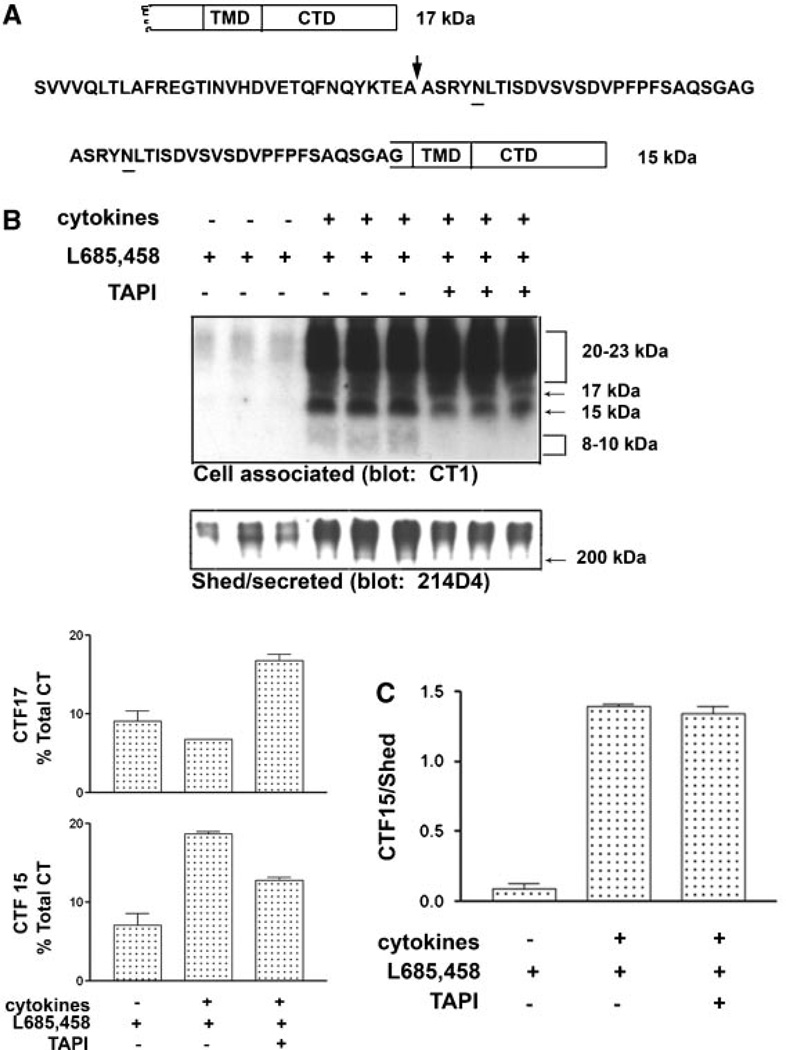
Under normal growth conditions with no stimulation of sheddase activity, virtually all detectable MUC1 C-terminal subunit in HES cells is associated with MUC1 ectodomain as a heterodimer which dissociates on SDS gels and migrates as a doublet between 20 and 23 kDa (see, e.g., Fig. 6). When HES cells are stimulated by the cytokines TNFα and IFNγ, MUC1F expression and shedding are elevated [Thathiah et al., 2004; Wang et al., 2008], and two additional C-terminal forms migrating at 17 and 15kDa are detected (Fig. 1). Since both forms are recognized by the CT-1 antibody, they must possess most, if not all, of the C-terminal sequence. We considered that conditions in which MUC1 is overexpressed, for example, in tumor cells or cytokine-stimulated cells, may exceed or alter the cellular capacity to post-translationally process MUC1. Because metabolic cleavage did not appear to be affected by cytokine stimulation processing events downstream of this initial cleavage event had to be responsible for the appearance of these fragments. These events would include sheddase cleavage and yet to be identified processing of the membrane-bound product of sheddase cleavage. Products of cleavage by TACE/ADAM17 can be substrates for γ-secretase cleavage [Medina and Dotti, 2003]. We initially applied the most commonly used approach to assess γ-secretase substrates, namely use of pharmacological inhibitors of this process. Inclusion of specific catalytic inhibitors of γ-secretase activity during cytokine stimulation of HES cells caused accumulation of the MUC1 15kDa C-terminal form (CTF15). The accumulation was both time and inhibitor concentration dependent (Fig. 1). The increased expression of MUC1F in response to cytokine stimulation was apparent by the increase in total CT forms which doubled by 37 h of treatment (Fig. 1B). Because these experiments were performed in the presence of serum and were of longer duration than performed previously, the magnitude of MUC1F expression reflected by total CT in Figure 1B would include any increase in cell number occurring at later time points. This was minimal since treatments were initiated on confluent cultures. The C-terminal doublet at 20–23 kDa and band at 17 kDa increased with time to levels that were not significantly different with respect to the presence of γ-secretase inhibitor. The 15 kDa C-terminal fragment (CTF15) was the only CT form recognized by the CT1 antibody that accumulated at a greater rate when cells were cytokine stimulated in the presence of γ-secretase inhibitor (Fig. 1B). By 26 h of inhibitor treatment, CTF15 represented approximately 7% of the total CT1- reactive species and continued to accumulate in the presence of inhibitor, reaching 18% of total C-terminal forms at 64 h.
Fig. 6.
Proteasomal inhibition causes accumulation of MUC1 CTF15. In panel A, HES cells were exposed to cytokines plus the reversible 7–secretase inhibitor, S2188 (10µM), for 56 h. S2188 then was removed and culture continued for the indicated times in the presence of cytokines and the absence (−) or presence (+) of the proteosomal inhibitor, MG132 (10µM), for the indicated times. CTF15 was detected by antibody CT1. Triplicate independent samples were analyzed in each case. Note that while CTF15 is lost upon removal of the S2188 it continues to accumulate in the presence of MG132. Panel B unstimulated or cytokine-stimulated HES cells were cultured with or without 10µM MG132 for 14 or 26 h. CT forms were detected in cell lysates by antibody CT1. Densitometric analysis demonstrates that within the treatment period used in panel A experiments, proteasomal inhibition by MG132 effectively blocked cytokine induction of all forms of CT (Total CT) and was capable of progressive reduction of constitutive expression. Note that CTF17 and CTF15 are undetectable in unstimulated HES cells. Densitometric analysis of CT forms expressed in response to cytokine stimulation reflect the relative concentrations of CT forms that would be produced during the same time frames.
Fig. 1.
Catalytic inhibition of presenillin-dependent γ-secretase causes accumulation of a 15 kDa MUC1 fragment containing the CTD (CTF15). Cytokine-stimulated HES cells were treated for 64 h with the indicated concentrations of the γ-secretase inhibitors, L685,458 or S2188 (panel A). Not shown are treatments with 0, 0.001, 0.01, and 0.1 µM L685,458 performed in the same experiment which were identical to results shown for 0.5 µM L685,458. Cell lysates were analyzed by SDS–PAGE and Western blotting with CT1 antibody as described in “Experimental Procedures Section.” Inhibitor treatment resulted in dose-dependent accumulation of a MUC1 peptide containing the CTD (panel A), migrating at 15 kDa. Migration of the larger CT subunit of the metabolic complex at approximately 20–23 kDa is indicated by a bracket to the right. Accumulation of the CTF15 was time-dependent in the presence of 10 µM L685,458 (panel B). Performed as described in “Experimental Procedures Section,” densitometric analyses of the accumulation of CTF15 relative to other CT forms are presented to the right of panel B. Total CT is defined as the sum of all bands recognized by CT1 within the sample.
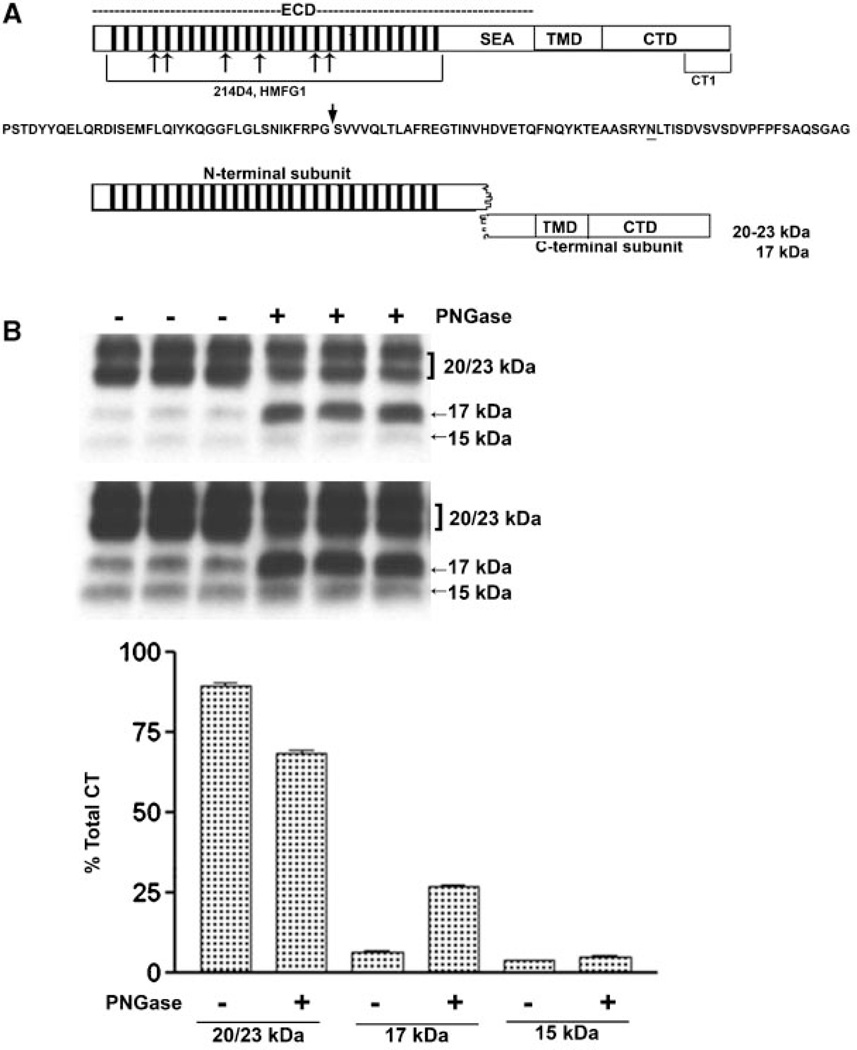
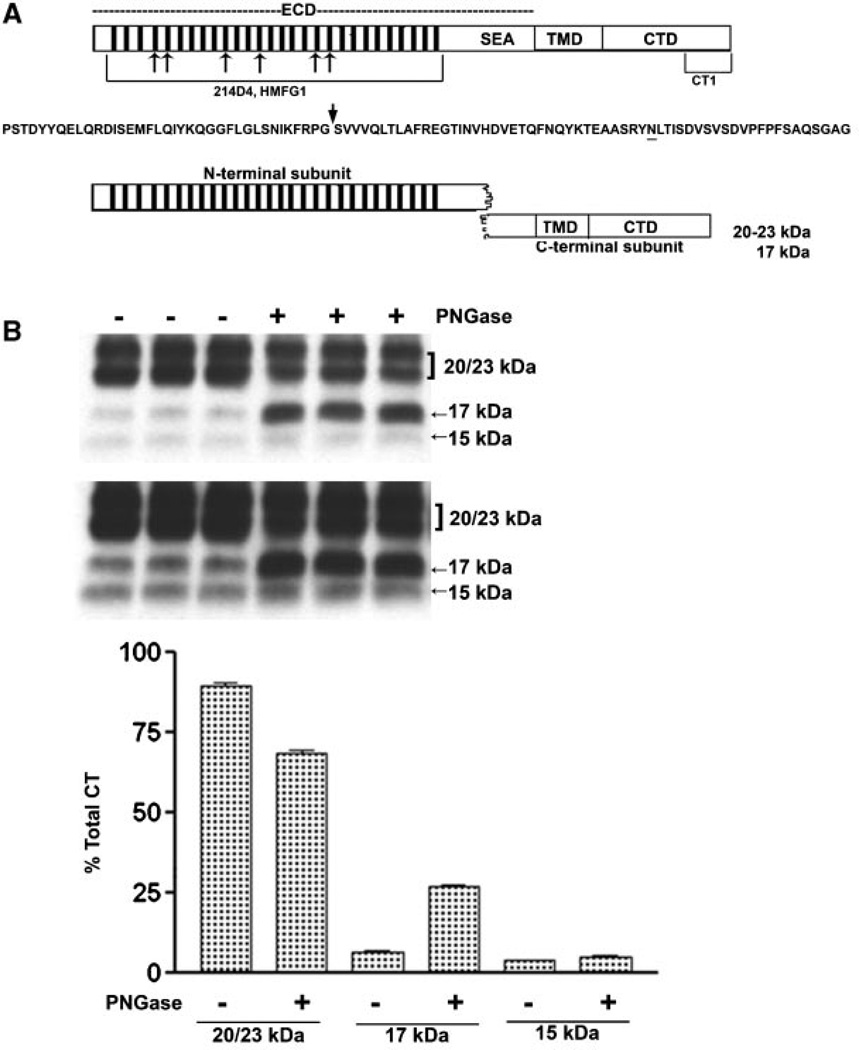
We concluded that CTF15 was the MUC1 substrate for γ-secretase. Figure 2A presents a diagram reflecting the first step of MUC1F processing. Post-translational modification also contributes to variably sized MUC1 C-terminal subunits. One consensus site for N-glycosylation resides within the 58 amino acid ectodomain portion of the C-terminal subunit between the metabolic cleavage site and the TMD (residue N36 described in [Ramasamy et al., 2007] and underlined in Fig. 2A). Mutation at this site previously shown to prevent N-glycosylation produced only the 17 and 15kDa C-terminal forms. The larger 20–23 kDa C-terminal forms treated only with N-glycanase were reduced to the 17 kDa form while the 15 kDa form was unchanged. It appears that N-glycosylatioin alone accounts for the larger forms with variations in the N-glycans accounting for the difference in the 20–23 kDa forms [Parry et al., 2006]. Application of a combination of enzymes including N-glycanase and O-glycanase produced similar results, reducing CTF 20–23 to CTF 17 [Mahanta et al., 2008]. We focused our studies in the N-glycosylation state of the C-terminal fragments. When lysates from cytokine-stimulated, γ-secretase inhibitor-treated HES cells were incubated with N-glycanase, the 17 kDa was increased (Fig. 2B). In contrast, the 20–23 kDa bands decreased in proportion to the increase in the 17 kDa band (Fig. 2B). CTF15 was unaffected by peptide: N-glycanase treatment and, therefore, also did not appear to be N-glycosylated (Fig. 2B,C). These results are in agreement with observations of others on the N-glycosylation state of various MUC1 C-terminal forms [Ren et al., 2004; Ramasamy et al., 2007; Mahanta et al., 2008]. The 20–23 kDa are N-glycosylated while the 17 and 15 kDa C-terminal forms appear to lack N-glycans.
Fig. 2.
N-Deglycosylation of MUC1 CTD fragments. The upper portion of panel A presents a diagram of intact full length MUC1 as it would exist immediately after translation and designates the location of various domains: ECD, extracellular domain containing the tandem repeats (bars) and SEA module; TMD, the transmembrane domain; and CTD, the cytoplasmic tail domain. Also indicated are the locations of epitopes recognized by various antibodies used in the present study. Mouse monoclonal antibodies, 214D4 and HMFG1, both recognize epitopes within the tandem repeat regions indicated by the hatched bars in the figure and the bracket underneath in the model. The number of bars used is arbitrary and is not meant to indicate the precise number of tandem repeat motifs. Both antibodies would recognize the MUC1 N-terminal subunit and potentially bind to multiple sites in a single MUC1 molecule. Recognition by these antibodies is impacted by glycosylation. The CT-1 antibody was generated against the C-terminal 17 amino acid peptide of MUC1 and, therefore, only recognizes fragments of MUC1 containing all or part of this C-terminal sequence. There is no reported evidence that recognition bytheCT-1 antibody is impacted by post-translational modifications. Beneath is the amino acid sequence of the SEA module within the ECD. An arrow indicates the location of autoproteolytic cleavage which produces the N- and C-terminal subunits which reassociate to form the non-covalent heterodimeric “metabolic complex” diagrammed below. The N-glycosylation site is underlined and distinguishes the 20–23 kDa C-terminal subunit (N-glycosylated) from the 17 kDa C-terminal subunit (not N-glycosylated). In panel B lysates from cytokine plus 1 µM L685,458 treated HES cells were incubated without (−) or with (+) PNGase and subsequently analyzed by SDS–PAGE and Western blotting with CT1 antibody as described in “Experimental Procedures Section.” Two exposures are included to demonstrate CTF15. Densitometric analysis was based on the shorter exposure. Note that PNGase treatment resulted in a partial reduction of 20–23 kDa signal and accumulation of 17 kDa signal, however, there was no change in CTF15.
The MUC1 γ-secretase substrate is membrane associated but is not associated with the MUC1 metabolic complex
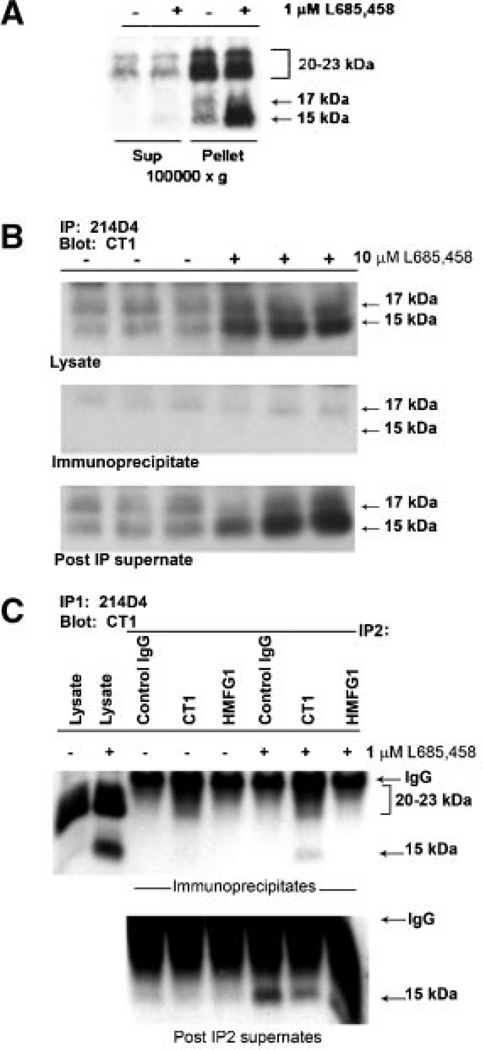
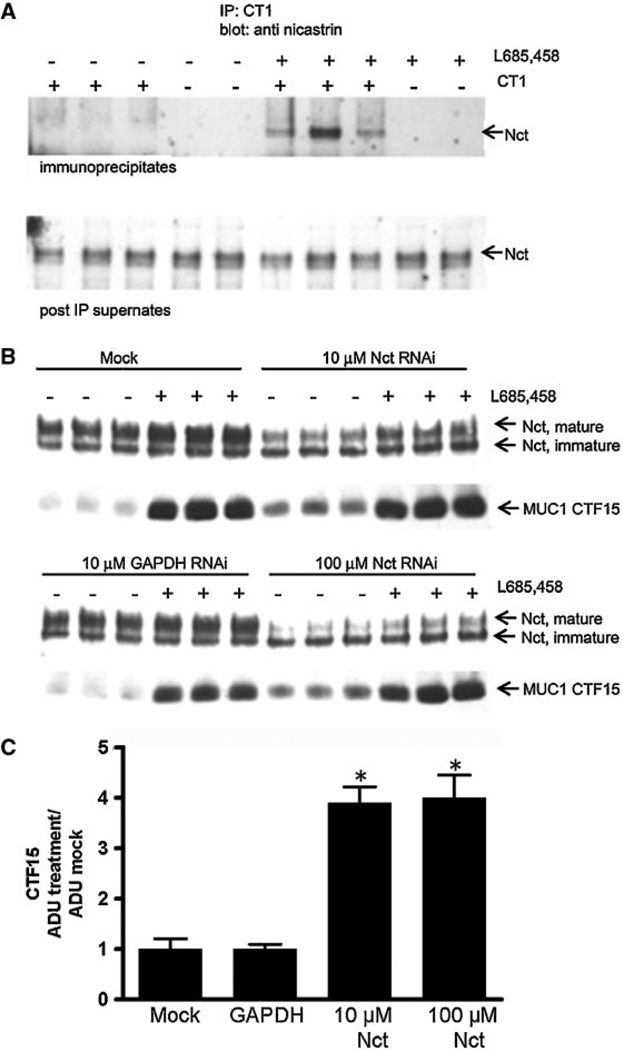
A substrate for γ-secretase should be membrane-associated until cleaved by the γ-secretase complex. Subcellular fractionation revealed that the CTF15 was associated with the 100,000g membrane enriched fraction and was depleted almost completely from the 100,000g supernatant (Fig. 3A). It was essential to include the irreversible inhibitor, L685,458, in the homogenization buffer to prevent loss of CTF15 during processing. Use of the reversible inhibitor, S2188, resulted in CTF15 loss (data not shown). The membrane anchored C-terminal subunits [Mahanta et al., 2008] of the MUC1F metabolic complex which serves as a substrate for TACE/ADAM17 also would be found in the 100,000g membrane fraction. Association with the N-terminal subunit would persist until released by TACE/ADAM17 cleavage. Thus, the substrates, but not the product of TACE/ADAM17, would be immunoprecipitated by antibodies recognizing the N-terminal subunit, that is, 214D4 and/or HMFG1. The MUC1 metabolic complex was immunoprecipitated from HES cell lysates with antibody 214D4 alone (Fig.3B), sequentially with 214D4 followed by HMFG1(Fig. 3C), or a combination of the two antibodies (not shown), which recognize distinct epitopes in the tandem repeats of the MUC1 ectodomain. Preliminary studies indicated that sequential immunoprecipitation with these antibodies removed almost all (>95%) of the MUC1 metabolic complex from these lysates [Wang et al., 2008; and data not shown]. In addition to the 20–23 kDa C-terminal subunit, the 17 kDa C-terminal subunit was associated with the heterodimeric metabolic complex (Fig. 3B). In contrast, MUC1 CTF15 was not immunoprecipitated by the antibodies recognizing the N-terminal subunit and, therefore, was not associated with the metabolic complex. We included immunoprecipitation by CT1 only to demonstrate that intact CTF15 remained in the first post-IP supernate (Fig. 3C) and could be recognized by CT1 in solution (not masked by association with other proteins).The ability of CT1 antibody to immunoprecipitate CTF15 and recognize it on Western blots indicated that the reduction in size relative to the metabolic complex-associated 20–23 kDa and 17 kDa CTD subunits must have occurred at the N-terminus of the C-terminal subunit and resulted from an additional cleavage within the ectodomain portion.
Fig. 3.
The MUC1 CTF15 is membrane associated but is not a component of the MUC1 “metabolic complex”. Panel A: Cytokine-stimulated HES cells were treated without or with 1 µM L685,458 for 54 h, homogenized and subjected to subcellular fractionation as described in “Experimental Procedures Section.” Resulting supernatants (Sup) and pellets (Pellet) were analyzed by SDS-PAGE and Western blotting with CT1 antibody as described in “Experimental Procedures Section.” MUC1 CTF15 as well as the 17 and 20–23 kDa components of the metabolic complex were associated with the 100,000g pellet. CTF15 was greatly enhanced in pellet fractions of cells treated with the γ-secretase inhibitor, L685,458. Equivalent proportions of each fraction were analyzed from both inhibitor treated and untreated cells. Panel B: Triplicate lysates from cytokine-stimulated HES cells treated without (−) or with (+) 10µM L685,458 were immunoprecipitated with MUC1 ectodomain antibody 214D4 and analyzed by SDS-PAGE and Western blotting with CT1 antibody. Only the region of the blot containing CTF17 and CTF15 in starting lysates (upper panel), immunoprecipitates (middle panel) or material not recognized by 214D4 (lower panel) is shown. Immunoblotting with CT1 antibody detected only the C-terminal MUC1 subunit (20–23 kDa doublet and CTF17) of the metabolic complex in the immunoprecipitate while the CTF15 remained in the post-IP supernatants. Panel C: To exclude the possibility that CTF15 might be associated with metabolic complex containing a glycoform not recognized by 214D4, ectodomain material in a post–214D4–immunoprecipitation supernatant subsequently was immunoprecipitated with control Ig (control), CT-1, or HMFG1 antibody. Immunoprecipitates (top panel) or IP supernatants (bottom panel) were analyzed by Western blotting with CT1 antibody. CTF15 was not present in HMFG1 immunoprecipitates. L, whole cell lysate.
The MUC1 γ-secretase substrate, CTF15, is the product of TACE/ADAM 17 cleavage
The MUC1 product of TACE/ADAM 17 cleavage would share the characteristics established above for the γ-secretase substrate, CTF15, that is, it would be membrane-associated by virtue of the TMD and would not be associated with the N-terminal subunit which would be released as a result of sheddase cleavage. The position of the sheddase cleavage site indicated in Figure 4A is based on the previously demonstrated ability of TACE/ADAM17 to cleave a MUC1 peptide in vitro [Thathiah and Carson, 2004] and would produce a product with a calculated MW of 13,511 if unmodified. The potential N-glycosylation site and TMD are retained in the CT product of sheddase cleavage. If CTF15 is both product of TACE/ADAM17 cleavage and substrate for γ-secretase, inhibition of TACE/ADAM17 cleavage should coordinately reduce both sheddase cleavage (release of MUC1 N-terminal subunit) and accumulation of CTF15 in response to γ-secretase inhibition. TACE/ADAM 17 cleavage mediates both constitutive and cytokine-induced shedding of MUC1F ectodomain in HES cells [Thathiah et al., 2003]. In these studies, to quantitate sheddase cleavage and cytokine induction of MUC1 expression in HES, cells were routinely withdrawn from serum for 24 h prior to treatment and treatment was conducted in serum-free medium. Since quantitation of response was based on detection of the MUC1 N-terminal subunit, initial experiments were performed to determine the effect of serum withdrawal on the relative expression of MUC1 C-terminal forms. Total CT was decreased by serum withdrawal. The magnitude of decrease was dependent on the duration of withdrawal. The magnitude of cytokine response in serum-free medium was a function of the expression threshold and period of treatment. Relative abundance of the various CT forms produced by cytokine stimulation in the presence of γ-secretase inhibitor approximated those produced in the presence of serum for the same period of treatment (Fig. 4B). An additional CTD form migrating as a diffuse band between 8 and 10 kDa was detected. This appears to be a product of the processing pathway involving sheddase activity since it was not detected in samples treated with TAPI. Cytokine stimulation in the presence of γ-secretase inhibitor increased accumulation of the CTF15 in conjunction with increased shedding. Inclusion of the TACE/ ADAM17 inhibitor, TAPI [Thathiah et al., 2003], proportionately reduced both the cytokine-stimulated shedding of MUC1 ectodomain and accumulation of CTF15. Consistent with this, the resulting ratios of the CTF15/shed ectodomain for the two conditions were not significantly different, 1.39 and 1.34 in the absence and presence of TAPI, respectively (Fig. 4C).
Fig. 4.
Inhibition of MUC1 cleavage by TACE/ADAM17 inhibits accumulation of MUC1 CTF15. A diagram of C-terminal subunit cleavage by TACE/ADAM17 is presented in panel A. Within the ectodomain amino acid sequence of C-terminal subunit the site of sheddase cleavage is indicated by an arrow and the position of the N-glycosylation site underlined. Cleavage by TACE/ADAM17 would produce CTF15 with an ectodomain “stub” of 27 amino acids and an intact TMD and CTD. After 48 h of serum withdrawal, HES cells were cultured 39 h in serum-free medium containing 1 µM L685458, with or without cytokine stimulation and 50 µM TAPI as indicated (panel B). Shed/secreted protein was probed with antibody 214D4 to quantify MUC1 ectodomain shedding. The immunoblot of cell-associated proteins was probed with antibody CT1 to quantify accumulation of the CTF15. Inclusion of TAPI produced a similar partial reduction in both MUC1 shedding and CTF15 accumulation. Densitometric analysis of the relative contribution of CTF17 and CTF15 to total C-terminal forms demonstrates the reduction of CTF15 accumulation in response to TAPI inclusion. Note the detection of the 8–10 kDa products in lanes from cells treated with cytokines and L685,458 in the absence of TAPI. In panel C, densitometric analyses and ratios of CTF15 signal to 214D4 signal for each indicated treatment in Panel B confirms that the reduction is proportionate. The bars represent the means ±s.d. of triplicate determinations in each case.
Interference with formation of active γ-secretase complex causes accumulation of MUC1 CTF15
The formation and activation of the γ-secretase complex is tightly regulated and dependent on the presence of all four components [Kimberly et al., 2002]. The nicastrin component functions in substrate recognition for the complex and has to undergo glycosylation and a conformational shift to attain a mature functional form [Kimberly et al., 2002; Shirotani et al., 2003]. Blocking catalytic activity of the complex permits detection of substrate recognition by nicastrin. Only in the presence of the irreversible catalytic inhibitor L685,458, did immunoprecipitation of MUC1 by CT1 antibody result in co-precipitation of nicastrin (Fig. 5A). Reduction in expression of presenillin or nicastrin coordinately reduces expression of other complex components and results in reduced formation of the active γ-secretase complex [Edbauer et al., 2002]. Another result is the accumulation of γ-secretase substrates in the absence of catalytic inhibition. Nicastrin siRNA was used to reduce the level of active γ-secretase complex as reflected by the level of mature nicastrin. A second transfection was necessary to maintain the reduced level of mature nicastrin during the period of cytokine treatment. When expression of mature nicastrin was reduced 50–75% by siRNA treatment, MUC1 CTF15 accumulated in the absence of catalytic inhibition three- to fourfold relative to the accumulation observed in mock-transfected cells or cells transfected with GAPDH siRNA (Fig. 5B,C). By comparison, catalytic inhibition by L685,458 increased accumulation eight- to ninefold indicating that the inhibitor was more effective in reducing catalytic activity than the siRNA treatment. The expression levels of the 20–23 and 17 kDa C-terminal forms were not significantly different in any of the samples, evidence that transfection had no effect on induction of MUC1F by cytokines (data not shown). Nor was the low expression level of immature nicastrin significantly affected by either the treatments or transfection. These data demonstrated that accumulation of the CTF15 was dependent upon the level of catalytically active γ-secretase complex.
Fig. 5.
siRNA knockdown of nicastrin inhibits CTF15 accumulation. Panel A: Cytokine-stimulated HES cells were incubated with or without 1 µM L685,458 for 72 h and subjected to a cold lysis. Lysates were immunoprecipitated with CT1 antibody and the blots probed with antibody recognizing both mature and immature forms of nicastrin (Nct) as described in “Experimental Procedures Section.” Mature nicastrin was present only in immunoprecipitates containing accumulated CTF15 (upper panel). Panel B: HES cells were transfected with nicastrin or GAPDH siRNA as described in “Experimental Procedures Section.” Cytokine treatment was initiated following the second transfection in the absence (−) or presence (+) of 1 µM L685,458 as indicated. Equal concentrations of total cell protein collected after 56 h of treatment were probed with antibody to nicastrin or CT1 (upper and lower panels, respectively). Densitometric analysis of Panel B samples not treated with L685,458 is presented in Panel C (ADU, arbitrary densitometric units). Treatment with nicastrin siRNA stimulated CTF15 accumulation relative to cells receiving mock or GAPDH siRNA treatment. Data show the means ±s.d. of triplicate determinations in each case. *P< 0.001 versus mock or GAPDH siRNA treated.
The product(s) of γ-secretase cleavage of MUC1 CTD-containing fragments are not detectable in the presence of proteasomal inhibitors
As an extension of the above studies, we examined the metabolic fate of CTF15. Proteasomal degradation has been proposed as the fate of the products of γ-secretase cleavage in normal cells, accounting for the inability to detect the end products of γ-secretase cleavage unless proteasomal activity is inhibited. We attempted to determine if the MUC1 product of γ-secretase cleavage was degraded by a proteasomal pathway. We used a reversible catalytic γ-secretase inhibitor to accumulate the substrate, CTF15. Subsequently, the γ-secretase inhibitor was removed and the proteasomal inhibitor, MG132, added to preserve any potential proteasome substrates generated by γ-secretase cleavage. Based on comparison of potential cleavage sites within the TMD of MUC1F with γ-secretase cleavage sites in TMDs of various other γ-secretase substrates (Table I), the expected size of a soluble CT product of γ-secretase cleavage would be 73–85 amino acids (ca. 8–10 kDa). As shown in Figure 6A, the γ-secretase substrate, CTF15, was lost following removal of the γ-secretase inhibitor in the absence of proteasomal inhibition, as expected. However, not only was CTF15 maintained in the presence of MG132, but continued to accumulate over the next 26h. Increasing the incubation period or substituting 10 µM MG101 or 10 µM lactacystin produced similar results (data not shown). Proteasomal inhibition of unstimulated or cytokine-stimulated cells by 10µM MG132 reduced constitutive MUC1 expression and blocked cytokine-induced expression (Fig. 6B), evidence that proteasomal activity was inhibited. Proteasomal activity is necessary to release activated NFkβ which drives both constitutive and cytokine-induced MUC1 expression in this cell context. Thus, although serum and cytokines were included during proteasomal inhibition (Fig. 6A), reduction in any CT form would reflect the balance of processing of existing CT involving proteasomal activity against a background of progressively reduced CT replacement. The reduction of the both 17 and 20–23 kDa CT forms implies that processing of these CT forms was unaffected, and thus do not involve proteasomal activity. Increased CTF15 would result from continued sheddase cleavage of the 17 kDa C-terminal form only if subsequent processing mechanisms were inhibited. Since no product of MUC1 cleavage by γ-secretase was detected, we concluded that the proteasomal inhibitors were inhibiting further γ-secretase-dependent processing, possibly by inhibiting maturation of active complex [Massey et al., 2005]. No smaller (8–10 kDa) form of MUC1 CT was detected during any experiment involving proteasomal inhibition in a whole cell context.
TABLE I.
Potential MUC1 γ-Secretase Cleavage Sites
| γ-secretase cleavage sitea |
||
|---|---|---|
| Substrate | P10-P1 | P1′-PX′b |
| SDC3 | VVGALFAAFL | VTLLIYRMKKK |
| NGFR | VYCSILAAVV | VGLVAYIAFKR |
| CD44 | ALALILAVCI | AVNSRRR |
| CDH1 | ILILLLLLFL | RRR |
| APP | VIATVIVITL | VMLKKK |
| APLP2 | AIATVIVISL | VMLRKR |
| APLP1 | GGGSLIVLSL | LLLRKKK |
| NOTCH1 | FVLLFFVGCG | VLLSRKRRR |
| MUC1c | ||
| I | IALLVLVCVL | VALAIVYLIA |
| II | LVCVLVALAI | VYLIALAVCQ |
| III | LAIVYLIALA | VCQCRRKNYG |
Sequences of substrates other than MUC1 are from references [Gu et al., 2001; Kopan and Ilagan, 2004]. Cleavage sites between P1 and P1′ are MEROPS style [Rawlings et al., 2008].
P1′–PX′ indicates cleavage site with C-terminal sequence arbitrarily included up to the nearest cluster of positively charged amino acids. In the case of the MUC1 sequences (I–III) 10 amino acids have been included.
These products are expected based on the preference for γ-secretase cleavage (39,52) and would generate CTD fragments (top to bottom) of 85 amino acids (9,222 MW), 80 amino acids (8,755 MW), and 73 amino acids(8,010 MW). The MW represent unmodified, average mass calculated in ExPASY program based on the 72 amino acid MUC1 cytoplasmic tail plus the portion of the sequence up to the predicted cleavage site.
MUC1 cleavage by γ-secretase occurs in multiple cell types
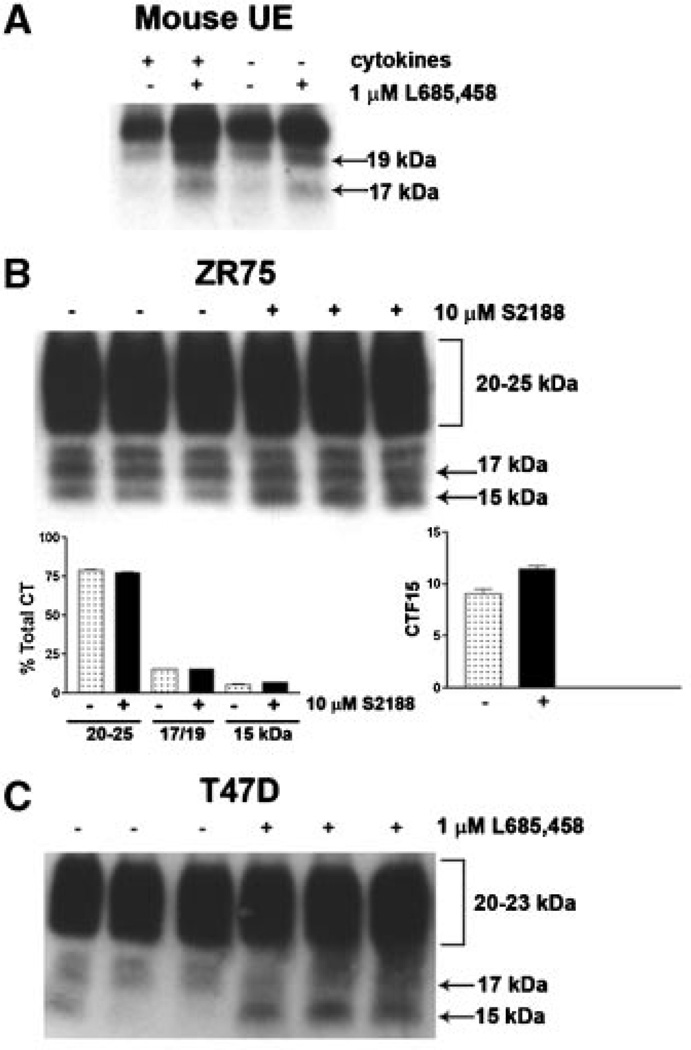
To address the question of whether MUC1 cleavage by γ-secretase is a common event, we examined endogenous MUC1F processing in normal mouse uterine epithelial cells and two human mammary tumor-derived cell lines, T47D and ZR75 (Fig. 7). In all cases, the MUC1 CTF15 accumulated in the presence of the γ-secretase catalytic inhibitor, implying that metabolic processing of endogenous full length MUC1 occurs similarly in normal and tumor cells and in mice as well as humans. We noted that the mouse fragment migrated slightly slower (ca. 17kDa) than the human fragment. All CT forms of mouse Muc1 migrated more slowly than the corresponding human forms when compared directly on the same gel. A further difference between the human and mouse uterine epithelial cell responses was noticed: cytokine treatment which induced MUC1 expression did not significantly elevate mouse Muc1, an observation which is partially explained by the lack of TNFaR1 in mouse UE at all phases of the cycle [Joswig et al., 2003]. Finally, constitutive Muc1 expression is not reduced by serum withdrawal since the cells were cultured in the absence of serum.
Fig. 7.
MUC1 processing by γ-secretase occurs in multiple cell contexts. Isolated mouse uterine epithelial cells (Mouse UE) were cultured to confluence in serum-free medium and treated with or without cytokines, ±1 µM L685,458 for 96 h as described in “Experimental Procedures Section.” Note that CTF15 ran slightly larger (ca. 17 kDa) in mouse uterine epithelia (all mouse C-terminal forms were about 2 kDa larger than the human CT forms). Human breast cancer cell lines, ZR75–1 (Panel B) and T47D (Panel C) were grown as described in “Experimental Procedures Section,” ±10µM S2188 or 1 µM L685458 for 14 and 56 h, respectively, without cytokine stimulation. Cell lysates were probed with CT1 antibody. Note that CTF15 was constitutively expressed in ZR75 cells, but enhanced slightly in the presence of S2188.
DISCUSSION
Collectively, these data describe a MUC1 processing pathway in which the MUC1F metabolic complex, formed in the endoplasmic reticulum by autoproteolytic cleavage, arrives at the plasma membrane where a subset of the membrane-associated subunit is cleaved by sheddases, releasing the ECD containing the tandem repeat region and producing a membrane-associated cleavage product containing a portion of membrane-proximal ECD, the TMD, and CTD. In the present study we have established a third step in the proteolytic cascade mediated by presenillin-dependent γ-secretase that occurs to the subset of MUC1 that is subjected to sheddase cleavage. Based on previous studies estimating a MUC1 half-life of 16 h [Pimental et al., 1996], it can be calculated that 87.5% of total cell-associated MUC1 should be turned over during the 48h period typically examined in the present studies; however, only 6% of MUC1 would be shed over the same period [Thathiah et al., 2003]. Therefore, only 6% of total MUC1 represents a potential pool of γ-secretase substrates. In good agreement, we found that the portion of CTF15 accumulated after 48 h of γ-secretase inhibition represented approximately 8% of the total CT forms. We used two independent approaches to inhibit γ-secretase activity: (1) reduction of catalytic activity by various specific pharmacologic inhibitors of γ-secretase and; (2) reduced expression of catalytically active γ-secretase complex by siRNA-mediated reduction of nicastrin expression. Both conditions resulted in accumulation of CTF15, a MUC1 C-terminal subunit fragment not observed in an unstimulated normal cell context. A third approach was attempted in which either TACE/ ADAM17 or presenillin null mouse embryonic fibroblasts (MEFs) were transiently transfected with MUC1; however, we were not able to obtain high enough levels of expression in wild-type MEFs in the presence of γ-secretase inhibitors to detect the CTF15, making results obtained with this cell context uninterpretable (data not shown). A direct association between MUC1 CTF15 and the γ-secretase complex was demonstrated by co-immunoprecipitation of nicastrin by CT1 antibody only from lysates in which CTF15 had accumulated. Thus, it appears that CTF15 must be generated for this association to occur.
MUC1 CTF15 was associated with the membrane as expected for a γ-secretase substrate and was not associated with the MUC1 metabolic complex. Both of these characteristics also distinguish the product of MUC1 cleavage by the sheddase, TACE/ADAM 17. Recognition of MUC1 CTF15 by the CT1 antibody, directed against the C-terminal 17 amino acids of MUC1, indicates that the truncation from the larger C-terminal forms associated with the metabolic complex to the 15kDa form must have occurred at the N-terminus of the C-terminal subunit of the MUC1 heterodimeric metabolic complex. The Mr of 15kDa is slightly higher than would be expected for the 127 amino acid C-terminal polypeptide expected to be generated by TACE/ADAM17 action, which has a calculated MW of 13511 when unmodified. Modification at the one consensus N-glycosylation site, or several potential O-glycosylation sites contained within CTF15 could account for the size differential. We did not observe a size shift in CTF15 after peptide:N-glycanase digestion indicating the N-glcosylation is unlikely. Inhibition of sheddase cleavage of MUC1F by the TACE/ADAM17 inhibitor, TAPI, resulted in a proportionate decrease in accumulation of the γ-secretase substrate, further evidence that shedding generates the substrate for catalytically active γ-secretase complex. MT1–MMP also mediates MUC1 shedding [Thathiah and Carson, 2004]; however, it is not clear whether the product produced by MT1– MMP cleavage of MUC1F is similarly recognized by γ-secretase. Although suitable substrates for γ-secretase cleavage are not limited to products of TACE/ADAM 17 cleavage, the size of the resulting ectodomain stub is critical. The probable cleavage site on MUC1 for MT1–MMP has not been identified and, thus, we cannot speculate on the suitability of the product of that cleavage as a substrate for γ-secretase.
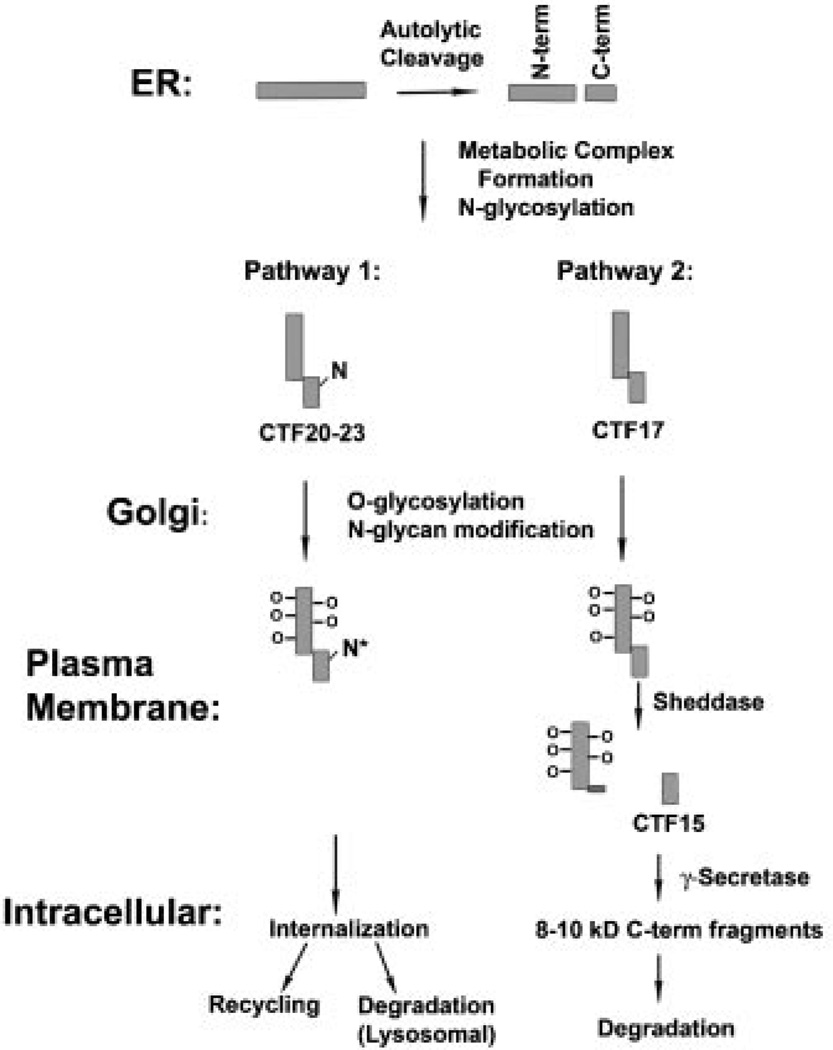
In conjunction with previous observations, the present study has established that a portion of endogenous full length MUC1 is subjected to a sequential series of proteolytic processing steps in normal epithelial cells. The results support a model (Fig. 8) in which the progression to the next step is dependent on successfully accomplishing the previous step in the proteolytic cascade. The first step, formation of the MUC1 metabolic complex from full length MUC1, mediated by an autoproteolytic cleavage within the SEA module of the ectodomain, takes place regardless of cell context, as long as the metabolic cleavage site is unaltered or the alteration allows folding to the stable conformation necessary for efficient autolytic cleavage [Macao et al., 2006]. A single amino acid mutation (Gly–Ser) which prevents the metabolic cleavage also prevents ectodomain shedding [Lillehoj et al., 2003]. Association of the two subunits depends on N-glycosylation of the N-terminal subunit, which contains four potential N-glycosylation sites, but not the C-terminal subunit [Baruch et al., 1999]. Heterodimeric association of the subunits assumes a conformation that exposes two hydrophobic patches on its surface that were suggested to be involved in protein/protein association [Macao et al., 2006]. The conformation attained through reassociation of the two subunits resulting from the autolytic cleavage may be necessary for recognition, association, and/or subsequent cleavage by sheddases. While N-glycosylation of the C-terminal subunit appears to have no affect on subunit association, it does affect recognition by TACE/ ADAM17, possibly by sterically blocking access to the cleavage site. In this regard, the N-glycosylation site in the C-terminal subunit lies four amino acids C-terminal to the TACE/ADAM17 cleavage site. We suggest that TACE/ADAM17 selectively targets the C-terminal subunit lacking N-glycosylation based on the following observations in the present study: (1) the product of TACE/ADAM17 cleavage (CTF15) was not modified by N-glycosylation, that is, it was resistant to peptide:N-glycanase treatment. If CTF15 is the product of TACE/ADAM17 cleavage, the TACE/ADAM17 substrate would have to be restricted to C-terminal forms lacking N-glycosylation (17 kDa); (2) when TACE/ADAM17 cleavage was inhibited by TAPI, although CTF15 accumulation was reduced in proportion to shedding, the 17 kDa C-terminal form was increased; (3) during proteasomal inhibition following release from γ-secretase inhibition, CTF15 continued to accumulate while the 17 kDa form disappeared. As noted above, only a fraction of MUC1F undergoes shedding (ca. 3–6%). Sheddase cleavage produces the substrate (CTF15) recognized by the γ-secretase complex. Accumulation of CTF15 was consistent with sheddase activity and was coordinately regulated.
Fig. 8.
A model of MUC1 processing. The model summarizes MUC1 processing pathways from a number of studies discussed in the text. Only the contribution of N-glycosylation to the variety of C-terminal forms detected when MUC1F (full length MUC1 precursor) is overexpressed in normal cells is considered in this model. N-glycosylation sites are indicated by an N on the C-terminal subunits. Within the endoplasmic reticulum (ER), MUC1F is translated as a single polypeptide which folds to a conformation inducing an autolytic cleavage within the SEA module to produce N- and C-terminal subunits (N-term and C-term). The resulting subunits remain stably associated as a heterodimer (metabolic complex) and undergo N-glycosylation. There are five potential N-glycosylation sites in human MUC1: four within the N-terminal subunit and one within the C-terminal subunit. For simplicity, only the latter site is indicated on the figure and the N-glycosylated forms of this C-terminal subunit are labeled CTF20–23 in the figure and the text. Forms of the C-terminal subunit lacking N-glycosylation are labeled CTF17 in the figure and the text. MUC1 heterodimers transit the Golgi complex in which extensive O-glycosylation of the tandem repeat region occurs (indicated by O – in the figure) as well as additional modification of the N-glycan (N*). The present data indicates that upon arrival at the plasma membrane, the subset of heterodimers lacking N-glycans on the C-terminal fragment are substrates for sheddases that generate CTF15. CTF15 is a substrate for γ-secretase and is cleaved within the transmembrane domain at one or more sites. The C-terminal products of γ-secretase generally were undetectable and, therefore, presumed to be rapidly degraded; however, we detected trace amounts of 8–10 kDa C-terminal fragments in our studies which presumably are the products of γ-secretase destined for rapid degradation. Heterodimers containing N-glycosylated C-terminal subunits appear to be processed by an alternate pathway, possibly involving internalization, recycling, or lysosomal degradation.
γ-Secretase has been proposed as the “membrane proteasome” [Kopan and Ilagan, 2004], functioning to either attenuate or facilitate signaling function of its various substrates depending on cell context. In normal cells, it would facilitate termination of the signaling function of MUC1 cytoplasmic tail in concert with and subsequent to shedding of the ectodomain if the ultimate fate of the CTD is degradation. The final step of this processing pathway in all cells we have examined is degradation of the γ-secretase cleavage product. We were unable to verify a proteasomal pathway for this step. This is possibly due to inhibition of γ-secretase complex assembly and activation by the proteasomal inhibitors [Massey et al., 2005], direct catalytic inhibition of γ-secretase by proteasomal inhibitors, or degradation by a proteasome-independent process [Del-Val and Lopez, 2002]. However, the observation of a CT form at the size predicted for the product of γ-secretase cleavage in cells withdrawn from serum suggests proteasomal involvement. Prolonged serum withdrawal of confluent cultures of normal cells has been reported to reduce proteasomal activity [Fuertes et al., 2003], and the 8–10 kDa CT form was observed only when serum withdrawal pretreatment exceeded 24h. Analogous to constitutive cleavage of the erbB4 receptor [Vecchi and Carpenter, 1997] proteasomal inhibitors resulted in slow accumulation of the membrane-associated product of TACE cleavage. In contrast to their effect in the present study, they appear to have no effect on generation of the soluble erbB4 ICD produced by phorbol ester induced cleavage which is executed sequentially by TACE/ADAM17 and PSI-dependent γ-secretase [Ni et al., 2001]. The sole distinction in the membrane-associated TACE/ADAM17 cleavage products accumulated in the two conditions was that those accumulated with proteasomal inhibitors were poly ubiquitinated while those produced by phorbol ester stimulation were not, implying that subset produced by unstimulated TACE cleavage are targeted for proteasomal mediated degradation. Although we were unable to detect ubiquitination of MUC1 CTF15 accumulated in response to proteasomal inhibitors, a non-ubiquitin-dependent proteasomal degradation following γ-secretase cleavage remains a possibility.
The cellular location for individual steps of the processing pathway initiated by sheddase cleavage is most likely the plasma membrane. The N-terminal subunit is released externally by sheddase cleavage. Proximity to substrate is critical for the activities of the sheddase and γ-secretase, both of which are membrane anchored. The site of mature active γ-secretase complex accumulation recently was established as the plasma membrane [Chyung et al., 2005], resolving the spatial paradox that the major concentration of γ-secretase complex components resided in intracellular compartments [Cupers et al., 2001]. A similar spatial paradox exists for TACE/ADAM17 [Schlondorff et al., 2000]. While presence of mature TACE/ADAM17 at the cell surface has been established in several cell contexts [Schlondorff et al., 2000; Srour et al., 2003], it was also detected in an unidentified intracellular compartment [Schlondorff et al., 2000]. Short term activation of cells by phorbol esters, which increase TACE/ADAM17 mediated shedding without increasing cell surface TACE/ADAM17 concentrations [Doedens et al., 2003], increased TACE/ADAM17 mediated shedding of MUC1 [Thathiah et al., 2003]. It is possible that this increase reflects a change in the membrane microdomain occupied by MUC1. Active γ-secretase was recently shown to associate with lipid rafts [Urano et al., 2005] and TACE/ADAM17–dependent shedding can be modulated by lipid rafts [Gil et al., 2007]. Whether all the interacting components occupy the same membrane microdomain, for example, lipid raft/non-raft in this cell context remains to be determined.
ACKNOWLEDGMENTS
The authors appreciate the many helpful discussions and critical reading of this manuscript by Dr. Mary C. Farach-Carson and Peng Wang. We appreciate the excellent secretarial assistance by Ms. Sharron Kingston. This work was supported by NIH (HD29963 to D.D.C.), University of Delaware (Dissertation Fellowship to N.D.).
Grant sponsor: NIH; Grant number: HD29963.
Abbreviations used
- ADAM
a disintegrin and metalloprotease
- CT
carboxy terminal
- CTD
cytoplasmic tail domain
- ECD
extracellular domain
- HRP
horseradish peroxidase
- CTF15
MUC1 C-terminal 15 kDa fragment
- PBS
phosphate buffered saline
- RIP
regulated intramembranous proteolysis
- SEB
sample extraction buffer
- TACE
tumor necrosis factor-alpha converting enzyme
- TCA
trichloroacetic acid
- TMD
transmembrane domain
REFERENCES
- Baruch A, Hartmann M, Yoeli M, Adereth Y, Greenstein S, Stadler Y, Skornik Y, Zaretsky J, Smorodinsky NI, Keydar I, Wreschner DH. The breast cancer-associated MUC1 gene generates both a receptor and its cognate binding protein. Cancer Res. 1999;59:1552–1561. [PubMed] [Google Scholar]
- Chyung JH, Raper DM, Selkoe DJ. Gamma-secretase exists on the plasma membrane as an intact complex that accepts substrates and effects intramembrane cleavage. J Biol Chem. 2005;280:4383–4392. doi: 10.1074/jbc.M409272200. [DOI] [PubMed] [Google Scholar]
- Cupers P, Bentahir M, Craessaerts K, Orlans I, Vanderstichele H, Saftig P, De Strooper B, Annaert W. The discrepancy between presenilin subcellular localization and gamma-secretase processing of amyloid precursor protein. J Cell Biol. 2001;154:731–740. doi: 10.1083/jcb.200104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del-Val M, Lopez D. Multiple proteases process viral antigens for presentation by MHC class I molecules to CD8(+) T lymphocytes. Mol Immunol. 2002;39:235–247. doi: 10.1016/s0161-5890(02)00104-9. [DOI] [PubMed] [Google Scholar]
- Doedens JR, Mahimkar RM, Black RA. TACE/ADAM-17 enzymatic activity is increased in response to cellular stimulation. Biochem Biophys Res Commun. 2003;308:331–338. doi: 10.1016/s0006-291x(03)01381-0. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Haass C, Steiner H. Presenilin and nicastrin regulate each other and determine amyloid beta-peptide production via complex formation. Proc Natl Acad Sci USA. 2002;99:8666–8671. doi: 10.1073/pnas.132277899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- Gil C, Cubi R, Aguilera J. Shedding of the p75NTR neurotrophin receptor is modulated by lipid rafts. FEBS Lett. 2007;581:1851–1858. doi: 10.1016/j.febslet.2007.03.080. [DOI] [PubMed] [Google Scholar]
- Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y. Distinct intramembrane cleavage of the beta-amyloid precursor protein family resembling gamma-secretase-like cleavage of notch. J Biol Chem. 2001;276:35235–35238. doi: 10.1074/jbc.C100357200. [DOI] [PubMed] [Google Scholar]
- Hanisch FG, Muller S. MUC1: The polymorphic appearance of a human mucin. Glycobiology. 2000;10:439–449. doi: 10.1093/glycob/10.5.439. [DOI] [PubMed] [Google Scholar]
- Joswig A, Gabriel HD, Kibschull M, Winterhager E. Apoptosis in uterine epithelium and decidua in response to implantation: Evidence for two different pathways. Reprod Biol Endocrinol. 2003;1:44. doi: 10.1186/1477-7827-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian J, Carson DD. Formation of MUC1 metabolic complex is conserved in tumor-derived and normal epithelial cells. Biochem Biophys Res Commun. 2002;293:1183–1190. doi: 10.1016/S0006-291X(02)00352-2. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Complex N-linked glycosylated nicastrin associates with active gamma-secretase and undergoes tight cellular regulation. J Biol Chem. 2002;277:35113–35117. doi: 10.1074/jbc.M204446200. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. Gamma-secretase: Proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- Li Y, Yu WH, Ren J, Chen W, Huang L, Kharbanda S, Loda M, Kufe D. Heregulin targets gamma-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 oncoprotein. Mol Cancer Res. 2003;1:765–775. [PubMed] [Google Scholar]
- Ligtenberg MJ, Kruijshaar L, Buijs F, van Meijer M, Litvinov SV, Hilkens J. Cell-associated episialin is a complex containing two proteins derived from a common precursor. J Biol Chem. 1992;267:6171–6177. [PubMed] [Google Scholar]
- Lillehoj EP, Han F, Kim KC. Mutagenesis of a Gly-Ser cleavage site in MUC1 inhibits ectodomain shedding. Biochem Biophys Res Commun. 2003;307:743–749. doi: 10.1016/s0006-291x(03)01260-9. [DOI] [PubMed] [Google Scholar]
- Lillehoj EP, Kim H, Chun EY, Kim KC. Pseudomonas aeruginosa stimulates phosphorylation of the airway epithelial membrane glycoprotein Muc1 and activates MAP kinase. Am J Physiol Lung Cell Mol Physiol. 2004;287:L809–L815. doi: 10.1152/ajplung.00385.2003. [DOI] [PubMed] [Google Scholar]
- Liu X, Yuan Z, Chung M. MUC1 intra-cellular trafficking is clathrin, dynamin, and rab5 dependent. Biochem Biophys Res Commun. 2008;376:688–693. doi: 10.1016/j.bbrc.2008.09.065. [DOI] [PubMed] [Google Scholar]
- Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- Mahanta S, Fessler SP, Park J, Bamdad C. A minimal fragment of MUC1 mediates growth of cancer cells. PLoS ONE. 2008;3:e2054. doi: 10.1371/journal.pone.0002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey LK, Mah AL, Monteiro MJ. Ubiquitin regulates presenilin endoproteolysis and modulates gamma-secretase components, Pen-2 and nicastrin. Biochem J. 2005;391:513–525. doi: 10.1042/BJ20050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Dotti CG. RIPped out by presenilin-dependent gamma-secretase. Cell Signal. 2003;15:829–841. doi: 10.1016/s0898-6568(03)00041-x. [DOI] [PubMed] [Google Scholar]
- Meyer EL, Strutz N, Gahring LC, Rogers SW. Glutamate receptor subunit 3 is modified by site-specific limited proteolysis including cleavage by gamma-secretase. J Biol Chem. 2003;278:23786–23796. doi: 10.1074/jbc.M301360200. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Parry S, Hanisch FG, Leir SH, Sutton-Smith M, Morris HR, Dell A, Harris A. N-Glycosylation of the MUC1 mucin in epithelial cells and secretions. Glycobiology. 2006;16:623–634. doi: 10.1093/glycob/cwj110. [DOI] [PubMed] [Google Scholar]
- Pimental RA, Julian J, Gendler SJ, Carson DD. Synthesis and intra-cellular trafficking of Muc-1 and mucins by polarized mouse uterine epithelial cells. J Biol Chem. 1996;271:28128–28137. doi: 10.1074/jbc.271.45.28128. [DOI] [PubMed] [Google Scholar]
- Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693–1701. doi: 10.1038/sj.onc.1209976. [DOI] [PubMed] [Google Scholar]
- Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Morton FK, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucl Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Becherer JD, Blobel CP. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE) Biochem J. 2000;347(Pt 1):131–138. [PMC free article] [PubMed] [Google Scholar]
- Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CEIII, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Shirotani K, Edbauer D, Capell A, Schmitz J, Steiner H, Haass C. Gamma-secretase activity is associated with a conformational change of nicastrin. J Biol Chem. 2003;278:16474–16477. doi: 10.1074/jbc.C300095200. [DOI] [PubMed] [Google Scholar]
- Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, Gendler SJ, Hollingsworth MA. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- Srour N, Lebel A, McMahon S, FournierI FugereM, Day R, Dubois CM. TACE/ADAM-17 maturation and activation of sheddase activity require proprotein convertase activity. FEBS Lett. 2003;554:275–283. doi: 10.1016/s0014-5793(03)01159-1. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382:363–373. doi: 10.1042/BJ20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386–3394. doi: 10.1074/jbc.M208326200. [DOI] [PubMed] [Google Scholar]
- Thathiah A, Brayman M, Dharmaraj N, Julian JJ, Lagow EL, Carson DD. Tumor necrosis factor alpha stimulates MUC1 synthesis and ectodomain release in a human uterine epithelial cell line. Endocrinology. 2004;145:4192–4203. doi: 10.1210/en.2004-0399. [DOI] [PubMed] [Google Scholar]
- Thompson EJ, Shanmugam K, Hattrup CL, Kotlarczyk KL, Gutierrez A, Bradley JM, Mukherjee P, Gendler SJ. Tyrosines in the MUC1 cytoplasmic tail modulate transcription via the extracellular signal-regulated kinase 1/2 and nuclear factor-kappaB pathways. Mol Cancer Res. 2006;4:489–497. doi: 10.1158/1541-7786.MCR-06-0038. [DOI] [PubMed] [Google Scholar]
- Urano Y, Hayashi I, Isoo N, Reid PC, Shibasaki Y, Noguchi N, Tomita T, Iwatsubo T, Hamakubo T, Kodama T. Association of active gamma-secretase complex with lipid rafts. J Lipid Res. 2005;46:904–912. doi: 10.1194/jlr.M400333-JLR200. [DOI] [PubMed] [Google Scholar]
- Vecchi M, Carpenter G. Constitutive proteolysis of the ErbB-4 receptor tyrosine kinase by a unique, sequential mechanism. J Cell Biol. 1997;139:995–1003. doi: 10.1083/jcb.139.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mensdorff-Pouilly S, Snijdewint FG, Verstraeten AA, Verheijen RH, Kenemans P. Human MUC1 mucin: A multifaceted glycoprotein. Int J Biol Markers. 2000;15:343–356. doi: 10.1177/172460080001500413. [DOI] [PubMed] [Google Scholar]
- Wang P, Julian JA, Carson DD. The MUC1 HMFG1 glycoform is a precursor to the 214D4 glycoform in the human uterine epithelial cell line, HES. Biol Reprod. 2008;78:290–298. doi: 10.1095/biolreprod.107.064584. [DOI] [PubMed] [Google Scholar]